Abstract
Cytosine-5 DNA methylation is a critical signal defining heritable epigenetic states of transcription. As aberrant methylation patterns often accompany disease states, the ability to target cytosine methylation to preselected regions could prove valuable in re-establishing proper gene regulation. We employ the strategy of targeted gene methylation in yeast, which has a naturally unmethylated genome, selectively directing de novo DNA methylation via the fusion of C5 DNA methyltransferases to heterologous DNA-binding proteins. The zinc-finger proteins Zif268 and Zip53 can target DNA methylation by M.CviPI or M.SssI 5–52 nt from single zinc-factor binding sites. Modification at specific GC (M.CviPI) or CG (M.SssI) sites is enhanced as much as 20-fold compared with strains expressing either the free enzyme or a fusion protein with the zinc-finger protein moiety unable to bind to DNA. Interestingly, methylation is also selectively targeted as far as 353 nt from the zinc-finger protein binding sites, possibly indicative of looping, nucleosomes or higher-order chromatin structure. These data demonstrate that methylation can be targeted in vivo to a potentially broad range of sequences using specifically engineered zinc-finger proteins. Further more, the selective targeting of methylation by zinc-finger proteins demonstrates that binding of distinct classes of factors can be monitored in living cells.
INTRODUCTION
Methylation of the C5 atom of cytosine in DNA (m5C) plays an important role in establishing correct patterns of gene expression in vertebrates, usually through repression of transcription. Mechanistically, one way DNA methylation can lead to transcriptional silencing is by decreasing the binding affinity of a transcriptional activator for its site (1). The introduction of m5C at sites adjacent to a factor binding site can also interfere with its binding (2). Perhaps more importantly, symmetrical methylation of CpG sequences (CG) serves as a signal for the recruitment of a family of methyl-CpG binding domain (MBD) proteins, such as MeCP2 and MBD2 (3). In turn, MBDs, either by themselves or as components of complexes, are known to recruit a variety of co-repressors, such as histone deacetylases (4–7), histone H3 lysine-9 methyltransferases (8) and heterochromatin coating factors like HP1 (9), which can function to establish a local, repressed region of chromatin (10–15). This silencing mechanism is also conserved in plants, as the DNA chromomethyltransferase CMT3, which methylates CNG residues, interacts with HP1 to facilitate heterochromatin formation (8).
While regions of m5C are often associated with hypoacetylation of histones H3 and/or H4 and altered chromatin structure (10–15), recent evidence suggests DNA methylation- and histone deacetylase-independent modes of silencing. First, trichostatin A, a specific inhibitor of histone deacetylation, fails to reactivate transcription from densely methylated DNA (2,11,12,15–17). Additionally, mbd2-null mice are viable and fertile (18) and Mecp2-null mice only display neurological abnormalities (19), questioning their global role in m5C-mediated silencing and cellular differentiation. Moreover, purified MeCP2 itself compacts reconstituted chromatin in the absence of DNA methylation (20).
Although the mechanisms are not yet fully understood, there is a strong correlation between promoter methylation and gene silencing (1,21–23). Moreover, once a methylation state is established, it is maintained heritably after many generations of replication (24) by the maintenance DNA methyltransferase (DMTase), Dnmt1 (25). An exception includes enhancer sequences that can be passively demethylated on replication and subsequent blockage of DMTase access by factor binding (26–30). However, this enhancer-specific loss of DNA methylation does not lead to derepression (26).
Proper regulation of gene expression is essential for normal cellular functions and the avoidance of disease states. DNA methylation, which occurs almost exclusively at CG dinucleotides in non-diseased cells, is localized to precise regions of the genome, usually in transposons and retroviral elements (25). In contrast, CG sites in euchromatic regions, most notably when concentrated in CpG islands, are generally unmethylated and are correlated with transcriptional activity. However, in cancer and other diseases, patterns of DNA methylation are frequently aberrant. For instance, the DNA in tumor cells is generally hypomethylated relative to that in normal cells (31), which may lead to genomic instability (23). In contrast, a number of tumor-suppressor genes, including BRCA1 and retinoblastoma (Rb), become hypermethylated and transcriptionally inactive (32). The presence of a single methylated CG site in a gene’s promoter is sufficient to repress its activation (21), although higher m5C density increases the probability of establishing gene repression (14,33–36). Thus, DNA methylation can be critical in defining the expression state of a gene.
Therefore, directing DNA methylation to improperly regulated loci could be used to re-establish proper gene expression through silencing. Previously, targeting of m5C has been demonstrated in vitro (37,38), however, selective enrichment of m5C was not observed in vivo (38). Recently, in yeast, using the dinucleotide-specificity DMTase M.CviPI (39) fused to the basic helix–loop–helix activator Pho4, we demonstrated specific targeting of cytosine methylation to promoters containing Pho4 binding sites [targeted gene methylation (TAGM)] (40). Methylation was efficiently targeted to GC sites in nucleosomes that were disrupted on promoter activation, as well as to histone-free regions.
In its present form, the TAGM strategy is limited to known factors that bind to well characterized DNA-binding sites, which are often present in multiple copies throughout the genome. Therefore, we have investigated the ability of zinc-finger proteins, which, in principle, may be selected to recognize one or a small subset of chromosomal regions (41), to target m5C in living cells. Whereas preferential targeting of 4 bp specificity MTases was not observed in vivo (38), we now show that, in yeast, both M.CviPI (GC methylation) and M.SssI (CG methylation) can be preferentially targeted by zinc-finger proteins to specific GC or CG sites neighboring their cognate binding sites. The potential to direct m5C at ∼20-fold increased resolution to a broad range of desired DNA sequences could lead to novel therapeutic approaches.
MATERIALS AND METHODS
Plasmids, yeast strains and growth conditions
All yeast strains used in this study were derived from the S288C background strain YPH500ΔL (MATα ade2-101 ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ63 lys2-Δ1) (26). Zinc-finger coding sequences were PCR amplified using the primers MKO46, 5′-gcactagttaggccagctgggccATGGCTGATATCGGATCTGG-3′, and MKO47, 5′-gaataattcgAGCGCTTTCAAGGTCATGGTGGATCCTAGGCCACCTCCACTCC-3′, and cloned between SfiI and AfeI restriction sites as in-frame fusions to either M.CviPI or M.SssI in pMPK1. The fusion proteins are expressed under the control of the GAL1 promoter after integration at LYS2 as previously described (26). Each N-terminal zinc-finger protein is separated from the DMTase by a G(SGGGG)2SGGGLGST (GS linker) peptide (37). As a free DMTase control, mutated Zif268 (mut Zif), which contains a single amino acid substitution (H58E) (42) that ablates DNA binding, was constructed by overlap site-directed mutagenesis using the primers MKO72, 5′-cagtcgtagtgacgAgcttaccacccac-3′, and MKO73, 5′-gtgggtggtaagcTcgtcactacgactg-3′ (mutated residues in upper case).
Cells were pre-grown in yeast extract (Difco)/peptone (Difco)/2% dextrose (YPD) medium and then washed and resuspended at an OD600 of 0.5 in YP/2% galactose (YPG). After resuspension in YPG, cells were incubated at 30°C for 16 h, or for the indicated times (Fig. 1C).
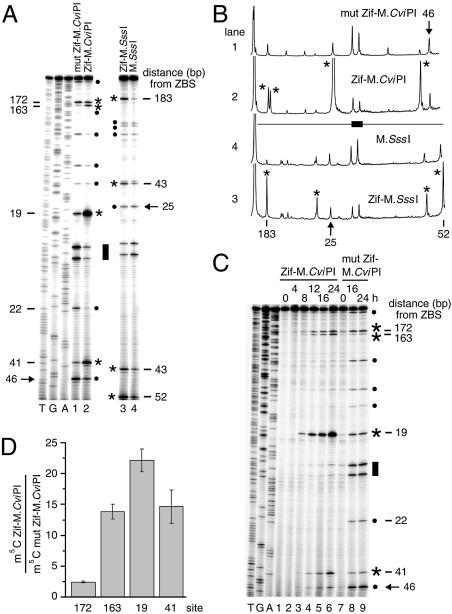
Figure 1.
Targeting C5 DMTases near a single Zif268 site. (A) Determination of m5C levels targeted by Zif268–DMTase fusions. Genomic DNA isolated from strains expressing wild-type Zif268–M.CviPI (Zif–M.CviPI, lane 2), Zif268–M.SssI (Zif–M.SssI, lane 3), or ‘free’ DMTase controls, a mutated Zif268 fused to M.CviPI (mut Zif–M.CviPI, lane 1) or M.SssI by itself (lane 4), was analyzed by modified bisulfite genomic sequencing (26) of CAR1 from +558 to +159. Distances (bp) of a subset of sites from the proximal edge of the Zif268 ZBS (filled bar; +438 to +446) are indicated at left and right of the gel. Sites of non-targeted methylation (filled circles). Sites 46 (M.CviPI strains) and 25 (M.SssI strains) (arrows) were chosen for normalization to enable lane-to-lane comparisons [see (B)]. Each DMTase was preferentially targeted to several CG and GC sites (asterisks) by Zif binding as compared with its respective control (compare lanes 1 with 2 and 3 with 4). For site 19, 41% (of all summed intensities) of the templates in the population are methylated. Lanes T, G and A (left) contain sequencing reactions with ddATP, ddCTP and ddTTP, respectively. (B) Quantitative scans of the phosphoimage in (A). See (A) for definitions of symbols. (C) Time course of targeting M.CviPI by Zif268. Expression of Zif–M.CviPI (lanes 1–6) and Zif–M.SssI (lanes 7–9) from the GAL1 promoter was initiated by transferring cells from YPD (dextrose) to YPG (galactose) medium. Genomic DNA was isolated at the indicated times and analyzed as in (A). Symbols are defined in (A). (D) Quantification of preferential targeting of M.CviPI by Zif268. Ratios of m5C for the indicated sites (normalized to site 46) for Zif–M.CviPI to mut Zif–M.CviPI are given (mean ± standard error; n = 3). Similar values are obtained if the ratios for each site are normalized to other sites of non-targeted methylation (A, filled circles) or calculated using absolute frequencies of methylation (see Materials and Methods).
Bisulfite genomic sequencing
Total genomic DNA was rapidly isolated by the phenol/chloroform lysis method (43) and analyzed by bisulfite genomic sequencing (44,45) as previously modified (26). PCR amplification from bisulfite-treated genomic DNA with the indicated primer pairs was performed with Jumpstart Taq DNA polymerase (Sigma) and the resulting products were subjected to primer extension using a 32P-labeled oligonucleotide as described previously using final concentrations of 5 µM dATP, dCTP and dTTP (dGTP omitted) as well as 50 µM ddGTP (26) (Figs 1 and 2), or with dNTPs (A, C, T) and ddGTP increased to 50 and 150 µM, respectively (Figs 3–5), as recently reported (40). Product intensities were determined by ImageQuaNT software (Molecular Dynamics) after subtracting the local background average. Absolute frequencies of cytosine methylation were obtained by dividing the intensity of a given band by all summed product intensities in a given lane, including the run-off product at the top of the gel generated by primer extension on templates lacking cytosine residues (i.e. templates not methylated in vivo). Oligo nucleotides used for PCR amplification of bisulfite-treated DNA are described in Table 1 using the original naming conventions of Frommer et al. (44).
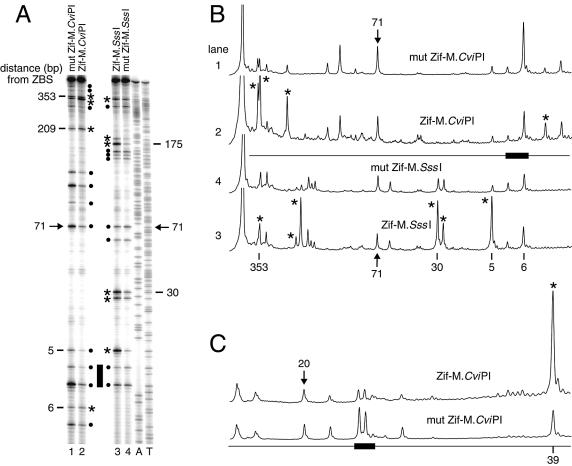
Figure 2.
Zif268 targets M.CviPI and M.SssI to additional ZBS. (A) Determination of m5C levels. A region of YBR108W (+1564 to +2163) spanning a single Zif268 site (+2067 to +2075; filled bar) was PCR amplified from the same bisulfite-treated samples analyzed in Figure 1A. Sequencing ladders (A, T) are at the right. Symbols are defined in the caption to Figure 1. (B) Scans of the phosphoimage in (A). The scanned lanes are indicated at the left. (C) Methylation targeted to a third Zif268 site (filled bar) near YOL019W (–397 to –389). Only scans of the phosphoimage resulting from the bisulfite genomic sequencing of the region from –509 to +254 are shown.
Figure 3.
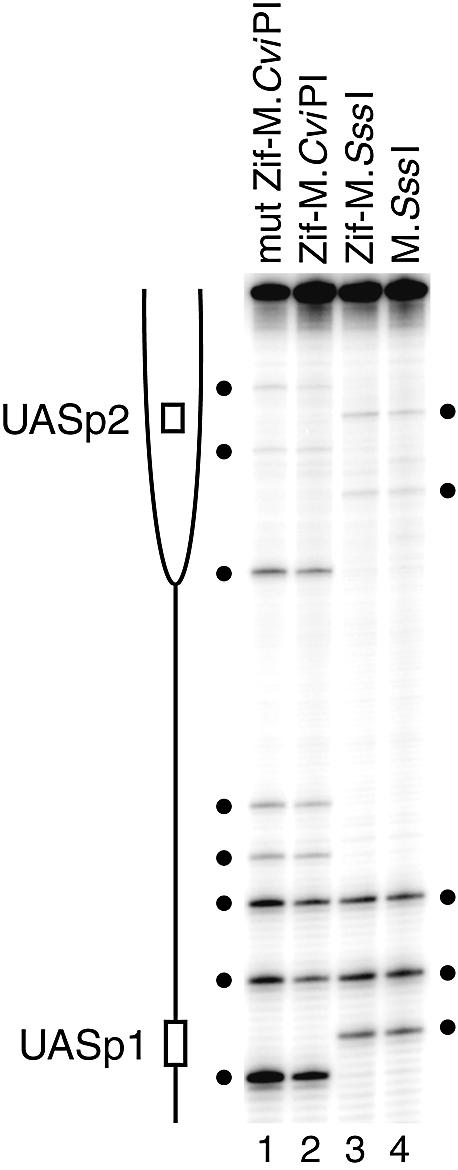
Absence of site-selective methylation at unlinked loci. The PHO5 promoter (–1009 to –205), lacking Zif268 sites, was PCR amplified from the same bisulfite-treated samples analyzed in Figures 1 and 2A to determine levels of m5C. The positions of the two known Pho4 transactivator binding sites, UASp1 and UASp2 (open bars), localized to a histone-free, DNase I-hypersensitive region and positioned nucleosome –2 (partial ellipse) (64), respectively, are indicated. GC (lanes 1 and 2) and CG (lanes 3 and 4) sites (filled circles). Note that, relative to the mut Zif–M.CviPI control (lane 1), the lower methylation frequencies in the Zif–M.CviPI strain (lane 2) at each GC site is consistent with the conclusion that it has reduced overall methylation activity. However, the similar ratios of site intensities within lanes 1 and 2 (M.CviPI) as well as within lanes 3 and 4 (M.SssI) demonstrate that m5C accumulates independent of the Zif (or mut Zif) fusion moiety.
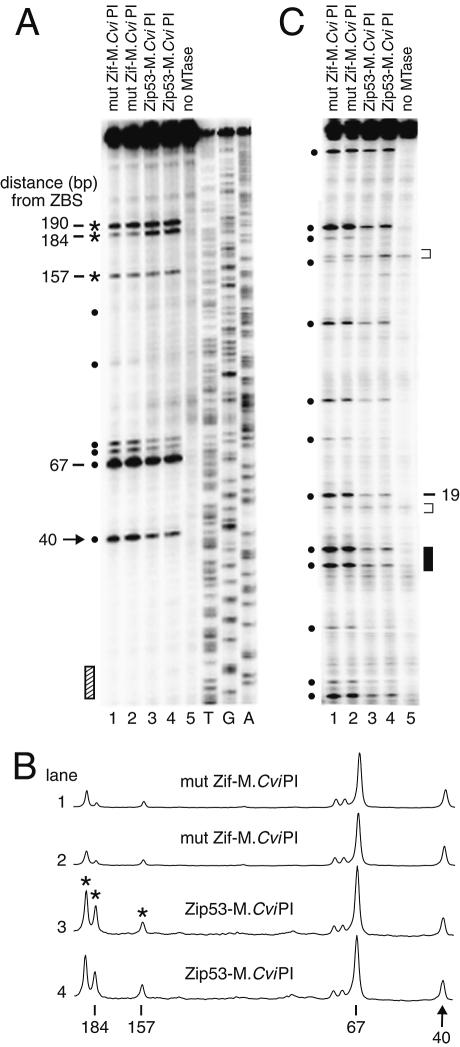
Figure 5.
Zip53-mediated targeting of m5C to YLR016C. (A) Determination of m5C levels at YLR016C (+418 to +28). Targeted methylation (asterisks), as normalized to site 40 (arrow), is detected at GC sites 157, 184 and 190 bp away from the Zip53 site (hatched bar) in two independent transformants (lanes 3 and 4) relative to mut Zif–M.CviPI (lanes 1 and 2). T, G and A sequencing ladders are at the right. Non-targeted methylation (filled circles). The sample in lane 5 was obtained from a Zip53–M.CviPI transformant harboring a non-functional DMTase. (B) Quantitative scans of the phosphoimage in (A). (C) Determination of m5C levels at the CAR1 locus (+558 to +159) that has a Zif268 binding site (filled bar) but no consensus Zip53 binding site. The PCR products analyzed in lanes 1–4 were amplified from the bisulfite-treated genomic analyzed in lanes 1–4, respectively, in (A). Lane 5 contains a sample from a parental strain that was not transformed with M.CviPI. Non-specific primer extension pauses that do not correspond to GC sites are marked with brackets. Symbols in (B) and (C) are as defined in (A).
Table 1. Bisulfite genomic sequencing primers.
| Oligonucleotides for PCR amplification | 32P-end-labeled oligonucleotides for primer extension | ||||
|---|---|---|---|---|---|
| Primera | Sequence | Figure | Primer | Sequence | Figure |
| CAR1b1–60 | CCATTTaAaaaACTCaaaACAATaTaaaAC | 1 and 5C | CAR1b1–60 | CCATTTaAaaaACTCaaaACAATaTaaaAC | 1 and 5C |
| CAR1b2–61 | TAtGGAATTAGAGtttTtAATGGAtGAG | 1 and 5C | YBR108Wa1–58 | aCTaTTaaaaaCATCACCCAAACaAC | 2A and B |
| YBR108Wa1–58 | aCTaTTaaaaaCATCACCCAAACaAC | 2A and B | YOL019Wb1–54 | CCaTAaaAATCAAATTTaCAACAAaTCTaC | 2C |
| YBR108Wa2–59 | AAAttTTTtAGAtATGGAtTTGATTtTG | 2A and B | PHO5b1–751 | TaTTTTCTCATaTAAaCaaACaTCaTCT | 3 |
| YOL019Wb1–54 | CCaTAaaAATCAAATTTaCAACAAaTCTaC | 2C | DED1a1–166 | AAaTaACTCTTTaTATTCTTTCATATTaTTT | 4 |
| YOL019Wb2-55 | ATAGAttTAGtGTTGtTATTGAGTTTttAGTTG | 2C | YLR016Ca1–1019 | CATTCATaAAaATaAaTTCaTAATCaTTATC | 5A and B |
| PHO5b1–922 | TTCAATTaCTAAATACAATaTTCCTTaaT | 3 | |||
| PHO5b2–924 | GAAAAtAGGGAttAGAATtATAAATTTAGTtT | 3 | |||
| DED1a1–166 | AAaTaACTCTTTaTATTCTTTCATATTaTTT | 4 | |||
| DED1a2–165 | ttTtAtTTAAGAGGAAAAttAAGAAGT | 4 | |||
| YLR016Ca1–1020 | TaACaaaAATTCAAAAaTTAACACACTaaAT | 5A and B | |||
| YLR016Ca2–1021 | GAtTATGTATGAAtTAGTGATATAtAG | 5A and B | |||
aPairs of ‘a’ (a1 and a2) or ‘b’ (b1 and b2) are PCR amplification primers for the upper and lower DNA strands, respectively, from bisulfite-treated DNA. Nucleotides in lower case represent either G to a or C to t transitions.
RESULTS
In vivo targeting of C5 DMTases near single, Zif268 binding sites
m5C has been selectively targeted in vitro by fusing C5 DMTases (M.HhaI, M.HpaII and M.SssI) to zinc-finger DNA-binding factors (37,38). However, attempts to use zinc-finger proteins as targeting entities in vivo have been unsuccessful (38). As a first step toward targeting DNA methylation in vivo using zinc-finger proteins, we tested whether we could increase cytosine methylation levels neighboring zinc-finger protein binding sites (ZBS) in the genetically tractable eukaryote, Saccharomyces cerevisiae. Yeast genomic DNA does not contain detectable levels of endogenous methylated residues (46) (Figs 4A, lane 4 and 5A and C, lane 5), enabling unambiguous detection of de novo DNA methylation. Also, low-level expression of C5 DMTases in yeast has no known effects on gene expression or growth (26,27,47).
Figure 4.
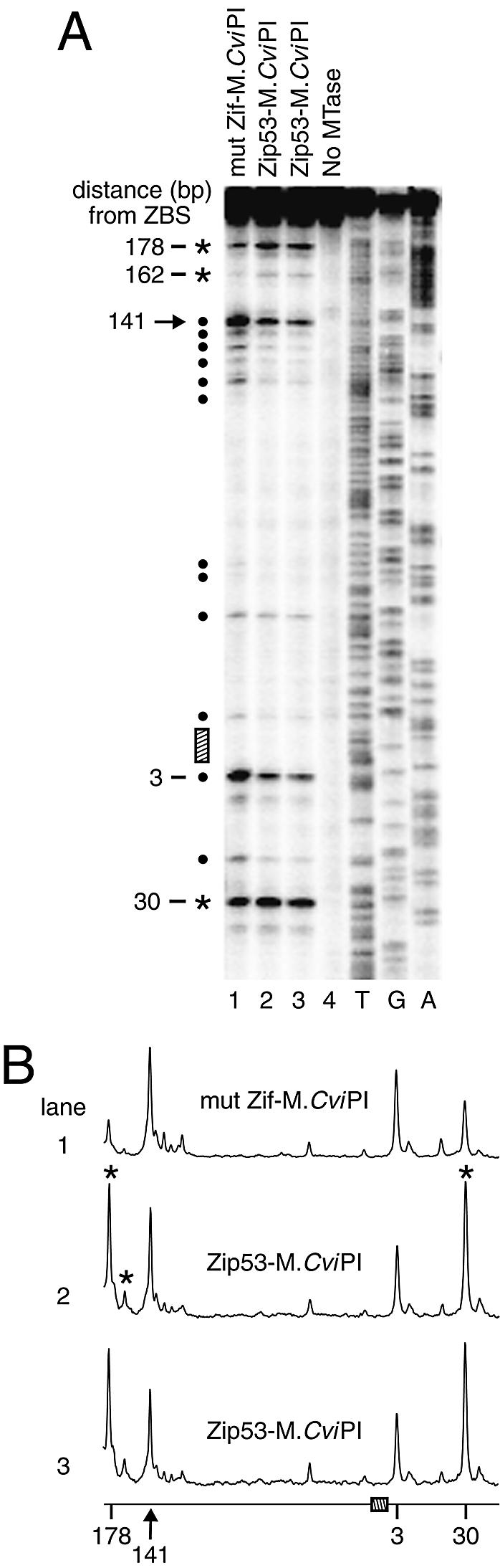
The engineered zinc-finger protein, Zip53, targets a DMTase to DED1. (A) Determination of m5C levels at DED1 (+475 to +67). Targeted methylation (asterisks), normalized to site 141 (arrow), is detected at GC sites 30, 162 and 178 bp away from the Zip53 binding site (hatched bar) in two Zip53–M.CviPI strains (lanes 2 and 3) that are representative of four independent strains containing the integrated Zip53–M.CviPI fusion gene. Filled circles indicate remaining CG and GC sites of non-targeted methylation (not selectively methylated) on expression of Zip53–M.CviPI. Lanes 1 and 4 contain bisulfite genomic sequencing results from the mut Zif–M.CviPI strain and a Zip53–M.CviPI transformant that contains a non-functional DMTase, respectively. Sequencing ladders (T, G, A) are at the right. (B) Quantitative scans of the phosphoimage in (A). Symbols are defined as in (A).
Since chromatin blocks access of DMTases to their target sites (26,27,47–49), our efforts to target m5C in vivo focus on the use of enzymes that methylate dinucleotide sites. This substantially increases the probability (∼20-fold) that DMTase target sites located in accessible, histone-free regions will be modified. Either of two C5 DMTases, M.CviPI (GC specificity) (39) or M.SssI (CG specificity) (50), was tethered to the archetypal zinc-finger protein, Zif268 (51) and expressed as a single-copy, integrated gene under the control of the galactose-inducible GAL1 promoter. The DNA-binding factor that is fused to the DMTase is designated the targeting factor. As a control, we expressed either the untethered DMTase or a fusion protein in which the DNA-binding activity of Zif268 was severely impaired (42). Strains expressing these ‘free’ DMTase controls establish the level of non-targeted methylation due to enzyme site preferences and accessibility in protein-free DNA and chromatin (26,27,47–49).
Endogenous yeast Zif268 binding sites (5′-GCGTGGGCG-3′) were identified by the PatMatch search engine (http://www.yeastgenome.org/). We determined the relative methylation frequencies at multiple GC (M.CviPI) and CG (M.SssI) sites at the CAR1 locus containing a single, consensus binding site for Zif268 by bisulfite genomic sequencing (see Materials and Methods) (40). Specific binding by the Zif268 moiety of each fusion protein is supported by protection of multiple CG and GC sites against methylation at the Zif268 site in strains expressing a wild-type Zif268 fusion as compared with its respective free DMTase (Fig. 1A–C, compare lanes 1 with 2 and 3 with 4, filled bar). Ratios of m5C among several sites in a given lane are similar, identifying sites at which non-targeted methylation occurs (filled circles), which enables normalization for differences in methylation activity between strains. By this criterion, the mut Zif–M.CviPI strain has ∼2-fold more methylation activity than cells expressing Zif–M.CviPI. The reason for this activity difference is unclear. DNA methylation increased substantially at several sites (asterisks) in cells expressing Zif–M.CviPI and Zif–M.SssI versus mut Zif–M.CviPI and M.SssI, respectively. Targeted modification sites (asterisks) are readily identifiable by normalizing to sites of non-targeted methylation (filled circles). In the case of Zif–M.CviPI, which targets m5C more efficiently, 41, 12.4, 2.3 and 2.6% of GC sites 19, 41, 163 and 172, were methylated, respectively. Since the methylation level at each of these sites exceeds that at the normalization site 46 over a time course of Zif–M.CviPI induction, different levels/duration of Zif–M.CviPI expression do not affect the relative efficiency of targeting m5C (Fig. 1C). The fold increases in m5C at each targeted site in strains expressing Zif–M.CviPI versus mut Zif–M.CviPI in three independent experiments are presented in Figure 1D.
M.CviPI is targeted most efficiently to a site located 19 bp from the ZBS (Fig. 1D), which correlates well with the optimal distance of 10–40 bp observed when methylating oligonucleotides with other DMTase fusion proteins in vitro (37,38) and in yeast (40). This optimal distance for introducing m5C is likely related to the length and amino acid sequence of the flexible peptide separating Zif268 and the DMTase (38). However, targeting methylation distal to the consensus ZBS (e.g. sites 163 and 183) is as or more efficient than to some proximal sites (e.g. sites 41, 43 and 52) (Fig. 1A–D). Preferential targeting of M.CviPI and M.SssI also occurs distally, at sites 163–183 nt from the ZBS (Fig. 1A–D). A single, DNA-bound monomer of Zif268 similarly targets both DMTases close to (5–30 bp) and at a considerable distance from (353 bp) a second consensus Zif268 site in YBR108W (+2067 to +2075; Fig. 2A and B). For a third Zif268 binding site (–397 to –389 of YOL019W), two GC sites are protected against methylation by Zif–M.CviPI bound at the ZBS, and m5C is targeted to an additional GC site 39 bp from the ZBS (Fig. 2C). In contrast, the relative levels of CG or GC site methylation at the PHO5 promoter, which lacks Zif268 sites, show no significant differences between the wild-type Zif268 fusion and its respective free DMTase control (Fig. 3, compare lanes 1 with 2 and 3 with 4). We conclude that the targeted methylation is due to site-specific DNA binding by Zif268.
Targeting M.CviPI via phage display-selected Zip53
The engineered zinc-finger protein Zip53, which specifically binds to a p53 consensus site (5′-GGGACATGT-3′; hereafter Zip53 binding site) (41), was previously fused to M.SssI and used in vitro to target methylation next to its cognate binding site in an oligonucleotide substrate (37). We tested if Zip53 could direct methylation by M.CviPI to regions containing a single Zip53 site in vivo. As above, the Zip53–M.CviPI fusion protein was integrated as a single copy at LYS2 and expressed from the GAL1 promoter. First, we analyzed m5C levels near the consensus Zip53 binding site located in the DED1 coding sequence (+284 to +276; Fig. 4). As expected, since yeast do not have endogenous cytosine DMTases, no modified cytosines are evident in a strain that does not contain a functional copy of M.CviPI (Fig. 4A, lane 4). Normalizing to site 141, relative to the ‘free’ DMTase control (mut Zif–M.CviPI), targeted methylation is detected 30 bp from the DED1 consensus Zip53 site on expression of Zip53–M.CviPI (Fig. 4A, compare lanes 2 and 3 with lane 1). Further, long-range methylation at sites 162 and 178 bp from the consensus ZBS is substantially enhanced. Lastly, there is reproducible, low-level protection of a GC site located 3 bp from the ZBS, indicative of Zip53 binding (Fig. 4A and B).
We also observed long-range targeting of m5C from a second consensus Zip53 site located in the YLR016C coding sequence (+298 to +306; Fig. 5A and B). Methylation was enhanced 5.5-fold at site 184, and somewhat less, but significantly (∼2.2-fold), at sites 157 and 190 in strains expressing Zip53–M.CviPI relative to mut Zif–M.CviPI. Protection against DNA methylation could not be observed because no GC sites are adjacent to or within the Zip53 binding site. To examine the specificity of the Zip53–DMTase fusion protein, we analyzed m5C levels at the CAR1 locus (Fig. 1), which contains a Zif268 site, but no Zip53 site (Fig. 5C). In each lane of the gel in Figure 5C, little to no change exists in the relative methylation levels of 13 GC sites at CAR1. In particular, methylation at site 19 of the CAR1 region, which shows >20-fold enrichment following expression of Zif–M.CviPI (Fig. 1), is not increased in the presence of Zip53–M.CviPI. This result demonstrates that Zip53 specifically binds its site, but not that of Zif268 (the two binding sites have 22% identity). We conclude that, as for Zif268, Zip53 is able to target M.CviPI and thereby significantly increase cytosine methylation at select GC sites near and distal to a cognate ZBS. The use of Zip53 to deliver m5C selectively further demonstrates that zinc-finger proteins engineered to recognize pre-determined sequences can be used to introduce de novo methylation essentially to any region of interest.
DISCUSSION
In this report, we demonstrate the ability to target m5C in vivo using two zinc-finger proteins, Zif268 and its artificially engineered derivative Zip53. First, significant targeting of m5C is observed at select sites both adjacent (5–52 bp) and distal (>150 bp) to a cognate, consensus ZBS (Figs 1, 2, 4 and 5A and B), whereas DNA methylation is not enriched at control loci lacking the ZBS (Figs 3 and 5C). Proximal and distal targeting of m5C was also observed in our previous studies using Pho4 as the DMTase targeting factor (40). The reasons for selective targeting of m5C to some sites as opposed to others in the same region are not currently understood. At least locally, the length of the peptide linker separating the DMTase and the targeting factor, the helical face of a particular CG or GC site relative to the DNA-bound targeting factor, and accessibility in chromatin each presumably contribute to the preferential targeting. Secondly, since DNA-bound factors impair access of DMTases to their target sites (26–30,40), the protection against methylation of CG or GC sites next to or within the ZBS provides further evidence of specific ZBS binding by each zinc-finger–DMTase fusion protein. Taken together, in addition to demonstrating selective enrichment of m5C near ZBS, TAGM provides a highly sensitive means for detecting protein–DNA interactions (40).
The occurrence of targeted m5C beyond distances of 40 nt suggests that two sites well separated in protein-free DNA are juxtaposed by looping, nucleosomes or higher-order chromatin structure (e.g. Fig. 2A, 353 bp away from the ZBS). While it is formally possible that the occurrence of distal methylation is due to binding at a secondary, non-consensus ZBS, we do not believe this to be the case. First, no footprints are observed near MTase target sites that are >150 bp from the primary Zif268 binding site, despite the fact that high-affinity sites contain two GC sites. Secondly, within 50 bp of each long-range targeted m5C site any potential ZBS has a minimum of four to five mismatches from consensus. Since two or more base substitutions result in background levels of binding (42,52–54), it is highly unlikely that any of these sites constitute a significant secondary ZBS. Thirdly, it is improbable that distal secondary ZFP binding sites would be present at all five loci that were analyzed. Finally, Dam MTase can also be targeted at a distance (55,56).
The design of multiple zinc-finger modules with desired specificities is proving a versatile platform for targeting a variety of protein moieties to accessible sites in vivo (57). For instance, engineered zinc-finger proteins have been fused to the catalytic domain of R.FokI endonuclease to direct site-specific double-stranded DNA cleavage, and hence homologous recombination, of desired regions (58). Designed zinc-finger proteins have also been used to target the catalytic domains of the histone methyltransferases G9A and SUV39H1 (59) as well as the VP16 activation domain (60–62), leading to repression and activation, respectively, of expression of the endogenous human erythropoietin (EPO), vascular endothelial growth factor A (VEGF-A) and other mammalian genes (57). This technology has also recently been extended to the regulation of gene expression in plants (63).
The targeting of DMTases by zinc-finger proteins selected to bind specific ZBS could provide an additional way to down-regulate the expression of desired genes. Moreover, since the DNA methylation state of a given promoter is maintained heritably through DNA replication by endogenous cellular mechanisms, an initial targeting event may be sufficient to establish stable silencing of improperly expressed genes. Therefore, heritable repression could also reduce the amount of treatment necessary to establish the proper regulation of a particular gene. In addition to providing a potentially powerful therapeutic tool, methylation-mediated repression of specifically targeted genes could yield an alternative to transgenic knockouts for studying loss-of-function phenotypes. Silencing genes through DNA methylation would be particularly valuable in the case of essential genes where tissue-specific knockouts of function are needed. Optimization of targeting factor occupancy at regions of interest will likely increase the efficacy of specific m5C targeting in vivo as well as minimize non-targeted methylation. The experimental system used herein provides a useful assay for pursuing such further investigations. Finally, the ability to target m5C specifically in vivo is likely to prove valuable in basic investigations of the biological roles and mechanistic consequences of DNA methylation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Guo-Liang Xu and Tim Bestor for providing plasmids containing Zif268 and Zip53 fused to M.SssI as well as helpful discussions. We also thank Tim Bestor, Mary Bryk and Archana Dhasarathy for their helpful comments on the manuscript and other members of the Kladde laboratory for useful insights throughout this study. This work was supported by awards from the American Heart Association—Texas Affiliate, the National Cancer Institute (CA095525) and the Texas Higher Education Coordinating Board.
REFERENCES
- 1.Attwood J.T., Yung,R.L. and Richardson,B.C. (2002) DNA methylation and the regulation of gene transcription. Cell Mol. Life Sci., 59, 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu W.G., Srinivasan,K., Dai,Z., Duan,W., Druhan,L.J., Ding,H., Yee,L., Villalona-Calero,M.A., Plass,C. and Otterson,G.A. (2003) Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21Cip1 promoter. Mol. Cell. Biol., 23, 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade P.A. (2001) Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene, 20, 3166–3173. [DOI] [PubMed] [Google Scholar]
- 4.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 5.Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 6.Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson J.P., Lindroth,A.M., Cao,X. and Jacobsen,S.E. (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 416, 556–560. [DOI] [PubMed] [Google Scholar]
- 9.Fuks F., Hurd,P.J., Deplus,R. and Kouzarides,T. (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res., 31, 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pikaart M.J., Recillas-Targa,F. and Felsenfeld,G. (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev., 12, 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubeler D., Lorincz,M.C., Cimbora,D.M., Telling,A., Feng,Y.Q., Bouhassira,E.E. and Groudine,M. (2000) Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure and histone acetylation. Mol. Cell. Biol., 20, 9103–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorincz M.C., Schubeler,D. and Groudine,M. (2001) Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol., 21, 7913–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvine R.A., Lin,I.G. and Hsieh,C.L. (2002) DNA methylation has a local effect on transcription and histone acetylation. Mol. Cell. Biol., 22, 6689–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorincz M.C., Schubeler,D., Hutchinson,S.R., Dickerson,D.R. and Groudine,M. (2002) DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation. Mol. Cell. Biol., 22, 7572–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutskov V.J., Farrell,C.M., Wade,P.A., Wolffe,A.P. and Felsenfeld,G. (2002) The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev., 16, 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cameron E.E., Bachman,K.E., Myohanen,S., Herman,J.G. and Baylin,S.B. (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet., 21, 103–107. [DOI] [PubMed] [Google Scholar]
- 17.Magdinier F. and Wolffe,A.P. (2001) Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl Acad. Sci. USA, 98, 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrich B., Guy,J., Ramsahoye,B., Wilson,V.A. and Bird,A. (2001) Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev., 15, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy J., Hendrich,B., Holmes,M., Martin,J.E. and Bird,A. (2001) A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nature Genet., 27, 322–326. [DOI] [PubMed] [Google Scholar]
- 20.Georgel P.T., Horowitz-Scherer,R.A., Adkins,N., Woodcock,C.L., Wade,P.A. and Hansen,J.C. (2003) Chromatin compaction by human MeCP2: assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem., 278, 32181–32188. [DOI] [PubMed] [Google Scholar]
- 21.Robertson K.D. (2001) Transcriptional activation of the Epstein–Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Oncogene, 20, 3139–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 23.Jones P.A. and Baylin,S.B. (2002) The fundamental role of epigenetic events in cancer. Nature Rev. Genet., 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 24.Stein R., Razin,A. and Cedar,H. (1982) In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc. Natl Acad. Sci. USA, 79, 3418–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 26.Kladde M.P., Xu,M. and Simpson,R.T. (1996) Direct study of DNA–protein interactions in repressed and active chromatin in living cells. EMBO J., 15, 6290–6300. [PMC free article] [PubMed] [Google Scholar]
- 27.Xu M., Simpson,R.T. and Kladde,M.P. (1998) Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol. Cell. Biol., 18, 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh C.L. (1999) Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol., 19, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin I.G. and Hsieh,C.L. (2001) Chromosomal DNA demethylation specified by protein binding. EMBO Rep., 2, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin I.G., Tomzynski,T.J., Ou,Q. and Hsieh,C.L. (2000) Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol., 20, 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feinberg A.P. and Vogelstein,B. (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature, 301, 89–92. [DOI] [PubMed] [Google Scholar]
- 32.Robertson K.D. (2001) DNA methylation, methyltransferases, and cancer. Oncogene, 20, 3139–3155. [DOI] [PubMed] [Google Scholar]
- 33.Boyes J. and Bird,A. (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell, 64, 1123–1134. [DOI] [PubMed] [Google Scholar]
- 34.Boyes J. and Bird,A. (1992) Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J., 11, 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh C.L. (1994) Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol., 14, 5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron E.E., Baylin,S.B. and Herman,J.G. (1999) p15INK4B CpG island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencing. Blood, 94, 2445–2451. [PubMed] [Google Scholar]
- 37.Xu G.L. and Bestor,T.H. (1997) Cytosine methylation targetted to pre-determined sequences. Nature Genet., 17, 376–378. [DOI] [PubMed] [Google Scholar]
- 38.McNamara A.R., Hurd,P.J., Smith,A.E. and Ford,K.G. (2002) Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res., 30, 3818–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M., Kladde,M.P., Van Etten,J.L. and Simpson,R.T. (1998) Cloning, characterization and expression of the gene coding for cytosine-5-DNA methyltransferase recognizing GpC sites. Nucleic Acids Res., 26, 3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvin C.D., Dhasarathy,A., Friesenhahn,L.B., Jessen,W.J. and Kladde,M.P. (2003) Targeted cytosine methylation for in vivo detection of protein–DNA interactions. Proc. Natl Acad. Sci. USA, 100, 7743–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greisman H.A. and Pabo,C.O. (1997) A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science, 275, 657–661. [DOI] [PubMed] [Google Scholar]
- 42.Nardelli J., Gibson,T.J., Vesque,C. and Charnay,P. (1991) Base sequence discrimination by zinc-finger DNA-binding domains. Nature, 349, 175–178. [DOI] [PubMed] [Google Scholar]
- 43.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 44.Frommer M., MacDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proffitt J.H., Davie,J.R., Swinton,D. and Hattman,S. (1984) 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol. Cell. Biol., 4, 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh J. and Klar,A.J.S. (1992) Active genes in yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev., 6, 186–196. [DOI] [PubMed] [Google Scholar]
- 48.Gottschling D.E. (1992) Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl Acad. Sci. USA, 89, 4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kladde M.P. and Simpson,R.T. (1994) Positioned nucleosomes inhibit Dam methylation in vivo. Proc. Natl Acad. Sci. USA, 91, 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renbaum P., Abrahamove,D., Fainsod,A., Wilson,G., Rottem,S. and Razin,A. (1990) Cloning, characterization and expression in Escherichia coli of the gene coding for the CpG DNA from Spiroplasma sp strain MQ-1 (M.SssI). Nucleic Acids Res., 18, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chavrier P., Zerial,M., Lemaire,P., Almendral,J., Bravo,R. and Charnay,P. (1988) A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J., 7, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nardelli J., Gibson,T. and Charnay,P. (1992) Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res., 20, 4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamieson A.C., Wang,H. and Kim,S.-H. (1996) A zinc finger directory for high-affinity DNA recognition. Proc. Natl Acad. Sci. USA, 93, 12834–12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller J.C. and Pabo,C.O. (2001) Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol., 313, 309–315. [DOI] [PubMed] [Google Scholar]
- 55.van Steensel B. and Henikoff,S. (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol., 18, 424–428. [DOI] [PubMed] [Google Scholar]
- 56.Lebrun E., Fourel,G., Defossez,P.-A. and Gilson,E. (2003) A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol. Cell. Biol., 23, 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urnov F.D. and Rebar,E.J. (2002) Designed transcription factors as tools for therapeutics and functional genomics. Biochem. Pharmacol., 64, 919–923. [DOI] [PubMed] [Google Scholar]
- 58.Bibikova M., Carroll,D., Segal,D.J., Trautman,J.K., Smith,J., Kim,Y.G. and Chandrasegaran,S. (2001) Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol., 21, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snowden A.W., Gregory,P.D., Case,C.C. and Pabo,C.O. (2002) Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol., 12, 2159–2166. [DOI] [PubMed] [Google Scholar]
- 60.Liu P.Q., Rebar,E.J., Zhang,L., Liu,Q., Jamieson,A.C., Liang,Y., Qi,H., Li,P.X., Chen,B., Mendel,M.C. et al. (2001) Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions: activation of vascular endothelial growth factor A. J. Biol. Chem., 276, 11323–11334. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Spratt,S.K., Liu,Q., Johnstone,B., Qi,H., Raschke,E.E., Jamieson,A.C., Rebar,E.J., Wolffe,A.P. and Case,C.C. (2000) Synthetic zinc finger transcription factor action at an endogenous chromosomal site: activation of the human erythropoietin gene. J. Biol. Chem., 275, 33850–33860. [DOI] [PubMed] [Google Scholar]
- 62.Rebar E.J., Huang,Y., Hickey,R., Nath,A.K., Meoli,D., Nath,S., Chen,B., Xu,L., Liang,Y., Jamieson,A.C. et al. (2002) Induction of angiogenesis in a mouse model using engineered transcription factors. Nature Med., 8, 1427–1432. [DOI] [PubMed] [Google Scholar]
- 63.Sanchez J.P., Ullman,C., Moore,M., Choo,Y. and Chua,N.H. (2002) Regulation of gene expression in Arabidopsis thaliana by artificial zinc finger chimeras. Plant Cell Physiol., 43, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 64.Almer A. and Hörz,W. (1986) Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J., 5, 2681–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]