Abstract
Following amputation, freshwater planarians properly regenerate a head or tail from the resulting anterior or posterior wound. The mechanisms that differentiate anterior from posterior and direct the replacement of the appropriate missing body parts are unknown. Here we report that RNA interference (RNAi) of β-catenin or dishevelled causes the inappropriate regeneration of a head instead of a tail at posterior amputations. Conversely, RNAi of the β-catenin antagonist adenomatous polyposis coli (APC) results in the regeneration of a tail at anterior wounds. In addition, the silencing of β-catenin is sufficient to transform the tail of uncut adult animals into a head. We suggest that β-catenin functions as a molecular switch to specify and maintain anteroposterior (A/P) identity during regeneration and homeostasis in planarians.
β-Catenin is a multi-functional protein that controls transcriptional output as well as cell adhesion. During embryonic development of both vertebrates and invertebrates β-catenin regulates a variety of cellular processes, including organizer formation, cell fate specification, proliferation, and differentiation (1–9). In adult animals, the Wnt/β-catenin pathway participates in regeneration and tissue homeostasis, while pathway misregulation can lead to degenerative diseases and cancer in humans (9–12). In response to upstream cues, such as Wnt ligands binding to Frizzled receptors, β-catenin accumulates in nuclei (Fig. 1A) and invokes transcriptional responses that direct the specification and patterning of tissues (13, 14). APC is an essential member of a destruction complex that phosphorylates β-catenin, resulting in its constitutive degradation. Hence, loss of APC leads to a rise in β-catenin levels that is sufficient to drive transcriptional responses (15). The intracellular protein Dishevelled has multiple functions but plays an essential role as a positive regulator of β-catenin by inhibiting the destruction complex (16).
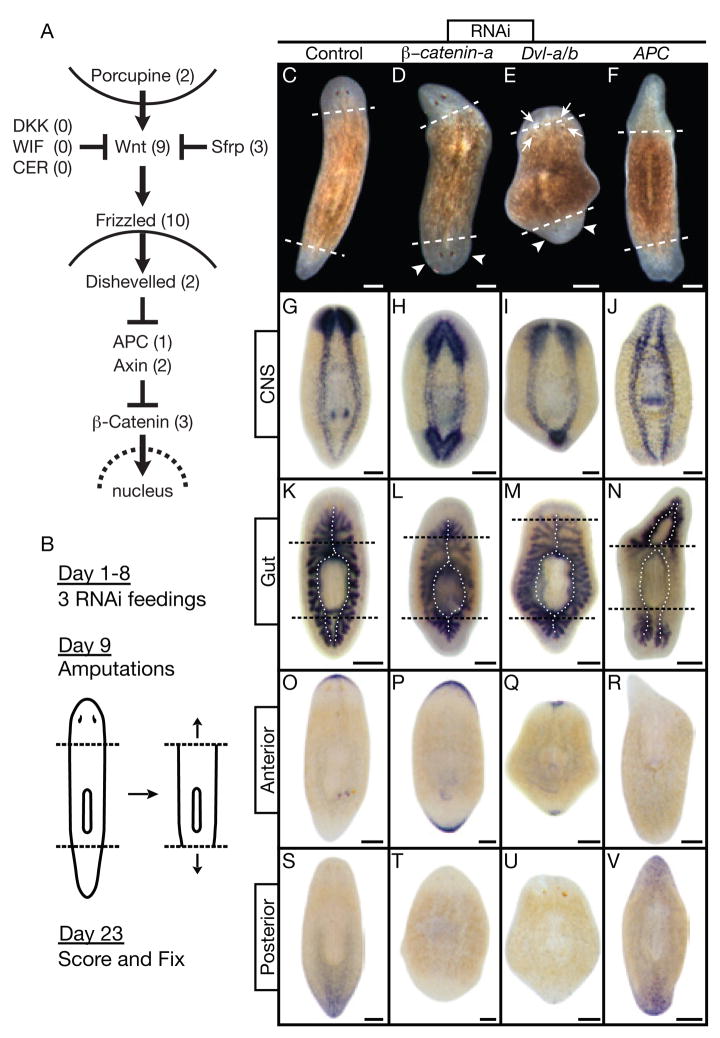
Figure 1.
Signaling through β-catenin defines head vs. tail during regeneration. (A) Canonical Wnt pathway. Numbers: S. mediterranea homologs identified and silenced. (B) Experimental strategy. (C through V) Trunk fragments 14 days post-amputation. Dashed lines: amputation planes. Control: unc-22(RNAi). (C through F) Live animals. (D, E) Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) posterior blastemas formed heads with photoreceptors (arrowheads). SmedDvl-a/b(RNAi) fragments also developed ectopic photoreceptors, often within old tissues (arrows). (F) SmedAPC(RNAi) anterior blastemas formed tails. (G through V) In situ hybridizations, n≥5 per marker. Markers: CNS, pro-hormone convertase 2 (PC2); Gut, Porcupine (SmedPorcn-a); Anterior, secreted Frizzled-related protein (SmedSfrp-a); Posterior, Frizzled (SmedFz-d). (G,K) Normal brain and ventral nerve cords; single anterior and dual posterior gut branches (dotted lines). (H,I,L,M) Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) posterior blastemas developed brain tissue and head-like gut branches. (J,N) SmedAPC(RNAi) anterior blastemas developed tail-like nerve cords and gut branches. (O,S) Normal anterior and posterior marker expression. (P,Q,T,U) Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) induced anterior marker expression at both ends while the posterior marker was virtually undetectable. (R,V) SmedAPC(RNAi) induced posterior marker expression at both ends while the anterior marker was virtually undetectable. Scale bars: 200 μm.
As part of a systematic effort to define the roles of signaling pathways in planaria, we analyzed the canonical Wnt signaling system in Schmidtea mediterranea. We cloned and determined the expression patterns of all identifiable homologs of core pathway components (Fig. 1A) and silenced them individually or in combinations based on the likelihood of redundancy gleaned from the expression data (Fig. S1–6; and table S1). RNAi-treated animals were then amputated to assess the role of the silenced genes during regeneration (Fig. 1B). After head and tail amputations of control worms, the remaining trunk formed one anterior and one posterior blastema, which then differentiated to replace the missing structures (Fig. 1C; n=28). However, RNAi of a single β-catenin (Smedβ-catenin-a), both Dishevelled homologs (SmedDvl-a/b), or APC (SmedAPC) caused dramatic alterations in the A/P identity of regenerating tissues (Fig. 1D–F). Both blastemas of Smedβ-catenin-a(RNAi) worms adopted an anterior fate, resulting in animals with two heads of opposite orientation (penetrance=100%, n=39). SmedDvl-a/b(RNAi) worms also regenerated heads from both blastemas but displayed additional phenotypes, including ectopic and supernumerary photoreceptors in the anterior region. This is consistent with the multiple roles of dishevelled in different pathways (Fig. 1E; penetrance=75%, n=20). On the other hand, SmedAPC(RNAi) animals regenerated tails from both amputation planes (Fig. 1F; penetrance=60%, n=43).
To address whether these changes were superficial or reflected a fate transformation of internal cell types and organ systems, we utilized anatomical and molecular markers of A/P identity. Anatomically, two organ systems with characteristic asymmetries along the A/P axis were examined: the central nervous system, composed of two anterior cephalic ganglia (brain) and two ventral cords projecting posteriorly (Fig. 1G); and the digestive system, consisting of a single anterior and two posterior gut branches (Fig. 1K). The “posterior” head of Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) animals contained a characteristically anterior nervous system and gut as did the “anterior” head (Fig. 1H,I,L,M). In contrast, the “anterior” tail of SmedAPC(RNAi) animals was devoid of discernible brain tissue and exhibited posterior structures as did the “posterior” tail (Fig. 1J,N).
To define the molecular extent of A/P mis-specification, we utilized two markers identified during our in situ analyses that are specifically expressed at the anterior and posterior ends of intact and regenerating animals (Fig. 1A,O,S). SmedSfrp-a, a homolog of Secreted frizzled-related proteins (Sfrp), was expressed in an arch of cells capping the anterior edge of the animal. In contrast, SmedFz-d, a homolog of Frizzled receptors, was expressed at the posterior edge in a posterior to anterior gradient. We refer to SmedSfrp-a and SmedFz-d as the “anterior marker” and the “posterior marker”, respectively, in all subsequent analyses.
In Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) worms, the “posterior” head expressed the anterior marker and the posterior marker was severely reduced or absent (Fig. 1P,Q,T,U). Conversely, the “anterior” tail of SmedAPC(RNAi) animals expressed the posterior marker whereas the anterior marker was severely reduced or absent (Fig. 1R,V). We noted that mis-specified heads and tails in RNAi-treated worms moved independently from the rest of the animal, demonstrating that this tissue was functioning autonomously (Movie S1–3). We conclude that silencing Smedβ-catenin-a, SmedDvl-a/b, or SmedAPC is sufficient to re-specify blastema identity.
The inappropriate regeneration of complete heads and tails in RNAi-treated animals suggested that regulators of β-catenin act early during regeneration. We therefore investigated the onset of blastema differentiation in control and RNAi-treated trunk fragments using the anterior and posterior markers (Fig. 2A,B). In control animals at 12 h post-amputation, the anterior marker was virtually undetectable whereas the posterior marker was clearly evident in the posterior blastema. 24 h post-amputation, the two markers recapitulated the A/P specificity seen later during regeneration and in adult animals (Figs. 1O,S; and 2A,B). However, in Smedβ-catenin-a(RNAi) trunks, the anterior marker was expressed at both ends by 12 h and maintained throughout the experiment whereas the posterior marker remained dramatically reduced (Fig. 2A,B). The reverse was observed in SmedAPC(RNAi) trunks (Fig. 2A,B). Consistent with the inferred time window for β-catenin signaling, Smedβ-catenin-a and SmedAPC were expressed at both ends in wild type animals by 12 h (fig. S7). These results indicate that β-catenin and APC act very early to determine blastema identity.
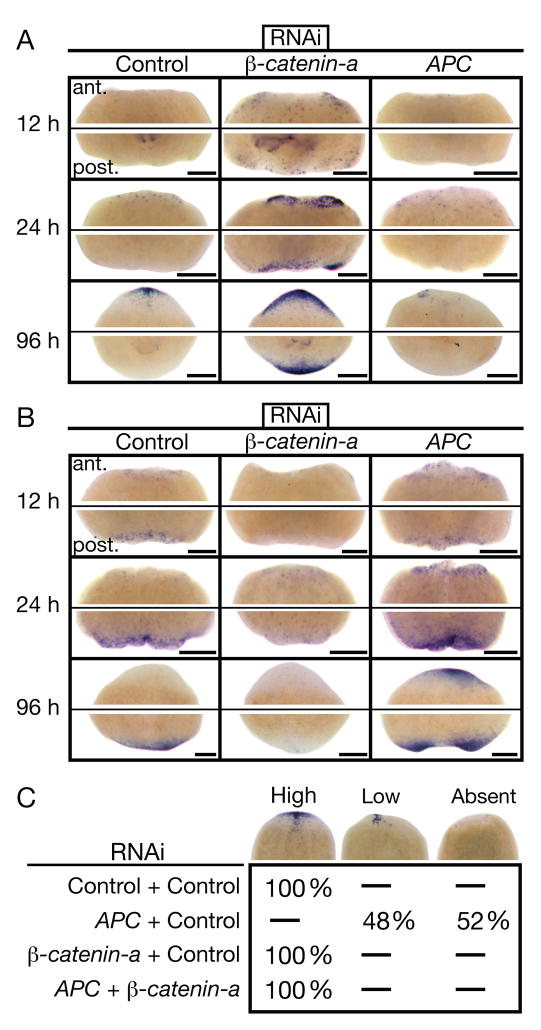
Figure 2.
Smedβ-catenin-a(RNAi) and SmedAPC(RNAi) phenotypes manifest early and the SmedAPC(RNAi) phenotype depends on Smedβ-catenin-a. (A) Anterior (SmedSfrp-a) and (B) posterior (SmedFz-d) marker expression time-course in regenerating trunk fragments. Anterior (ant.) and posterior (post.) blastemas from the same representative fragments are shown, n≥15 per condition. Scale bars: 200 μm. (C) Double-silencing experiments assaying SmedSfrp-a expression in anterior trunk blastemas 4 days post-amputation; scored as High, Low, or Absent (see examples, top row). Percent of scored animals are listed for each category. n≥24 per condition. Control: unc-22(RNAi).
We next used animals silenced for both Smedβ-catenin-a and SmedAPC to test whether the SmedAPC(RNAi) phenotype results from increased β-catenin activity. Under these conditions anterior blastemas were properly fated, indicating that the mis-specification phenotype of SmedAPC(RNAi) depends on Smedβ-catenin-a (Fig. 2C). Additionally, posterior blastemas adopted an anterior fate, indicating that the Smedβ-catenin-a(RNAi) phenotype does not depend on APC activity. The combined data show that signaling through β-catenin occurs at posterior amputations and is necessary and sufficient to specify tail fate. In contrast, signaling through β-catenin is blocked or never occurs at anterior amputations and this is necessary and sufficient to specify head fate. Additionally, the premature expression of the anterior marker in Smedβ-catenin-a(RNAi) worms may indicate that in wild type planarians, β-catenin inhibition does not immediately follow amputation (Fig. 2A). We suggest that β-catenin activity acts as a molecular switch to specify “head” versus “tail” fate in planarians.
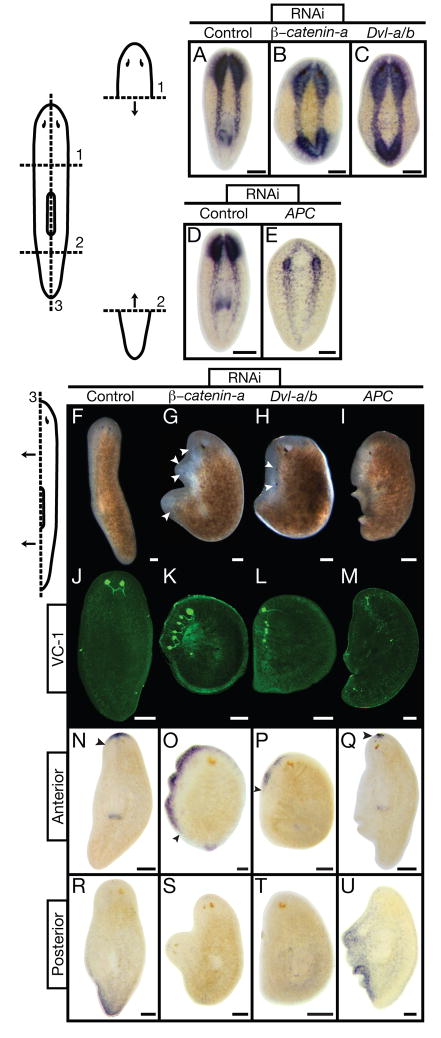
We then explored whether the β-catenin switch plays a role in blastema identity regardless of the A/P location or angle of amputation. Indeed, the head fragments of Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) worms regenerated a head from the posterior wound (penetrance=79%, n=24; penetrance=82%, n=33), and the tail fragments of SmedAPC(RNAi) worms regenerated a tail from the anterior wound (penetrance=67%, n=27; Fig. 3A–E; Movie S4). Following longitudinal amputation along the midline, control animals formed a blastema along the A/P axis and regenerated medio-laterally (Fig. 3F,J,N,R). In contrast, Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) worms regenerated anterior tissue and developed multiple ectopic heads along the lateral edge (Fig. 3G,H,K,L,O,P,S,T). The most severely affected SmedAPC(RNAi) worms regenerated posterior tissue and failed to replace the lost head structures (Fig. 3I,M,Q,U). Thus, the laterally regenerating tissue in RNAi-treated animals was mis-specified. Our data indicate that β-catenin activity is regulated during lateral regeneration and the β-catenin switch can dominantly mis-specify regenerating tissues regardless of A/P position or amputation angle.
Figure 3.
Smedβ-catenin-a, Dvl-a/b, APC (RNAi) phenotypes do not depend on position or orientation of amputation. Fragments shown: 14 days post-amputation. Control: unc-22(RNAi). (A through E) PC2 in situ hybridization (CNS), n≥5. Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) animals regenerated posterior brain tissue; SmedAPC(RNAi) animals failed to regenerate anterior brain tissue. (F through M) During lateral regeneration, Smedβ-catenin- a(RNAi) and SmedDvl-a/b(RNAi) animals developed supernumerary heads and photoreceptors; SmedAPC(RNAi) prevented proper regeneration of these structures, n≥15. Photoreceptors visualized with VC-1 antibody (Kind gift from Dr. K. Agata) (25). (N through U) In situ hybridization, n≥5 per marker. Anterior marker (SmedSfrp-a) expression expanded posteriorly along the blastema in Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) animals, while SmedAPC(RNAi) restricted expression to pre-existing tissues. Posterior marker (SmedFz-d) expression was severely reduced in Smedβ-catenin-a(RNAi) and SmedDvl-a/b(RNAi) animals, but expanded throughout regenerated tissues in SmedAPC(RNAi) animals. (F through I) Live animals. White arrowheads: photoreceptors. Black arrowheads: extent of anterior marker expression. Scale bars: 200 μm.
Smedβ-catenin-a(RNAi) animals also displayed striking changes in thenon-regenerating portions of regenerating fragments. Smedβ-catenin-a(RNAi) tail fragments at 24 and 48 h post-amputation expressed the anterior marker in cell clusters around the circumference of the fragment (Fig. 4A,B,D,E). By day 14, in addition to a new head, moving head-like protrusions developed from the periphery (Fig. 4C,F; Movie S5). Similar protrusions also developed in trunk fragments (Movie S2).
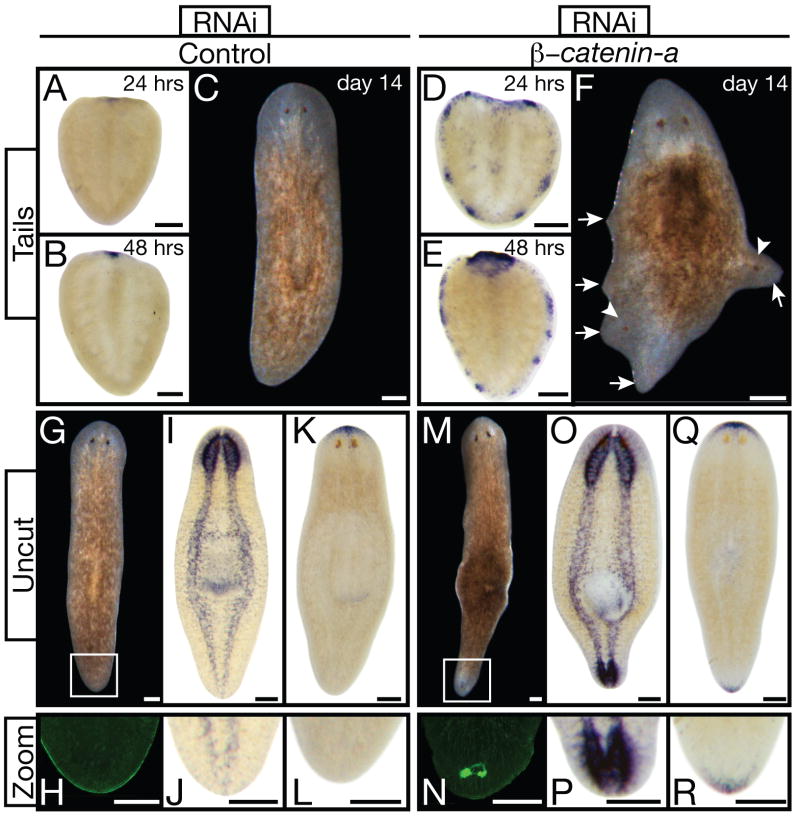
Figure 4.
Smedβ-catenin-a(RNAi) transforms non-regenerating tissues. (A through F) Tail fragments. (A,B,D,E) Anterior marker (SmedSfrp-a) analyses 24 and 48 hours post-amputation demonstrated early ectopic expression in Smedβ-catenin-a(RNAi), n≥5 per condition. (C,F) Live tail fragments 14 days post-amputation. Smedβ-catenin-a(RNAi) caused lateral ectopic protrusions (arrowheads, ectopic photoreceptors; arrows, abnormal protrusions). (G through R) Transformation of tail into head tissue in uncut Smedβ-catenin-a(RNAi) animals 14 days after final RNAi-feeding. (G, M) Living animals. (I, O) PC2 in situ hybridization (CNS), n≥4. (K, Q) Anterior marker expression, n≥4. (H,J,L,N,P,R) Magnification of tail tips (boxes in G, M). (H, N) VC-1 antibody staining (photoreceptors). Control: unc-22(RNAi). Scale bars: 200 μm.
Such fate changes may have been initiated by amputation. We therefore observed un-amputated (intact) worms 14 days after the final RNAi feeding. Ectopic photoreceptors were visible in the tail of all (n=20) Smedβ-catenin-a(RNAi) animals and none of the control animals (Fig. 4G,H,M,N). Intact RNAi-treated animals also exhibited ectopic lateral protrusions, formed a brain in the tail region, and expressed the anterior marker posteriorly (Fig. 4I–L, O–R; Movie S6). The molecular basis for such a change of A/P polarity in an adult organism is previously unknown.
Together, our data demonstrate the fundamental importance of β-catenin in the maintenance of polarity and cell fate during tissue regeneration and homeostasis in planarians (Fig. S8). Our findings reveal a dynamic control of β-catenin in adult animals that is not readily apparent during the progression of embryogenesis: the precise quantity and location of regenerating tissue is different for each individual and for each regeneration event, newly regenerated tissues must integrate with the old, and ongoing homeostatic cell-turnover may require sustained instructive cues. It is interesting that we did not observe any head or tail mis-specification phenotypes for any of the upstream components of canonical Wnt signaling (Wnts, Frizzleds or Porcupines, see Table S1). Although we cannot rule out protein perdurance, incomplete gene silencing, or redundancy with unidentified components, the intracellular components of β-catenin signaling may be regulated by an unconventional upstream mechanism to specify polarity during regeneration and/or homeostasis. Indeed, β-catenin regulation can be Wnt-independent in vertebrate cells and dishevelled remains the most upstream known β-catenin regulator during early sea urchin development (14, 17, 18). With respect to putative downstream effectors, planarians can regenerate double heads after pharmacological gap junction inhibition and β-catenin is implicated in gap junction formation and function (19–21). Finally, whether specification and maintenance of the planarian A/P axis via β-catenin is or is not independent of Hox proteins remains to be determined.
Over 100 years ago, T. H. Morgan reported that fragments with closely-spaced anterior and posterior amputation planes occasionally regenerate two-headed animals (22, 23). He termed these animals “Janus heads” and suggested that “something in the piece itself determines that a head shall develop at the anterior cut surface and a tail at the posterior cut surface” (24). Here we demonstrate that β-catenin activity is a key target of polarity specification in planarians, providing mechanistic insight into the old, unanswered question of how blastema fate is controlled. Reminiscent of its role during metazoan embryogenesis (6, 8), we propose that the evolutionarily ancient β-catenin protein acts as a molecular switch in adult planarians and that it may play a similar role in the adult tissues of other animals.
Supplementary Material
Acknowledgments
We thank B. Pearson and G. Eisenhoffer for optimization of in situ protocols, R. Dorsky, T. Piotrowski, and all Sánchez lab members for comments. K.A.G. is supported by NIH-NIGMS grants F32GM082016, T32CA093247. J.C.R. is funded by the European Molecular Biology Organization. A.S.A. is a Howard Hughes Medical Institute Investigator. Supported by NIH-NIGMS RO-1 GM57260 to A.S.A. For S. mediterranea genes, Genbank accession numbers are EU130785, EU130786, EU130787, EU130788, EU130789, EU130790, EU130791.
References and Notes
- 1.Schneider S, Steinbeisser H, Warga RM, Hausen P. Mech Dev. 1996 Jul;57:191. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 2.Larabell CA, et al. J Cell Biol. 1997 Mar 10;136:1123. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Development. 1999 Jan;126:345. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 4.Wikramanayake AH, et al. Nature. 2003 Nov 27;426:446. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 5.Orsulic S, Peifer M. J Cell Biol. 1996 Sep;134:1283. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocheleau CE, et al. Cell. 1997 Aug 22;90:707. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 7.Rocheleau CE, et al. Cell. 1999 Jun 11;97:717. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 8.Schneider SQ, Bowerman B. Dev Cell. 2007 Jul;13:73. doi: 10.1016/j.devcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 10.Stoick-Cooper CL, et al. Development. 2007 Feb;134:479. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama H, Ogino H, Stoick-Cooper CL, Grainger RM, Moon RT. Dev Biol. 2007 Jun 1;306:170. doi: 10.1016/j.ydbio.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi C, Saito Y, Ogawa K, Agata K. Dev Biol. 2007 Jun 15;306:714. doi: 10.1016/j.ydbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Stadeli R, Hoffmans R, Basler K. Curr Biol. 2006 May 23;16:R378. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 14.Willert K, Jones KA. Genes Dev. 2006 Jun 1;20:1394. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 15.Gordon MD, Nusse R. J Biol Chem. 2006 Aug 11;281:22429. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 16.Wallingford JB, Habas R. Development. 2005 Oct;132:4421. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 17.Brembeck FH, et al. Genes Dev. 2004 Sep 15;18:2225. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ettensohn CA. Sci STKE. 2006 Nov 14;:pe48. doi: 10.1126/stke.3612006pe48. [DOI] [PubMed] [Google Scholar]
- 19.Nogi T, Levin M. Dev Biol. 2005 Nov 15;287:314. doi: 10.1016/j.ydbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Shaw RM, et al. Cell. 2007 Feb 9;128:547. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guger KA, Gumbiner BM. Dev Biol. 1995 Nov;172:115. doi: 10.1006/dbio.1995.0009. [DOI] [PubMed] [Google Scholar]
- 22.Morgan TH. Arch Entw Mech Org. 1898;7:364. [Google Scholar]
- 23.Reddien PW, Sanchez Alvarado A. Annu Rev Cell Dev Biol. 2004;20:725. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 24.Morgan TH. Am Nat. 1904;38:502. [Google Scholar]
- 25.Sakai F, Agata K, Orii H, Watanabe K. Zool Sci. 2000;17:375. doi: 10.2108/jsz.17.375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.