Abstract
The bidentate RNase III Dicer cleaves microRNA precursors to generate the 21–23 nt long mature RNAs. These precursors are 60–80 nt long, they fold into a characteristic stem–loop structure and they are generated by an unknown mechanism. To gain insights into the biogenesis of microRNAs, we have characterized the precise 5′ and 3′ ends of the let-7 precursors in human cells. We show that they harbor a 5′-phosphate and a 3′-OH and that, remarkably, they contain a 1–4 nt 3′ overhang. These features are characteristic of RNase III cleavage products. Since these precursors are present in both the nucleus and the cytoplasm of human cells, our results suggest that they are generated in the nucleus by the nuclear RNase III. Additionally, these precursors fit the minihelix export motif and are thus likely exported by this pathway.
INTRODUCTION
In the last few years, microRNAs have emerged as a major new RNA family (for reviews see 1–3). This family contains hundreds of members, each possibly regulating the expression of several cellular mRNAs at various developmental stages. MicroRNAs thus have an unprecedented importance in the regulation of gene expression and they are the focus of very intense interest, to understand both their functions and their biogenesis.
One remarkable feature of microRNA biogenesis is its link with the RNA interference pathway (4–7). Indeed, both microRNAs and siRNAs are generated by the RNase III Dicer and the two types of RNAs are incorporated into RNP complexes able to perform similar functions, either in the cleavage of mRNA targets or in their translational repression (8,9). The end result appears to be mainly determined by the type of base pairing between the small RNA and its target.
Despite the extremely strong interest in the biogenesis of these small RNA species, little is known beside the role of Dicer, and this is especially true in the case of microRNAs. One remarkable feature of microRNAs that is conserved in worms, flies and mammals is the presence of relatively large amounts of precursors of about 60–80 nt in length and which fold into a characteristic stem–loop structure (1–3). For clarity, these precursors will be referred to as the minihelix microRNA precursors.
While it is not known how these minihelix precursors are generated, it has been demonstrated in a number of model systems that they are the substrates for Dicer, which cuts them to generate the mature microRNAs (4,5). A recent study that used human cell extracts to analyze the processing of microRNAs in vitro showed that large precursors of a few hundred nucleotides are cleaved in nuclear extracts to generate the minihelix precursors (10). Consistent with the fact that Dicer is localized in the cytoplasm (11), these precursors are processed in cytoplasmic extracts into microRNAs (10). The endoribonuclease generating the minihelix precursors is unknown, but based on genetic and localization data, a role for Dicer in this process is unlikely.
It is well established that RNA cleavage by endoribonucleases generates characteristic termini. In order to define which enzyme could generate the minihelix precursors, we have determined the precise nature of their 5′ and 3′ ends. This is especially important because these precursors have been proposed to be exported from the nucleus by the so-called minihelix pathway, which depends on exportin-5 (12,13). Indeed, export by this pathway requires that the RNA substrates fold into a stem–loop structure containing the RNA 5′ end, and harboring only a few nucleotides of 3′ overhang (12). As little as three unpaired bases at the 5′ end is deleterious for export by this pathway, but a single unpaired base at the 5′ end still leads to RNA export (12). The determination of the extremities of minihelix microRNA precursors might therefore validate or refute the possibility that they are exported by the exportin-5 pathway.
MATERIALS AND METHODS
Determination of let-7 RNA ends
Aliquots of 10 µg of total RNA from HeLa cells were self-ligated with T4 RNA ligase as described (14,15), except that the reaction volume was increased to 100 µl to enhance self-ligation of RNA molecules. Total RNA were either used directly, treated with T4 polynucleotide kinase or decapped with tobacco acid pyrophosphatase (15). Following ligation, RNAs were phenol/chloroform extracted, precipitated with ethanol and let-7 RNA precursors were amplified by RT–PCR with the following oligonucleotides: Set1, 5′-AACTATACAATCTACTGTCTT and 5′-AACTATACAACCTACTACCT; Set2, 5′-CTACTACCTCACCCCAAA and 5′-CTA CTGTCTTTCCTGAAGT. PCR products were resolved by gel electrophoresis and each band was purified and cloned by AT cloning with an A/T cloning kit (Promega). Individual clones were sequenced.
Cell fractionation and RNA analysis
Cytoplasmic and nuclear RNA were prepared by lysing cells in 0.5% Triton X-100 at 4°C for 5 min in 50 mM Tris pH 7.4, 150 mM NaCl. Cells were then centrifuged for 10 min at 3500 g on a cushion containing 10% sucrose and the cytoplasmic RNAs present in the supernatant were purified with Trizol. The nuclear fraction was purified a second time by resuspending the first pellet in the same buffer and centrifuging it again on a sucrose cushion. Nuclear RNA were finally extracted by resuspending the pellet in Trizol and sonicating it for 10 s. To avoid contamination of the nuclear fraction by cytoplasmic RNA, purification of nuclei was performed in the presence of 3 mM EDTA, previously shown to remove cytoplasmic remnants otherwise attached to nuclei (16). Finally, RNA was analyzed by northern blot on 10% acrylamide–urea gels with oligonucleotide probes specific for let-7, U6 or 18S rRNA.
RESULTS AND DISCUSSION
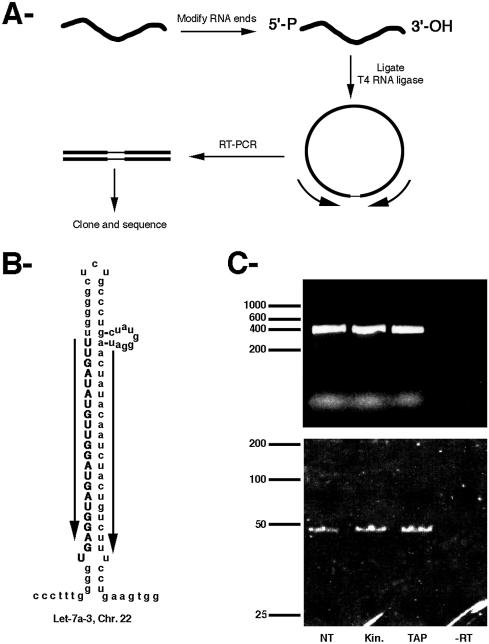
To characterize both the RNA 5′ and 3′ ends in a reliable way and in a single experiment, we took advantage of a previously developed technique (17) (Fig. 1A). This technique starts by circularizing the RNA with T4 RNA ligase under dilute conditions, such that most of the RNA molecules self-ligate. The RNA species of interest are then amplified by RT–PCR with oligonucleotides designed to amplify the 5′–3′ junction and the PCR products are resolved by gel electrophoresis and cloned. For the analysis of microRNA precursors, we focused on the human let-7 species. Indeed, these RNA species are abundant, ubiquitous and generated from multiple genes, allowing us to characterize a number of RNA species simultaneously. To obtain insights into the chemical nature of the RNA ends, several treatments were applied to the RNA before circularization. Indeed, RNA ligation with T4 RNA ligase requires a 5′-phosphate and a 3′-OH and RNA containing different types of extremities would not be circularized (18). To remove potential 2′,3′-cyclic phosphate or 2′- and 3′-phosphate groups, RNA was treated with T4 polynucleotide kinase (18). This enzyme also converted 5′-OH into 5′-phosphate groups (14). To convert 5′-cap structures into 5′-phosphates, RNAs were treated with tobacco acid pyrophatase (15). Total RNA from HeLa cells were thus pre-treated with each of these treatments or used directly and PCR products resulting from the self-ligated let-7 precursors were amplified with specific oligos (set 1, Fig. 1B) and resolved by gel electrophoresis (Fig. 1C).
Figure 1.
(A) Schematic of the amplification strategy used to characterize let-7 minihelix precursors. (B) Secondary structure of human let-7a-3. Arrows indicate the oligonucleotides used for the RT–PCR. (C) Amplification products were resolved on a 1% agarose gel (upper panel) or a 12% acrylamide gel (lower panel) and stained with ethidium bromide. The RNA was treated before ligation as follows. Lane NT, no pre-treatment; lane Kin, T4 polynucleotide kinase; lane TAP, tobacco acid pyrophosphatase; lane –RT, reverse transcriptase was omitted.
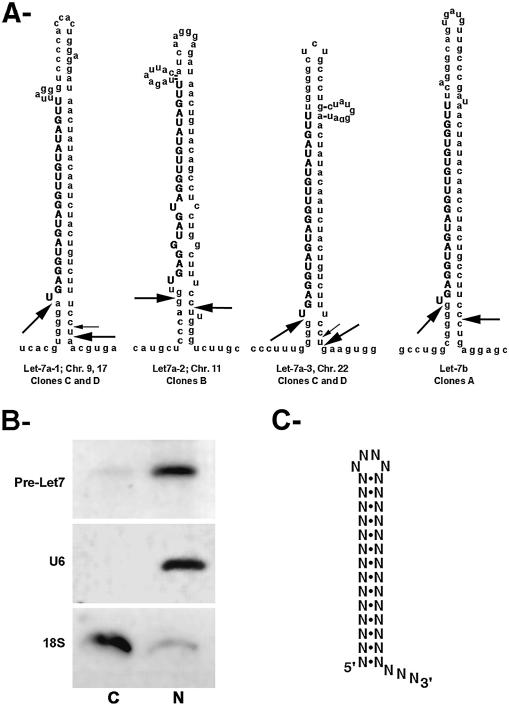
Identical results were obtained regardless of the pre-treatment of the RNA and two bands were observed, of about 400 and 50 nt long (Fig. 1C). Sequence analysis of individual molecules cloned from the longest band revealed that it contained a single RNA species, arising from amplification of the intergenic region between two let-7 genes located 360 nt apart on chromosome 9. This result demonstrated the specificity of the technique and also showed that let-7 RNAs are indeed transcribed from long, polycistronic transcripts, as proposed previously (19). Sequence analysis of the molecules contained in the shortest band revealed that it arose from minihelix let-7 precursors (Fig. 2A). First, the sequence contained the two correct nucleotides at the 5′ end of the mature let-7 (UG), which were absent in the sequence of the oligonucleotide used for PCR amplification. Second, the rest of the sequence could be assigned to the various let-7 minihelix precursors, as deduced from the human genome sequence (Fig. 2A). Four types of clones were observed, which arose from several of the known let-7 genes. Due to the sequence similarity between these genes, each clone could originate from several loci. Clones of type A could arise from let-7b or let-7f-1, clones of type B could arise from let-7a-2 or let-7c and clones C and D could arise from either let-7a-1, let-7a-2, let-7a-3 or let-7c. The corresponding extremities are represented in Figure 2A on let-7a and let-7b precursors, which appear to be the most abundant species (19).
Figure 2.
(A) Structure of several of the human let-7 precursors. Arrows indictate the extremities of the stem–loop precursors characterized in this study. Clones of type A arose from let-7b (14% of the clones), clones B from let-7a-2 (14% of the clones) and clones C (large arrows, 58% of the clones) and D (small arrows, 14% of the clones) from either let-7a-1 or let-7a-3. (B) Nucleo-cytoplasmic distribution of let-7 stem–loop precursors. C, cytoplasmic RNAs; N, nuclear RNAs. Similar amounts of RNA were loaded in each lane. The RNA species probed is indicated on the left. (C) Consensus for the minihelix export motif (12). The length of the stem should be longer than 12 nt. Also note that the motif can tolerate many mismatches and bulges; the first base at the 5′ end can also be left unpaired.
Remarkably, the extremities of the minihelix precursors shared a similar structure regardless of the let-7 loci they originated from. Clones A, C and D had a 5′ end identical to that of the mature let-7 RNA, while clones B had their 5′ ends 2 nt upstream. Importantly, all clones had a 1–4 nt overhang at their 3′ end. Furthermore, the clones originating from various RNA pre-treatments were identical to those not pre-treated, This demonstrated that the initial RNA species contained 5′-phosphate and 3′-OH ends. This also ruled out the possibility that the products amplified from the various pre-treatments contained minor variants unresolved in the gel.
RNase III are universally conserved RNases that cleave double-stranded RNAs (20). RNase III cleavage products contain 5′-phosphate and 3′-OH ends and have a few nucleotides of 3′ overhang. This overhang is usually of 2 nt on regular RNA helices, but it can vary from 1 to 4 nt when the helix is distorted (see 21 for a characterization of many natural sites for yeast RNase III). The let-7 minihelix precursors we have characterized have all the signatures of RNase III cleavage products: they are double-stranded RNAs with the expected 5′ and 3′ ends. Because the minihelix precursors are generated by cleavage of longer precursors, our data strongly suggest that they are generated by an RNase III. Interestingly, the yeast RNase III recognizes its substrates by binding to AGNN tetraloops (21). However, the enzyme involved here appears to be different in this respect, since no clear consensus can be drawn from the loops of the let-7 fold-back precursors.
Several experiments were conducted to confirm that the RNA species we had characterized indeed corresponded to the bona fide let-7 minihelix precursors. First, the size of the precursors observed on northern blots corresponded to the sizes of the predicted sequences. Indeed, in human cells, microRNA minihelix precursors co-migrate with a 70 base RNA marker (10) and our sequences predict a size of 70–71 nt for let-7a-1 and let-7a-3, 69 nt for let-7a-2 and 67 nt for let-7b. Second, to rule out the possibility that a second, longer precursor could have escaped detection, we repeated the experiment, but during the gel purification the material above the 50 nt band and shorter than 100 nt (24 nt above the maximal size of the let-7 minihelix precursors determined by gel electrophoresis) was purified, re-amplified (because it was undetectable by ethidium bromide staining; Fig. 1C), cloned and sequenced. The majority of the clones obtained turned out to be non-specific PCR products and a minority of clones was identical to those previously characterized. Third, we repeated the experiments with a slightly different set of oligonucleotides (set 2), designed to initiate the PCR elongation 4 nt downstream of the let-7a-3 3′ end determined here. In this case, no specific PCR products could be recovered.
It has been proposed previously that the minihelix microRNA precursors were generated in the nucleus and exported into the cytoplasm for processing by Dicer (3,9). To gain support for this possibility, we analyzed the localization of let-7 minihelix precursors by cell fractionation. As shown in Figure 2B, cytoplasmic RNAs contained the minihelix let-7 precursors and it was also detected in the nuclear RNAs. Controls with U6 and 18S probes showed that both signals were significant and that the RNA was thus present in both the nucleus and the cytoplasm. Additionally, the nuclear and cytoplasmic let-7 precursors migrated at the same size, demonstrating that these RNA species were indeed likely the substrate of the export machinery.
Given the role of the RNase III Dicer in the generation of mature microRNAs (4,5,7), the finding that the minihelix let-7 precursors harbored the hallmarks of RNase III cleavage products raised the possibility that Dicer itself was responsible for their generation. Several observations, however, argue against this hypothesis. First, Dicer is strictly confined to the cytoplasm of human cells (11) and, as shown here, the minihelix let-7 precursors are present in both nuclear and cytoplasmic extracts. Second, these precursors can be generated in vitro from longer precursors by nuclear, but not cytoplasmic, extracts (10). Third, depletion of Dicer by RNAi in worms and flies leads to the disappearance of mature microRNAs with a concomitant increase in the amount of minihelix precursors (4,5). Besides Dicer, there is a second gene in the human genome that encodes a RNase III homolog (22,23). This gene is conserved in mammals, flies and worms and it has been shown to encode a protein localized in the nucleus and showing RNase III activity (22–24). Thus, our results suggest that this enzyme could be responsible for the nuclear processing of long polycistronic microRNA precursors into the minihelix species.
A comparison of the stem–loop let-7 precursors with the consensus for RNA export by exportin-5 (Fig. 2C) reveals that these RNAs fit the consensus and are thus likely exported by the so-called minihelix pathway (12,13). Interestingly, exportin-5 also binds and exports proteins containing a double-stranded RNA-binding domain (25) and, in the case of tRNA export, exportin-5 was shown to form ternary export complexes with tRNA bound to eEF1α (26,27). By analogy, it is possible that exportin-5 recognizes the minihelix precursors still bound by the nuclear RNase III. The double-stranded RNA-binding domain of the nuclear RNase III may help to promote the assembly of a ternary export complex and, in the case of let-7, such a ternary export complex may provide the stability required to compensate for the lack of 1 bp at the 5′ end.
Finally, the fact that let-7 minihelix precursors are present in the cytoplasm suggests that these are not processed directly, but first stored. Such a delay between minihelix production and processing into mature microRNA has been previously observed (4) and could participate in the regulation of these fascinating RNAs.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the MNRT (ACI to E.B.) and the ARC. E.B. was supported by a fellowship from Sidaction. F.S. was supported by a fellowship from A.N.R.S.
REFERENCES
- 1.Moss E. (2002) MicroRNAs: hidden in the genome. Curr. Biol., 12, R138–R140. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. (2001) MicroRNAs: tiny regulators with great potential. Cell, 107, 823–826. [DOI] [PubMed] [Google Scholar]
- 3.Grosshans H. and Slack,F. (2002) Micro-RNAs: small is plentiful. J. Cell Biol., 156, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutvagner G., McLachlan,J., Pasquinelli,A., Balint,E., Tuschl,T. and Zamore,P. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- 5.Ketting R., Fischer,S., Bernstein,E., Sijen,T., Hannon,G. and Plasterk,R. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein E., Caudy,A.A. and Hammond,S.M. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 7.Grishok A., Pasquinelli,A., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D., Fire,A., Ruvkun,G. and Mello,C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Y., Wagner,E. and Cullen,B. (2002) Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell, 9, 1327–1333. [DOI] [PubMed] [Google Scholar]
- 9.Cullen B. (2003) Nuclear RNA export. J. Cell Sci., 116, 587–597. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y., Jeon,K., Lee,J., Kim,S. and Kim,V. (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J., 21, 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billy E., Brondani,V., Zhang,H., Muller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwizdek C., Bertrand,E., Dargemont,C., Lefebvre,J., Blanchard,J., Singer,R. and Doglio,A. (2001) Terminal minihelix, a novel RNA motif that directs polymerase III transcripts to the cell cytoplasm. J. Biol. Chem., 276, 25910–25918. [DOI] [PubMed] [Google Scholar]
- 13.Gwizdek C., Ossareh-Nazari,B., Brownawell,A., Doglio,A., Bertrand,E., Macara,I. and Dargemont,C. (2003) Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem., 278, 5505–5508. [DOI] [PubMed] [Google Scholar]
- 14.Bertrand E., Fromont-Racine,M., Pictet,R. and Grange,T. (1993) Visualization of the interaction of a regulatory protein with RNA in vivo. Proc. Natl Acad. Sci. USA, 90, 3496–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromont-Racine M., Bertrand,E., Pictet,R. and Grange,T. (1993) A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res., 21, 1683–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weil D., Boutain,S., Audibert,A. and Dautry,F. (2000) Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA, 6, 962–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez T., Sidrauski,C., Dorfler,S. and Walter,P. (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J., 18, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 20.Conrad C. and Rauhut,R. (2002) Ribonuclease III: new sense from nuisance. Int. J. Biochem. Cell Biol., 34, 116–129. [DOI] [PubMed] [Google Scholar]
- 21.Chanfreau G., Legrain,P. and Jacquier,A. (1998) RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- 22.Filippov V., Solovyev,V., Filippova,M. and Gill,S. (2000) A novel type of RNase III family proteins in eukaryotes. Gene, 245, 213–221. [DOI] [PubMed] [Google Scholar]
- 23.Wu H., Xu,H., Miraglia,L. and Crooke,S. (2000) Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem., 275, 36957–36965. [DOI] [PubMed] [Google Scholar]
- 24.Fortin K., Nicholson,R. and Nicholson,A. (2002) Mouse ribonuclease III. cDNA structure, expression analysis and chromosomal location. BMC Genomics, 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownawell A. and Macara,I. (2002) Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol., 156, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnsack M., Regener,K., Schwappach,B., Saffrich,R., Paraskeva,E., Hartmann,E. and Gorlich,D. (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J., 21, 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calado A., Treichel,N., Muller,E., Otto,A. and Kutay,U. (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J., 21, 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]