Abstract
The importance of MAPK signaling in melanoma is underscored by the prevalence of activating mutations in N-Ras and B-Raf; yet, clinical development of inhibitors of this pathway has been largely ineffective, suggesting that alternative oncogenes may also promote melanoma. Notch is an interesting candidate that has only been correlated with melanoma development and progression; a thorough assessment of tumor-initiating effects of activated Notch on human melanocytes would clarify the mounting correlative evidence and perhaps identify a novel target for an otherwise untreatable disease. Analysis of a substantial panel of cell lines and patient lesions demonstrated that Notch activity is significantly higher in melanomas than their non-transformed counterparts. The use of a constitutively-active, truncated Notch transgene construct (NIC) was exploited to determine if Notch activation is a ‘driving’ event in melanocytic transformation or instead a ‘passenger’ event associated with melanoma progression. NIC-infected melanocytes displayed increased proliferative capacity and biological features more reminiscent of melanoma such as dysregulated cell adhesion and migration. Gene expression analyses supported these observations and aided in the identification of MCAM, an adhesion molecule associated with acquisition of the malignant phenotype, as a direct target of Notch transactivation. NIC-positive melanocytes grew at clonal density, proliferated in limiting media conditions, and also exhibited anchorage-independent growth suggesting that Notch, alone, is a transforming oncogene in human melanocytes, a phenomenon not previously described for any melanoma oncogene; this new information yields valuable insight into the basic epidemiology of melanoma and launches a realm of possibilities for drug intervention in this deadly disease.
Keywords: Notch, melanoma, transformation, MCAM, therapy
Introduction
The Notch signaling pathway is an evolutionarily conserved signaling cascade that impacts cell fate decisions and many differentiation processes during both embryonic and postnatal development (1). Notch signaling has also been implicated in neoplastic malignancies; a potential role for aberrant Notch signaling was first observed in T-cell acute lymphoblastic leukemia (T-ALL), where a chromosomal translocation resulted in the liberation of a truncated and constitutively-activated form of the Notch1 receptor (NIC) (2). Since this original report, the link of the Notch signaling pathway to tumorigenesis has been well-established. Aberrant Notch signaling has been linked to prostate carcinoma (3), mouse mammary epithelial cell tumors (4), small cell lung cancer (5), neuroblastoma (6), cervical carcinoma (7) and most recently Kaposi's sarcoma (8). Activated Notch can transform primary Schwann cells (9) and, in collaboration with the adenovirus E1a protein, cultured rat embryonic epithelial cells (10).
Metastatic melanoma is a highly invasive tumor derived from epidermal melanocytes that is refractory to most therapies. Melanocytes are melanin-producing cells that reside along the basement membrane in the basal layer of the epidermis interspersed amongst surrounding keratinocytes. Keratinocytes in turn play a major role in regulating the growth and differentiation of melanocytes. Under normal conditions, E-cadherin is expressed on the surface of both melanocytes and keratinocytes and is the critical cell-adhesion protein between these two cell types in the human epidermis (11, 12). Loss of E-cadherin expression is a well documented step in melanoma development and progression (13-15). Conversely, upregulation of other cell adhesion molecules, such as MCAM (MelCAM, MUC18, CD146), N-cadherin and αvβ3 integrins have been strongly correlated with melanoma progression and metastasis (16-19). Expression of MCAM, a highly glycosylated membrane protein, confers metastatic potential in experimental murine models to MCAM-negative melanoma cells (18, 20).
Recent data suggest that Notch activation may play a role in melanoma progression. We have previously demonstrated that activation of Notch signaling promotes the progression of early stage melanoma cell lines in a β-catenin-dependent manner both in vitro and in vivo (21). Furthermore, microarray profiling comparing the gene expression of normal human melanocytes to human melanomas revealed upregulation of Notch target genes in melanoma cells, suggesting activation of the Notch signaling pathway in melanoma (22). Based on these observations, we sought to determine the effects of Notch activation on primary human melanocytes. The data described herein define an oncogenic role for Notch signaling in melanocytes and highlight the potential for Notch inhibition to be utilized as a therapeutic approach for the treatment of melanoma.
Materials and Methods
For further details on reagents and methodology, refer to supplementary Materials and Methods.
Cell Culture
Normal human primary melanocytes were isolated from human epidermal foreskin and cultured as previously described (23). All human melanoma cells lines were isolated and cultured as described elsewhere (23).
RNA Extraction and Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
RT-PCR analyses were carried out as described previously (23). Primer sequences are listed in Supplementary Table 1.
Real-time RT-PCR analysis of Notch receptor expression
One μg of total RNA was used to generate cDNA using the Taqman Reverse Transcription Kit (PE Applied Biosystems, Foster City, CA). The SYBR Green I assay and the GeneAmp 5700 Sequence Detection System (PE Applied Biosystems, Foster City, CA) were used for detecting real-time PCR products. Primers are detailed in Supplementary Table 1. HEMN, a representative melanocyte cell line, was used to determine the relative fold induction for each sample relative to these cells.
Sequencing of Notch1 Heterodimerization, Transcriptional Activation and PEST Domains
All PCRs were performed as described elsewhere (24). Primer sequences were as published by Weng et al. (24).
Microarray-based gene expression analysis
10 μg of total high-quality RNA was transcribed, labeled and U133A chips were hybridized and scanned according to the standard protocol recommended by Affymetrix (Santa Clara, California). GeneSpring was then used perform fold-change restriction analyses on the filtered lists.
Immunoblotting
Standard western blotting procedures were performed, as previously described (23).
Immunohistochemistry
Paraffin-embedded, formalin fixed tissue sections were subjected to staining procedures, as described elsewhere (25).
Cell growth assays
Cell proliferation was measured by either cell counting or MTT assays, as described previously (23).
Cell adhesion and migration assays
Cell adhesion and migration (Boyden Chamber) assays were performed as previously described (26).
Colony-formation assay
Colony formation in soft agar was conducted, as described previously (23).
Recombinant lentiviruses
Lentiviral vectors were constructed for gene transfer, viral particles were produced, and cell infections were performed, as previously described (23).
Immunofluorescence Microscopy
Primary human melanocytes were seeded onto glass coverslips in 12-well plates and incubated overnight. Cells were then fixed in 4% formaldehyde solution and stained using appropriate primary and secondary antibodies.
Chromatin Immunoprecipitation (ChIP) assay
ChIP was performed using ChIP assay kits (Upstate Biotechnology) following the manufacturer's recommendations.
Statistical Analyses
Data (from triplicate experiments) are presented as mean ± SD and were analyzed by 2-tailed Student's t test. A P value of < 0.05 was considered significant.
Results
The Notch signaling pathway is activated in human melanoma compared to primary human melanocytes
To assess the potential role of Notch signaling in human melanocytic transformation, the activation status of the Notch pathway was assayed through several approaches. Using an antibody against activated Notch1, immunohistochemical staining was performed on melanoma patients' samples (Figure 1A). Active Notch1 staining co-localized with that of HMB45, a marker for cells of melanocytic origin. Normal tissue immediately adjacent to the lesion did not stain positively for active Notch1 (Figure 1A, far right panel). Quantitative RT-PCR was performed on RNA from fresh melanoma lesions and established cell lines to determine levels of Notch1 expression (Figure 1B); from 9 lesions and 14 cell lines, every sample tested expressed at least 5-fold more Notch1 than the representative primary human melanocytic control. The expression of Notch target genes in laser-microdissected melanoma lesions, melanoma cell lines, and melanocyte cell lines demonstrated that Hes1 and Hey1, but not Hey2, were dramatically upregulated in both melanoma tissue and cell lines when compared to primary melanocytes (Figure 1C, top). Microarray analyses on a panel of melanoma cell lines displayed similar patterns of gene expression (Figure 1C, bottom). RT-PCR analysis confirmed these data, as Notch-target transcripts were elevated in melanoma lines when compared to melanocytes (data not shown). Immunoblots depicted absent or low levels of the activated Notch1 protein in melanocytes, but higher levels in melanoma lines suggesting that the Notch signaling pathway is only active in malignant cells (Figure 1D).
Figure 1. Notch signaling pathway activation status in melanocytes and melanoma cells.
A, Immunohistochemical analysis of active Notch1 expression in primary human melanoma lesions. Anti-HMB45 was employed to identify cells of melanocytic origin. Darker staining depicts melanin production whereas active Notch1 and HMB45 are indicated by red staining. Unaffected epidermal and dermal tissue immediately adjacent to the primary melanoma lesion was also photographed for control purposes.
B, Quantitative RT-PCR analysis of Notch1 expression in a panel of melanoma lesions and cell lines. Fold change is reflected against a representative human melanocyte cell line, HEMN.
C, (Top) Microarray analysis of Hes1, Hey1 and Hey2 expression in four laser microdissected melanoma lesions, four melanoma cell lines, and four primary melanocyte cell lines. (Bottom) Microarray analysis of Hes1, Hey1 and Hey2 expression in a panel of human melanoma cell lines.
D, Western blot analysis of activated Notch1 protein in melanocytes and melanoma cells from C.
Mechanism of Notch Activation in Melanoma
Based on the endogenous overexpression of active Notch1 protein in melanoma tissue and cell lines, melanoma cell lines were screened in search of activating mutations within the Notch1 locus similar to those reported in at least 50% of T-ALLs (24). 17 human melanoma and 3 T-ALL cell lines were sequenced within exon 26 (heterodimerizations; HD-N1, HD-N2 and HD-C) and exon 34 [transcriptional activation domain (TAD) and PEST domain] of Notch1. Although no activating Notch1 genetic alterations were identified in the melanomas, insertional and missense mutations were confirmed in the three T-ALL cell lines (Supplementary Figure 1). Approximately 70% of the melanoma cell lines displayed the T allele of the C/T SNP previously reported at nucleotide 5097 (24). Despite the absence of Notch1 activating mutations in the melanoma cell lines tested, dramatic upregulation of the Notch1 transcript was detected by real-time RT-PCR in 14 melanoma cell lines and 9 fresh tumor specimens but not normal melanocytes (Figure 1B), suggesting the mechanism for enhanced Notch activation in melanoma may be due to Notch overexpression. Melanoma tumor specimens and cell lines showed significantly higher levels of Notch1 expression, with an average 7.9- and 37.8-fold increase over a representative melanocyte cell line HEMN, respectively. The levels of Notch2/4 transcripts were also robustly upregulated in melanoma lesions and cell lines (Supplementary Figure 2).
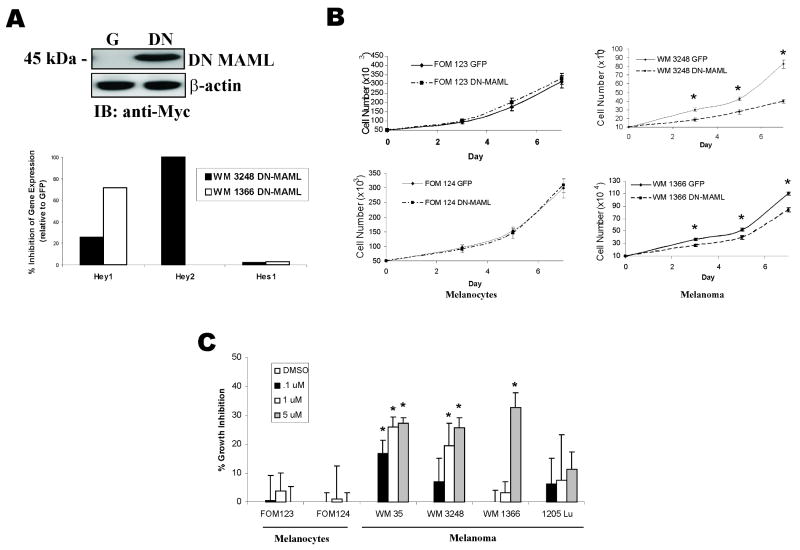
Inhibition of Notch1 Activity Elicits Anti-Melanoma Activities
The data thus far suggested strong Notch activation in melanoma, therefore, the effects of Notch inhibition on melanocyte and melanoma growth was tested in vitro. To suppress the Notch signaling cascade, a dominant negative mutant of the Mastermind-like (MAML) protein was utilized; this construct acts as a pan-Notch inhibitor and prevents transcription of downstream target genes by binding to NIC and preventing the recruitment of co-activator proteins to the Notch enhanceosome. The myc-tagged DN-MAML vector was lentivirally-infected into two melanocyte cell lines and two melanoma cell lines; after selection, these lines were analyzed for DN-MAML expression by probing for the myc tag (Figure 2A, top panel). Quantitative RT-PCR analysis of Hey1 and Hey2 revealed suppression of these Notch targets in DN-MAML infected melanoma cell lines; Hes1 levels were initially low and were generally unaffected by DN-MAML expression (Figure 2A, bottom panel). As depicted in Figure 2B, the growth rate of DN-MAML infected melanoma cell lines was significantly reduced when compared to GFP-expressing cells, but was unaffected in two primary melanocyte cell lines. Consistent with these data, suppression of Notch signaling activation via pharmacological inhibition selectively decreased the growth of melanoma cell lines but not melanocytes, in a dose-dependent manner (Figure 2C). A γ-secretase inhibitor, GSI X, at the highest concentration of 5 μM inhibited melanoma growth by as much as 32%, as determined by MTT analysis. These results are compatible with previous work indicating that γ-secretase inhibitors preferentially decrease the growth of human melanoma cells but not melanocytes (27).
Figure 2. Inhibition of Notch signaling suppresses melanoma, but not melanocyte growth in vitro.
A, Expression of DN-MAML protein in melanoma cell line WM 3248 was detected by immunoblotting for the myc-tagged DN-MAML. Effect of DN-MAML expression on Notch1 targets Hes1, Hey1 and Hey2 was determined by quantitative RT-PCR in 2 melanoma cell lines, WM3248 and WM1366.
B, 7 day growth curves of melanocytes and melanoma cells infected with DN-MAML or GFP lentiviruses. * p<0.05 (Student's T-test).
C, Growth inhibition in melanocytes and melanoma cells in the presence of increasing concentrations of a γ-secretase inhibitor. Cell growth was determined by MTT analysis. Results are percentage of growth inhibition compared with untreated controls (adjusted to 0%). * p<0.005 (Student's t-test).
Expression of human activated Notch1 (NIC) in human melanocytes
Based on the above data, the potential role of Notch signaling in melanocyte transformation and melanoma development was investigated. To determine the effects of active Notch1 expression in primary human melanocytes, the NIC gene was introduced into primary human melanocytes. First, lentiviral vectors encoding either GFP or NIC-GFP were used for stable, intracellular expression of NIC. To verify expression of the lentiviral-derived NIC, Notch1 mRNA and protein expression was assessed via semi-quantitative RT-PCR and western blotting (Figure 3A). Immunofluorescence of GFP and NIC-infected melanocytes was performed with a Notch1 antibody to assess the localization of the lentiviral-derived construct (Figure 3B). NIC protein was appropriately detected in the nucleus and was also capable of activating expression of downstream Notch1 target genes as evidenced by increased expression of the Notch-regulated genes, Hey1 and Hey2 (Figure 3C).
Figure 3. Constitutive activation of the Notch1 pathway enhances melanocyte growth in vitro.
A, (Left) mRNA analysis of Notch1 expression in GFP- and NIC-infected FOM 117 cells by RT-PCR. (Right) Western blot analysis of Notch1 protein expression in GFP and NIC-infected FOM 117 cells.
B, Immunofluorescence analysis of NIC localization in FOM 117 GFP and NIC-infected cells.
C, mRNA expression of Hey1 and Hey2 in FOM 117 GFP and NIC-infected cells as determined by quantitative RT-PCR.
D, ∼ 2 week growth curve of GFP- and NIC-infected FOM 123 and 124 cells. *p<0.005 (Student's T-test).
Since the NIC oncoprotein has been shown previously to promote cellular growth (28, 29), the effects of constitutive Notch1 activation on primary melanocyte growth were ascertained in vitro. The growth rate of NIC-infected melanocytes was significantly increased compared to that of corresponding GFP control cells (Figure 3D). Collectively, the data suggest that Notch is activated in melanoma and that this is sufficient to increase melanocytic growth in vitro.
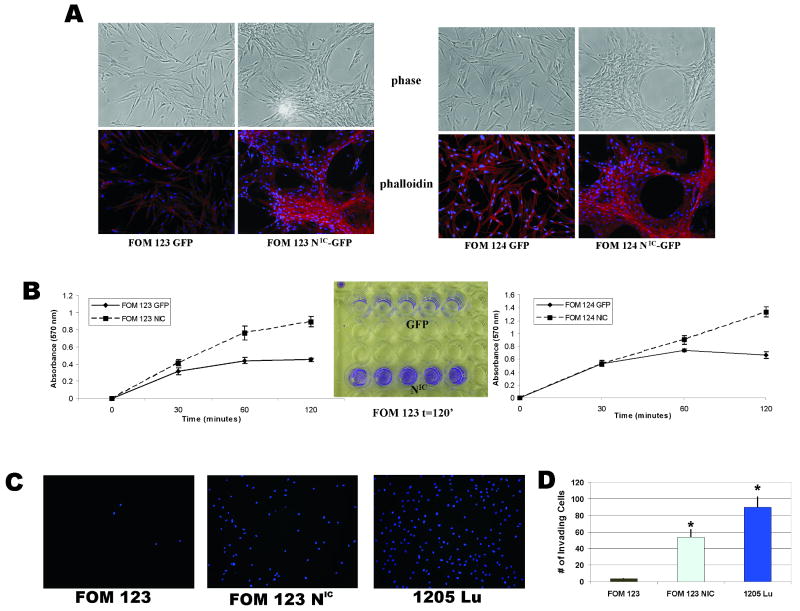
Activated Notch1 promotes cytoskeletal changes, increased adhesion, and migration in primary human melanocytes
In addition to increased growth rates, at 72-96 hours post-infection, NIC-expressing melanocytes exhibited marked morphological changes compared to GFP (Figure 4A) or uninfected parental controls (data not shown). The cells exhibited shorter dendrites, grew in large clusters, and formed endothelial-like networks reminiscent of vasculogenic mimicry (30). These changes were consistent across all 6 melanocyte lines infected with the NIC lentivirus. In addition to these morphological changes, the NIC transgene promoted increased adhesion in melanocytes (Figure 4B). Lastly, migration assays using Boyden chambers demonstrated that NIC-infected melanocytes possess increased invasive capacity compared to GFP-infected controls (Figure 4C & 4D). Each of these biological properties supports a role for Notch activity in acquisition of a malignant phenotype, although the downstream effector molecules of Notch1 which may contribute to the observed phenotypes are uncertain.
Figure 4. Constitutive Notch1 activation induces cytoskeletal, adhesion, and migratory changes in primary melanocytes.
A, Phalloidin immunostaining reveals morphological changes induced by active Notch1 expression. FOM 123 and 124 NIC cells form networks of capillary like structures when grown in monolayer as compared to the normal dendritic growth pattern of normal GFP-infected cells.
B, Cell adhesion assay demonstrated that NIC-infected FOM cells possess increased adhesive properties compared to GFP-infected controls within 2 hours post-plating. *p<0.005 (Student's t-test).
C & D, NIC- and GFP-infected melanocytes were subject to Boyden chamber assays to test their ability to migrate through matrigel. After 24 hours, membranes stained with DAPI demonstrated enhanced ability of NIC-infected FOM cells to migrate; 1205Lu melanoma cells acted as a positive control.
Identification of MCAM as a Target of Notch Signaling in Melanocytes
Microarray analyses were performed to aid in the identification of genes differentially-expressed between Notch-infected and control melanocytes [microarray data is available at GEO (GSE15040)]. Using GeneSpring software gene ontology classification analysis, subgroups with specified biological functions were identified, including those within cell adhesion and invasion families; a small fraction of those genes are depicted in Supplementary Table 1. Notably, MCAM, an important mediator of melanoma progression, was significantly upregulated in NIC-infected cells, as well as the N-cadherin (CDH2) (25).
Quantitative RT-PCR was performed on a panel of genes selected from the gene ontology groups to confirm the observed changes in gene expression (data not shown). Of these, E-cadherin and MCAM were differentially expressed and therefore chosen for further analysis because loss of E-cadherin and upregulation of MCAM expression are well-characterized events in melanoma development and progression. Immunoblotting analyses demonstrated that NIC expression induced robust upregulation of MCAM and downregulation of E-cadherin in the melanocyte cells (Figure 5A). The gene expression changes induced by NIC are consistent with a shift towards a malignant phenotype. Expression levels of β3 integrin, another adhesion protein implicated in melanoma development and progression, however, was unaffected by NIC overexpression (Figure 5A).
Figure 5. Identification of a CSL binding sequence within the human MCAM promoter.
A, Immunoblot analysis of E-cadherin, MCAM, β3-integrin, and active Notch1 protein levels in primary melanocytes infected with GFP (G) or NIC (N) lentiviruses.
B, FOM 117 melanocytes were plated on Jagged1-Fc or control Fc plates for 72 hours and subsequently assayed for active Notch1 and MCAM expression by immunoblotting.
C, Enhanced MCAM protein expression in NIC-infected WM 3248 melanoma cells (left). Interruption of Notch signaling by treatment with 5 μM GSI X decreased MCAM expression in uninfected WM 3248 cells, as determined by immunoblotting for MCAM (right).
D, (Top) Multiple consensus binding sequences for CSL, the hexamer TGGGAA, were identified within the human MCAM promoter at positions -477 and -3500 (see text for details). (Bottom) Chromatin immunoprecipitation (ChIP) analysis demonstrating association of CSL with regions of the MCAM promoter. Hes1 acts as a positive control for binding activity, while MCAM -1.5K is a negative control.
MCAM is a direct Notch1 target
To begin to test the hypothesis that MCAM is a direct Notch target, melanocytes were plated on tissue culture dishes anchored with Jagged-1 to induce ligand-mediated Notch activation. Cell extracts confirmed that MCAM expression was induced in response to Notch activation, indicating that MCAM upregulation can occur through induction of endogenous Notch1 receptor activity (Figure 5B). Moreover, in an MCAM-expressing melanoma cell line, WM 3248, inhibition of Notch activation with a γ-secretase inhibitor suppressed MCAM expression; likewise, expression of NIC increased MCAM levels, as predicted (Figure 5C). Knockdown of MCAM in NIC-expressing melanocytes had no bearing on their ability to grow in soft agar, suggesting that upregulation of MCAM in this system is a contributory, rather than an initiating event in melanomagenesis (data not shown).
Based on the robust upregulation of MCAM in NIC-expressing melanocytes, we sought to further define the mechanism of MCAM induction. Multiple CSL/Notch-binding sequences were identified within the MCAM promoter including a non-conserved sequence at -477, as well as three conserved sites in the proximity of -3.5K (-3487, -3510, and -3522) (Figure 5D, top). The consensus motif, hexamer TGGGAA, has been shown to bind CSL (31) and is present in the human p21 promoter (32), the murine Hes1 promoter (33), and the human Skp2 promoter (34). Chromatin immunoprecipitation (ChIP) assays demonstrated that NIC strongly enhanced CSL binding at the conserved -3500 sites on the MCAM promoter; the non-conserved sequence at -477, however, yielded less binding (Figure 5D, bottom). The results from ChIP assays were subsequently confirmed through electrophoretic mobility shift assay (EMSA) analysis (data not shown). Together, these data underscore a direct role for Notch in upregulation of MCAM and, likely, disease progression.
NIC confers transforming properties to melanocytes in vitro
Loss of contact inhibition is a common phenotype of transformed cells. After selection, NIC-infected cells formed discrete foci, while GFP control cells did not (Figure 6A). These foci were adherent and viable, as determined by trypan blue exclusion (data not shown). The changes in cellular morphology and the loss of contact inhibition in NIC-transduced melanocytes were reminiscent of transformed cells. In addition, GFP- and NIC-infected cells were plated and their survival was assayed over the span of 72 hours in limiting media conditions (Figure 6B). NIC-infected cells display enhanced survival in media conditions that otherwise kill normal human melanocytes.
Figure 6. Notch1 activation transforms primary melanocytes in vitro.
A, Constitutive Notch1 activation induces focus formation in NIC-infected melanocytes grown in monolayer on tissue culture dishes. *p<0.005 (Student's T-test).
B, NIC-infected melanocytes display increased survival in limiting growth factor conditions, as depicted. 100% media indicates normal melanocytes media containing bFGF, SCF, and ET3. 10% media is a 10-fold decrease in FBS; the growth factor(s) present in each respective medium is described beneath each panel. *p<0.005 (Student's T-test).
C, NIC-infected melanocytes display anchorage-independent growth in soft agar. *p<0.005 (Student's T-test).
Anchorage-independent growth is a hallmark of malignant cell transformation that highly correlates with neoplasia; thus, anchorage-independent colony-forming capacity of GFP and NIC-infected melanocytes was analyzed. After plating GFP and NIC-infected melanocytes in soft agar with normal melanocyte media, GFP cells failed to establish viable colonies, while NIC-infected cells readily formed colonies (Figure 6C). To assess neoplastic transformation potential in vivo, 2 × 106 GFP or NIC-expressing melanocytes were injected subcutaneously into NOD-SCID mice. Twelve weeks post-injection, tumors were not detected (data not shown) suggesting that additional genetic events may be required to achieve growth in animal models.
Discussion
Here, we demonstrate that Notch1 signaling is activated in melanoma cells but not melanocytes and that constitutive Notch1 activation confers transforming properties to primary melanocytes in vitro. Notch receptors 1, 2 and 4 are overexpressed in melanoma cell lines and lesions, particularly when compared against primary melanocytes or normal human skin. Notch and Notch-target genes are upregulated in both melanoma lesions and melanoma cell lines. Ectopic NIC expression induced gross morphological changes, increased growth, adhesion, migration, survival, and resulted in the loss of E-cadherin expression and upregulation of MCAM, two well-characterized events in melanoma development. We identify MCAM as a direct Notch target due to the presence of two high-affinity CSL binding sites present in the MCAM promoter. The NIC oncoprotein conferred anchorage-independent growth, increased survival, and loss of contact inhibition; suppression of Notch signaling decreased the growth of melanoma cell lines while primary melanocytes were unaffected. Taken together, these data suggest that deregulation of Notch signaling plays a specific role in promoting a transformed phenotype in human melanocytes and define the importance of Notch signaling in human melanoma.
Our microarray data describing Notch pathway activity is underscored by a recent report by Hoek et al. that revealed upregulation of Notch2 and Hey1 in a separate, but distinct panel malignant melanoma cell lines, suggesting a role for Notch activation in the transformation of melanocytes (22). Previous immunohistochemical studies on early phase melanoma lesions have demonstrated overexpression of full-length Notch1 protein in melanoma tissue compared against benign human nevi (21) and normal human skin (8). In our current study, we examined active Notch1 levels and found overexpression of this protein by immunohistochemical analysis of paraffin-embedded melanoma lesions and western blotting of melanoma cell lines. Furthermore, suppression of Notch signaling via a dominant negative Mastermind-like construct or γ-secretase inhibition did not affect melanocyte growth but inhibited melanoma proliferation in vitro as well as melanoma tumorigenicity in SCID mice (21). Our study focused on Notch1 because of its overexpression in melanomas; however, Notch2 and Notch4 transcripts were also increased in melanoma tissues and cell lines. Therefore, it is likely that other Notch receptors play a role in mediating the oncogenic effect of Notch1 signaling activation. Further studies will be useful in determining the contribution of the individual Notch receptors to melanocyte transformation and melanoma development.
In light of studies highlighting novel activating mutations in T-ALL (24), it might be expected that such mutations exist in melanomas as well. However, sequencing of a panel of 17 melanoma lines did not reveal any genetic alterations within the heterodimerization or PEST domains of Notch1 that have previously been shown to harbor activating mutations in 50% of human T-ALLs. In the absence of genetic mutations, another mechanism must exist to account for enhanced Notch signaling in human melanoma. We favor a scenario in which overexpression of Notch receptors in melanoma cells results in robust Notch signaling activation in human melanoma. Indeed, we observed significant upregulation of Notch receptors 1, 2 and 4 at the mRNA level by real-time RT-PCR in melanoma cell lines and fresh melanoma specimens when compared to normal melanocytes. There are likely upstream factors that account for the increased transcriptional activity at the Notch receptor loci. One possibility was Ras, as it has been previously shown that oncogenic Ras activates Notch signaling and the wild-type Notch1 receptor is required to maintain the neoplastic phenotype of Ras-transformed cells (35). Studies performed in our laboratory, however, do not implicate MAPK signaling in transcriptional regulation of Notch1, as inhibitors of both Raf and MEK were unable to abrogate expression of Notch1 (data not shown). Certainly, unraveling the mechanistic details responsible for Notch1 upregulation in melanoma will be of immense value in the near future.
E-cadherin is the key adhesion molecule expressed by keratinocytes and melanocytes that permits keratinocytes to communicate with and exert regulatory control over melanocytic cellular processes (12, 15, 36). Loss of E-cadherin expression allows epidermal melanocytes to regulate their growth and adhesion independent of keratinocytes and is a key event in melanoma development (12, 15). Here, NIC downregulated E-cadherin expression in melanocytes and also promoted robust upregulation of MCAM, a cell-adhesion molecule whose protein levels highly correlate with aggressive invasive behavior of melanoma cells in vitro and in vivo (17-19). Our data suggest that MCAM is a direct Notch target based on the identification of 2 high-affinity CSL binding sites within the MCAM promoter. We propose that in early melanoma tumorigenesis, Notch activation results in MCAM expression and may ultimately contribute to melanoma progression. The significance of these data is underscored because loss of E-cadherin and upregulation of MCAM are consistent with changes in gene expression that occur during the development of malignant melanoma.
There is only one report of active Notch1 protein acting alone to fully transform primary cells (9). Forced NIC expression in primary rat Schwann cells resulted in transformation and loss of Schwann cell differentiation markers. Active Notch1 induced transformation in rat kidney embryo (RKE) cells; however, this was in cooperation with adenoviral protein E1A. Synergy of activated Notch1 and papillomavirus oncogenes E6 and E7 has also been reported in the transformation of immortalized epithelial cells (37). While transformation of immortalized melanocytes subsequent to overexpression of a single oncogene has been reported (38, 39), full transformation of primary melanocytes, as defined by inducing tumorigenicity in animal models, generally requires disruption of several pathways including Rb and p53 (40, 41). Oncogenic Ras is capable of promoting growth in soft agar as well as tumor formation in SCID mice in primary melanocytes, but only in the presence of the Simian Virus 40 early region (SV40ER), which encodes the viral large T (LT) and small T (st) oncoproteins, and the catalytic subunit of the telomerase holoenzyme (hTERT) (40). Thus, although in our primary melanocyte cell lines NIC overexpression alone was capable of inducing a transformed phenotype in vitro, it is not alarming that NIC-infected cells failed to form tumors in NOD-SCID mice; therefore, Notch1 overexpression alone is not sufficient for full neoplastic transformation due to a lack of in vivo growth. However, our findings are nonetheless of significant importance in the consideration of signaling pathways that are deregulated and cooperate in the process of melanocyte transformation and melanoma development.
Our current studies strongly suggest that constitutive Notch signaling is associated with melanocyte transformation and melanoma tumorigenesis. Of particular significance, is the ability of a γ-secretase inhibitor to selectively inhibit the growth of melanoma cell lines. These findings are consistent with a recent report identifying a γ-secretase inhibitor that induced effective apoptosis in human melanoma cells while sparing melanocytes (27). Multiple γ-secretase inhibitors have been developed and are presently in trials for use in the treatment of Alzheimer's patients (42). Based on the recent identification of activating Notch mutations in roughly 50% of human T-ALLs (24), therapies designed to interfere with Notch activation, such as γ-secretase inhibitors, will undoubtedly be explored as a treatment option. Further extending the potential of Notch inhibitors for the treatment of human disease, these experiments suggest that targeting the Notch signaling may be a viable strategy in the therapy of melanoma.
Supplementary Material
Acknowledgments
We thank Drs. B. Keith and L. Brass for helpful discussions. We thank J. Hayden for imaging assistance. We express our gratitude to Ms. Sherry Yang for assistance with Notch receptor real-time PCR experiments. We also express sincere gratitude to Gao Zhang for his expertise and aid in processing the microarray data. This work was supported through funding from The National Institutes of Health (CA76674, CA25874, CA10815, CA93372, CA47159, CA80999, CA098101, CA117881, GM071695).
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science (New York, NY. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 3.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291–7. [PubMed] [Google Scholar]
- 4.Raafat A, Bargo S, Anver MR, Callahan R. Mammary development and tumorigenesis in mice expressing a truncated human Notch4/Int3 intracellular domain (h-Int3sh) Oncogene. 2004;23:9401–7. doi: 10.1038/sj.onc.1208187. [DOI] [PubMed] [Google Scholar]
- 5.Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–5. [PubMed] [Google Scholar]
- 6.Grynfeld A, Pahlman S, Axelson H. Induced neuroblastoma cell differentiation, associated with transient HES-1 activity and reduced HASH-1 expression, is inhibited by Notch1. Int J Cancer. 2000;88:401–10. [PubMed] [Google Scholar]
- 7.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U S A. 1995;92:6414–8. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry CL, Reed LL, Golde TE, Miele L, Nickoloff BJ, Foreman KE. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene. 2005 doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Rao PK, Wen R, et al. Notch and Schwann cell transformation. Oncogene. 2004;23:1146–52. doi: 10.1038/sj.onc.1207068. [DOI] [PubMed] [Google Scholar]
- 10.Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–73. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu MY, Wheelock MJ, Johnson KR, Herlyn M. Shifts in cadherin profiles between human normal melanocytes and melanomas. J Investig Dermatol Symp Proc. 1996;1:188–94. [PubMed] [Google Scholar]
- 12.Tang A, Eller MS, Hara M, Yaar M, Hirohashi S, Gilchrest BA. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J Cell Sci. 1994;107 (Pt 4):983–92. doi: 10.1242/jcs.107.4.983. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Fukunaga M, Herlyn M. Reversal of melanocytic malignancy by keratinocytes is an E-cadherin-mediated process overriding beta-catenin signaling. Exp Cell Res. 2004;297:142–51. doi: 10.1016/j.yexcr.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene. 2001;20:8125–35. doi: 10.1038/sj.onc.1205034. [DOI] [PubMed] [Google Scholar]
- 15.Hsu MY, Meier FE, Nesbit M, et al. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am J Pathol. 2000;156:1515–25. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann JM, Holzmann B, Breitbart EW, Schmiegelow P, Riethmuller G, Johnson JP. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with a molecular weight of 76,000. Cancer Res. 1987;47:841–5. [PubMed] [Google Scholar]
- 17.Luca M, Hunt B, Bucana CD, Johnson JP, Fidler IJ, Bar-Eli M. Direct correlation between MUC18 expression and metastatic potential of human melanoma cells. Melanoma Res. 1993;3:35–41. doi: 10.1097/00008390-199304000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Xie S, Luca M, Huang S, et al. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57:2295–303. [PubMed] [Google Scholar]
- 19.Johnson JP. Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 1999;18:345–57. doi: 10.1023/a:1006304806799. [DOI] [PubMed] [Google Scholar]
- 20.Schlagbauer-Wadl H, Jansen B, Muller M, et al. Influence of MUC18/MCAM/CD146 expression on human melanoma growth and metastasis in SCID mice. Int J Cancer. 1999;81:951–5. doi: 10.1002/(sici)1097-0215(19990611)81:6<951::aid-ijc18>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Balint K, Xiao M, Pinnix CC, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–76. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 23.Balint K, Xiao M, Pinnix CC, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. The Journal of clinical investigation. 2005;115:3166–76. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (New York, NY. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 25.Liu ZJ, Xiao M, Balint K, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer research. 2006;66:4182–90. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer research. 2001;61:3819–25. [PubMed] [Google Scholar]
- 27.Qin JZ, Stennett L, Bacon P, et al. p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas. Mol Cancer Ther. 2004;3:895–902. [PubMed] [Google Scholar]
- 28.Jundt F, Anagnostopoulos I, Forster R, Mathas S, Stein H, Dorken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398–403. doi: 10.1182/blood.v99.9.3398. [DOI] [PubMed] [Google Scholar]
- 29.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–34. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniotis AJ, Folberg R, Hess A, et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. The American journal of pathology. 1999;155:739–52. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–71. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangarajan A, Talora C, Okuyama R, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. Embo J. 2001;20:3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 34.Sarmento LM, Huang H, Limon A, et al. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med. 2005;202:157–68. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa F, Fujii K, Horiguchi Y, et al. Roles of E- and P-cadherin in the human skin. Microsc Res Tech. 1997;38:343–52. doi: 10.1002/(SICI)1097-0029(19970815)38:4<343::AID-JEMT2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology. 2001;286:23–30. doi: 10.1006/viro.2001.0867. [DOI] [PubMed] [Google Scholar]
- 38.Dotto GP, Moellmann G, Ghosh S, Edwards M, Halaban R. Transformation of murine melanocytes by basic fibroblast growth factor cDNA and oncogenes and selective suppression of the transformed phenotype in a reconstituted cutaneous environment. The Journal of cell biology. 1989;109:3115–28. doi: 10.1083/jcb.109.6.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson RE, Dooley TP, Hart IR. Induction of tumorigenicity and lack of in vitro growth requirement for 12-O-tetradecanoylphorbol-13-acetate by transfection of murine melanocytes with v-Ha-ras. Cancer research. 1989;49:711–6. [PubMed] [Google Scholar]
- 40.Gupta PB, Kuperwasser C, Brunet JP, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 42.Citron M. Strategies for disease modification in Alzheimer's disease. Nat Rev Neurosci. 2004;5:677–85. doi: 10.1038/nrn1495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.