Abstract
Skeletal muscle is one of the largest tissues in the human body. Changes in mRNA and protein abundance in this tissue are central to a large number of metabolic and other disorders, including, commonly, insulin resistance. Proteomic and microarray analyses are important approaches for gaining insight into the molecular and biochemical basis for normal and pathophysiological conditions. With the use of vastus lateralis muscle obtained from two groups of healthy, nonobese subjects, we performed a detailed comparison of the muscle proteome, obtained by HPLC-ESI-MS/MS, with the muscle transcriptome, obtained using oligonucleotide microarrays. HPLC-ESI-MS/MS analysis identified 507 unique proteins as present in four out of six subjects, while 5193 distinct transcripts were called present by oligonucleotide microarrays from four out of six subjects. The majority of the proteins identified by mass spectrometry also had their corresponding transcripts detected by microarray analysis, although 73 proteins were only identified in the proteomic analysis. Reflecting the high abundance of mitochondria in skeletal muscle, 30% of proteins detected were attributed to the mitochondrion, as compared to only 9% of transcripts. On the basis of Gene Ontology annotations, proteins assigned to mitochondrial inner membrane, mitochondrial envelope, structural molecule activity, electron transport, as well as generation of precursor metabolites and energy, had more corresponding transcripts detected than would be expected by chance. On the contrary, proteins assigned to Golgi apparatus, extracellular region, lyase activity, kinase activity, and protein modification process had fewer corresponding transcripts detected than would be expected by chance. In conclusion, these results provide the first global comparison of the human skeletal muscle proteome and transcriptome to date. These data show that a combination of proteomic and transcriptic analyses will provide data that can be used to test hypotheses regarding the pathogenesis of muscle disorders as well as to generate observational data that can be used to form novel hypotheses.
Keywords: human skeletal muscle, proteomic analysis, HPLC-ESI-MS/MS, microarrays, transcriptome, tissue profiling, Gene Ontology
Introduction
The diagnosis and management of disorders of human muscle remains a challenging task. There is a lack of comprehensive knowledge regarding the molecular basis of changes in protein expression that are part of normal physiologic adaptations, such as those that occur during exercise training or age-related sarcopenia, or pathophysiological changes associated with disease processes, including atrophy (caused by disuse, spinal cord injury, or weightlessness), ischemic damage, muscular dystrophies, and insulin resistance. Insulin resistance in skeletal muscle is a condition that is common to obesity, type 2 diabetes, and cardiovascular disease. Between 25 and 50 million individuals in the United States are affected by these diseases. During the past decades, many studies have been carried out to define the molecular abnormalities underlying insulin resistance.1–3 On the basis of such studies, an increasing number of genes and proteins in several signaling and metabolic pathways have been implicated in skeletal muscle insulin resistance.1–5 However, a majority of studies have focused on only a limited number of genes and proteins.
There is a clear need for approaches capable of evaluating global changes in protein expression and modification. High-throughput gene expression technologies such as microarrays are powerful tools for the study of physiological and pathological conditions with complex or multifactorial underlying mechanisms, proving useful in studies of age-related sarcopenia,6 muscle insulin resistance,7–10 muscular dystrophy,11 denervation, 12 and endurance training.13 However, expression of muscle mRNA may not accurately reflect the abundance of proteins and can give no information regarding their post-translational modifications,14,15 indicating the need to understand the relationships and differences between global gene expression experiments and large-scale protein identification experiments.
A number of previous proteomic studies of human skeletal muscle have been accomplished using protein separation by 2D-gel electrophoresis with subsequent identification using MALDI-TOF/MS and/or HPLC-ESI-MS/MS analyses.16–21 Most are comparative studies reporting only proteins that are differentially regulated in conditions such as type 2 diabetes,16 obesity,17 Tibetans living at high altitude,21 and aging,18 or in comparisons between different muscle fiber types.19 The proteins reported in those studies represent mainly highly abundant structural or metabolic proteins. Furthermore, those studies focus primarily on differentially regulated proteins and, thus, many other proteins that are not altered in abundance or are present at too low a level to be easily quantified by spot analysis often were not identified. A number of transcriptome studies of human skeletal muscle have been reported. Those studies also focused primarily on differentially expressed transcripts associated with a disease or intervention, such as the neuromuscular disorder facioscapulohumeral muscular dystrophy,11 insulin resistance and diabetes,7 insulin treatment, 9 lipid infusion,10 endurance training,13 and muscle denervation.12
A number of large-scale proteome–transcriptome comparison studies have been reported. Most have used cell lines,15,22–29 plant30,31 and animal models,32–36 and only a few have studied in vivo human tissue samples14,37–39 due to the limited availability of human samples and lack of sensitive analytical approaches. To our knowledge, no large-scale proteome–transcriptome comparison studies on human skeletal muscle have been reported.
In a previous study, we used microarrays to study the effect of lipid infusion on healthy lean subjects and found that lipid infusion could decrease or increase the expression of some genes. Analysis of a portion of the results has been published.10 In a separate study, we used 1D gel electrophoresis and HPLC-ESI-MS/MS to characterize the proteome of human skeletal muscle obtained using percutaneous needle biopsies of the vastus lateralis muscle in healthy volunteers.40 In the present study, we applied proteomics analysis to six lean healthy volunteers and performed detailed comparison of the obtained proteome and transcriptome (also obtained from six lean healthy volunteers10). Moreover, for an additional subject, we compared the results of proteomics analysis to the results obtained from transcriptomics where the same muscle biopsy was used in both cases. To our knowledge, this is the first large-scale proteome and transcriptome comparison for human skeletal muscle.
Experimental Section
Subjects
The skeletal muscle samples used for proteomics analysis in this study were obtained from six healthy, nonobese volunteers (age, 20–47 years, 34.7 ± 3.7; body mass index, 25.7 ± 1.3 kg/m2; percent body fat, 23.5 ± 2.6%) with normal glucose tolerance and no family history of type 2 diabetes. Skeletal muscle samples used for mRNA analysis were from another set of six lean, healthy subjects described in ref 10, and a portion of the mRNA expression data (from the Affymetrix “A” chip) was reported previously.10 The subject for whom we had both proteome and transcriptome data was a 63-year old female with body mass index at 27.6 kg/m2 and with normal glucose tolerance and no family history of type 2 diabetes. The purpose, nature and potential risks of the study were explained to the participants, and written consent was obtained before participation. The protocols were approved by the Institutional Review Boards of Arizona State University or the University of Texas Health Science Center at San Antonio.
Muscle Preparation, Electrophoresis, and Staining
A percutaneous needle biopsy of the vastus lateralis muscle was obtained under local anesthesia, and the muscle biopsy specimen was immediately blotted free of blood, frozen, and stored in liquid nitrogen until use. For protein analysis, the muscle biopsy specimens were homogenized while still frozen in an ice-cold buffer (10 µL/mg tissue) consisting of (final concentrations): 50 mM HEPES, pH 7.6; 150 mM NaCl; 20 mM sodium pyrophosphate; 20mM β-glycerophosphate; 10mMNaF; 2mM sodium orthovanadate; 2 mM EDTA; 1% Triton; 10% glycerol; 2 mM phenylmethylsulfonyl fluoride; 1 mM MgCl2; 1 mM CaCl2; 10 µg/mL leupeptin; and 10 µg/mL aprotinin. A Polytron homogenizer (Brinkman Instruments, Westbury, NY) set on maximum speed for 30 s was used for homogenization. The homogenate was cooled on ice for 20 min and then centrifuged at 10 000g for 20 min at 4 °C; the resulting supernatant was frozen until use. Protein concentrations were determined by the method of Lowry.41 Sixty micrograms of muscle lysate proteins from subjects were separated on 4–20% gradient SDS polyacrylamide gels (Bio-Rad Laboratories, Hercules, CA); proteins were visualized with Coomassie blue (Bio-Rad Laboratories, Hercules, CA).
In-Gel Digestion
The gel lane resulting from each experiment was cut into 20 slices of approximately equal size. Each slice was cut into 1 mm cubes prior to digestion. The gel pieces were placed in a 0.6-mL polypropylene tube, washed with 400 µL of water, destained twice with 300 µL of 50% acetonitrile (ACN) in 40 mM NH4HCO3 and dehydrated with 100% ACN for 15 min. After removal of the ACN by aspiration, the gel pieces were dried in a vacuum centrifuge at 62 °C for 30 min. Trypsin (250 ng; Sigma Chemical Co., St. Louis, MO) in 30 µL of 40 mM NH4HCO3 was added and the samples were maintained at 4 °C for 15 min prior to the addition of 50 µL of 40 mM NH4HCO3. The digestion was allowed to proceed at 37 °C overnight and was terminated by the addition of 10 µL of 5% formic acid (FA). After incubation at 37 °C for an additional 30 min and centrifugation for 1 min, each supernatant was transferred to a clean polypropylene tube. The extraction procedure was repeated using 80 µL of 0.5% FA, and the two extracts were combined. The sample volume was reduced to approximately 5 µL by vacuum centrifugation, and 20 µL of 0.05% heptafluorobutyric acid (HFBA)/1% FA/2%ACN was added.
Mass Spectrometry
HPLC-ESI-MS/MS was performed on a hybrid linear ion trap (LTQ)-Fourier Transform Ion Cyclotron Resonance (FTICR) mass spectrometer (LTQ FT; Thermo Fisher, San Jose, CA) fitted with a PicoView nanospray source (New Objective, Woburn, MA). The mass spectrometer was calibrated weekly according to manufacturer’s instructions, achieving mass accuracy of the calibrants within 2 ppm. Online capillary HPLC was performed using a Michrom BioResources Paradigm MS4 micro HPLC (Auburn, CA) with a PicoFrit column (New Objective; 75 µm i.d., packed with ProteoPep II C18 material, 300Å). Samples were desalted using an online Nanotrap (Michrom BioResources, Auburn, CA) before being loaded onto the PicoFrit column. HPLC separations were accomplished with a linear gradient of 2–27% ACN in 0.1% FA in 70 min, a hold of 5 min at 27% ACN, followed by a step to 50% ACN, hold 5 min and then a step to 80%, hold 5 min; flow rate, 300 nL/min. A “top-10” data-dependent tandem mass spectrometry approach was utilized to identify peptides in which a full scan spectrum (survey scan) was acquired followed by collision-induced dissociation (CID) mass spectra of the 10 most abundant ions in the survey scan. The survey scan was acquired using the FTICR mass analyzer in order to obtain high resolution, high mass accuracy data.
Data Analysis and Bioinformatics
Tandem mass spectra were extracted from Xcalibur ”RAW” files and charge states were assigned using the Extract_MSN script (Thermo Fisher, San Jose, CA). Charge states and monoisotopic peak assignments were then verified using DTA-SuperCharge, part of the MSQuant suite of software (msquant.sourceforge.net),42 before all ”DTA” files from each gel lane in an experiment were combined into a single Mascot Generic format file. The fragment mass spectra were then searched against the IPI-_HUMAN_v3.28 database (68 020 entries, http://www.ebi-.ac.uk/IPI/) using Mascot (Matrix Science, London, U.K.; version 2.2). The false discovery rate was determined by selecting the option to search the decoy randomized database. The search parameters that were used were 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine and tyrosine. Probability assessment of peptide assignments and protein identifications were made through use of Scaffold (version Scaffold-01_06_19, Proteome Software, Inc., Portland, OR). Only peptides with ≥95% probability were considered. Criteria for protein identification included detection of at least 2 unique identified peptides and a probability score of ≥95%. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped. Multiple isoforms of a protein were reported only if they were differentiated by at least one unique peptide with ≥95% probability, based on Scaffold analysis.
Gene Ontology annotation of human proteins was down-loaded from Gene Ontology Annotation (GOA) Databases (http://www.ebi.ac.uk/GOA, version 55.0). This GOA human database contains 33 731 distinct proteins and 172 661 GO associations. In addition, GO hierarchy information (version: 52) was downloaded from www.geneontology.com. Human GO associations and GO hierarchy information were assembled into a new database by an in-house Script written using MATLAB. IPI IDs, gene names, UniProt and Swiss-Prot IDs of identified proteins were input into the database to obtain GO associations and GO hierarchy information.
Microarray Analysis: Target Preparation, Hybridization, taining, Scanning, and Analysis of Image
For mRNA analyses, muscle biopsy specimens were homogenized directly in RNAStat solution (Tel-Test, Inc., Friendswood, TX), using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY). RNA pellets were stored in ethanol/sodium chloride solution at −80 °C. Prior to use, total RNA was purified with RNeasy and DNase I treatment (Qiagen, Chatsworth, CA).
RNA was prepared for hybridization to Affymetrix (Santa Clara, CA) HG-U133A arrays according to the manufacturer’s instructions. Total RNA was used as a template for double-stranded cDNA synthesis (Superscript Double-Stranded cDNA Synthesis kit, Invitrogen, Carlsbad, CA), which was used as a template for biotin-labeled cRNA synthesis (Enzo BioArray High Yield RNA Transcription Labeling Kit, Affymetrix). Purified (RNeasy kit, Qiagen, Chatsworth, CA), fragmented (35–200 nucleotides) biotinylated cRNA was hybridized to the HG-U133A Gene Chips overnight for 16 h at 45 °C in a rotating incubator. Following hybridization, the probe arrays were washed and stained using the Gene Chip Fluidics station protocol EukGE-ES2. The protocol consisted of nonstringent and stringent washes followed by a staining procedure whereby the hybridized cRNA was fluorescently labeled using antibiotin antibodies and streptavidin-phycoerythrin solution (SAPE). The intensity of bound dye was measured with an argon laser confocal scanner (GeneArray Scanner, Agilent). The probe arrays were scanned twice and the stored images were aligned and analyzed using the GeneChip software Microarray Analysis Suite (MAS) 5.0 (Affymetrix, Santa Clara, CA).
Microarray Data Expression and Analysis
The Affymetrix data acquisition programs in MAS 5.0 automatically generate a cell intensity file (CEL) from the stored images that contain a single intensity value for each probe cell on the array. The CEL files were imported into the R software package (http://www.r-project.org) and the probe level data were converted to expression measures using the Affy package43 from Bioconductor. Expression values for each mRNA were obtained by the Robust Multiarray Analysis (RMA) method of Irizarry44 which adjusts for background on the raw intensity scale, carries out a nonlinear quantile normalization of the perfect match values, log-transforms the background-adjusted perfect match values and carries out a robust multichip analysis of the quantilenormalized, log-transformed values. The mRNA corresponding to a particular protein was deemed to be present if it was called present by Affymetrix software for the majority (four out of six) subjects. For comparison of the proteome with mRNA expression data from the two groups of subjects, it was required that a protein be detected from four out of six subjects based on the rigorous criteria described above. Gene names and Swiss-Prot IDs of identified transcripts were input into the in-house GO database described above to obtain GO information.
Comparison of the Proteome and Transcriptome
There are three major categories of GO information: Cellular component (abbreviation: C), biological process (abbreviation: P) and molecular function (abbreviation: F). Some GO terms belong to another GO term, that is, they are child terms of the latter (parent term). On the basis of the GO hierarchy information obtained from www.geneontology.org, proteins assigned to a child GO term were also assigned to its parent term. For example, mitochondrial matrix (GO ID 0005759) and mitochondrial envelope (GO ID 0005740) both belong to mitochondrion (GO ID 0005739). Therefore, mitochondrion (GO ID 0005739) is the parent term. All proteins assigned to the mitochondrial matrix and mitochondrial envelope also were assigned to mitochondrion. Another example is ion binding (GO ID 0043167). It has 3 direct child terms: anion binding (GO ID 0043168), cation binding (GO ID 0043169) and metal ion binding (GO ID 0046872). All protein assigned to anion binding, cation binding and metal ion binding also were assigned to ion binding.
Bootstrap analysis, a widely used statistical tool, was used to study the relationship between the human muscle proteome and transcriptome based on their GO associations.45,46 For each given GO term, i, a count was made of the number of proteins identified and associated with that GO term, Num-Pi, and the corresponding number of transcripts detected and associated with that GO term, Num-Ti•.The experimental fraction was calculated as Num-Ti/Num-Pi•. In order to determine the confidence that this fraction is significantly greater or less than the fraction that would be expected by chance alone, proteins were randomly selected and it was found how many of them had corresponding transcripts detected. This process was repeated 10 000 times in order to determine the chance that a set of proteins would have a corresponding set of transcripts detected. The fifth and 95th percentile of those 10 000 percentages serve as the upper and lower confidence boundaries of the distribution. If the experimental fraction is within the fifth and 95th percentile of the 10 000 percentages, it means % of transcripts present for proteins with a given GO term is about the same as would be expected by chance. When the fraction of transcripts found for a set of proteins is greater than the 95th percentile, it indicates that % of transcripts present for these proteins is more than would be expected by chance. When the fraction is less than the fifth percentile, % of transcripts present is less than would be expected by chance.
Results and Discussion
Human Skeletal Muscle Proteome
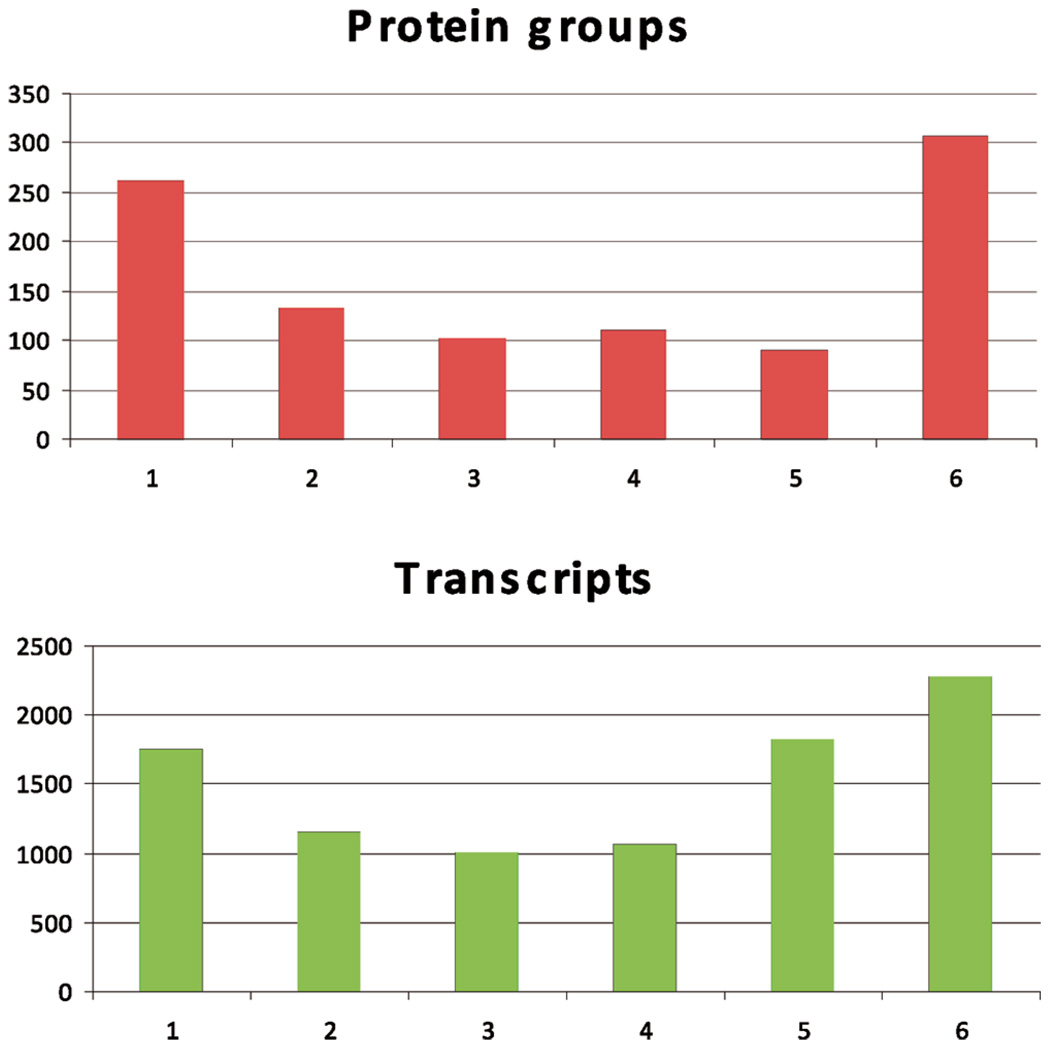
To obtain a comprehensive proteomic characterization of human vastus lateralis muscle, we carried out HPLC-ESI-MS/MS-based analysis of lysates of whole muscle from which proteins were first fractionated by 1D gel electrophoresis. We performed HPLC-ESI-MS/MS analyses on proteins isolated from the muscle of 6 healthy, nonobese subjects. In subjects 1–6, 635, 573, 619, 582, 494, and 664 unique proteins were identified, respectively. The average number of identified proteins from the 6 subjects was 594 ± 59. A total of 1003 unique proteins were identified in at least one subject after reduction for redundancy. There were at least two unique peptides (≥95% confidence) assigned to each identified protein, all with a confidence level ≥95% based on the Scaffold analysis. The false discovery rates, as assessed by Mascot searching of a randomized database, were less than 6%. Among these proteins, 306 were identified in all 6 subjects, 91 in 5 subjects, 110 in 4 subjects, 102 in 3 subjects, 133 in 2 subjects and 261 were identified in only one subject (Figure 1, top panel). A detailed list of all proteins identified in this study together with their IPI ID, molecular weight, sequence coverage, and number of unique peptides assigned to each protein are provided as Supporting Information (Supplemental Table 1). Among the proteins identified in this study, a number of entries derived from the protein identification searches had multiple IPI IDs. In many cases, assignment of multiple IDs results from the potential presence of protein isoforms that could not be distinguished on the basis of unique peptides. Proteins with multiple IDs were assigned to a “protein group” (see Supplemental Table 1). Proteins that were attributed a unique IPI ID are listed as a single-entry protein group. As can be seen from Supplemental Table 1, when the analysis was perfomed in this manner, there were 1003 protein groups. For protein groups with multiple IPI IDs, the minimum, maximum and mean of molecular weights (MW) and the number of amino acids in each protein sequence are listed in Supplemental Table 1.
Figure 1.
Number of protein groups (A) and transcript groups (B) detected in 1–6 individuals. (Top panel) For the 6 individuals who participated in the proteomics study, 1003 unique protein groups were identified in at least one individual. A total of 507 out of the 1003 protein groups were detected in 4 out of 6 subjects. (Bottom panel) For the 6 individuals who participated in the microarray study, 9113 distinct transcript groups were called present by the Affymetrix software in at least one individual. A total of 5193 out of the 9113 transcript groups were called present in 4 out of 6 subjects.
Human Skeletal Muscle Transcriptome
We used microarray analysis on 6 lean, healthy subjects to obtain characterization of the human vastus lateralis muscle transcriptome. There are many probes on the chip which have the same gene names or Swiss-Prot IDs, and they were grouped into one transcript set. Affymetrix software called 2286 transcripts “present” in all 6 subjects, 1834 in 5 subjects, 1073 in 4 subjects, 1008 in 3 subjects, in 1155 in 2 subjects and 1757 identified in only one subject (Figure 1, bottom panel). As mentioned in the experimental section, the mRNA corresponding to a particular protein was deemed to be present if it was called present by Affymetrix software for the majority (four out of six) subjects. All 5193 “present” transcript groups with their gene names and Swiss-Prot IDs were listed in Supplemental Table 2.
Comparison of the Muscle Proteome and Transcriptome from Two Groups of Subjects
We compared the skeletal muscle proteome obtained in this study (n = 6) with mRNA expression data obtained from healthy human skeletal muscle using oligonucleotide microarray analysis (n = 6). Since the mRNA corresponding to a particular protein was deemed to be present if it was called present by Affymetrix software for four out of six subjects, we also required that a protein needed to be detected from four out of six subjects based on the rigorous criteria for the comparison of the proteome with transcriptome. A total of 507 out of 1003 protein groups were identified in four out of six subjects. The majority of these, 448 (88%), had corresponding transcripts detected by microarray analysis. Among the 5193 present transcripts, 437 had their corresponding proteins detected by mass spectrometry. Even though the number of detected transcripts identified by microarray was 10 times greater than the number of proteins identified using HPLC-ESI-MS/MS, there were 59 proteins (12%) identified by mass spectrometry that did not have their corresponding transcripts detected by microarray analysis (see Table 1). The transcripts for 1 out of these 59 proteins were not present as probes in the microarray chips. Thus, 58 of the 507 identified proteins did not have corresponding transcripts detected even though the transcripts were present as probes on the chips. Some of these proteins were present in relatively high abundance. For example, Apolipoprotein A-1, which is transported into muscle,47,48 was found in all 6 subjects with the maximum sequence coverage of 42%. It is known that this protein is synthesized primarily in the liver and circulates in the plasma to peripheral tissues such as muscle, where it is cleared by a well-described endocytosis process.47,48 The presence of this protein in skeletal muscle of all six healthy individuals suggests that proteomic analysis may be useful for studying apolipoprotein A1 metabolism in human muscle. Some other proteins that clearly derive from muscle, such as myosin-binding protein H, which was detected in 5 out of 6 subjects by proteomic analysis, had no detectable mRNA in any of the 6 individuals. This latter type of conflicting result likely is due to either rapid degradation of transcripts specific to those proteins or poor binding of mRNA for those proteins to the complementary oligonucleotide on the array.
Table 1.
Proteins identified by mass spectrometry (≥ 4 out of 6 Subjects) but not detected by microarray (≥ 4 out of 6 Subjects)
| protein group | IPI IDs | gene name | max sequence coverage (%) | max unique peptides | protein name(s) |
|---|---|---|---|---|---|
| 9 | IPI00001539 | ACAA2 | 13 | 4 | 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL. |
| 37 | IPI00005158; IPI00334291 | LONP1 | 11 | 9 | LON PROTEASE HOMOLOGUE, MITOCHONDRIAL PRECURSOR; CDNA FLJ39307 FIS, CLONE OCBBF2013208, HIGHLY SIMILAR TO MITOCHONDRIALLON PROTEASE. |
| 60 | IPI00009253; IPI00445144 | NAPA | 19 | 4 | ALPHA-SOLUBLE NSF ATTACHMENT PROTEIN; CDNA FLJ44555 FIS, CLONE UTERU3007640, HIGHLY SIMILAR TO HOMO SAPIENSN-ETHYLMALEIMIDE-SENSITIVE FACTOR ATTACHMENT PROTEIN, ALPHA. |
| 78 | IPI00011229 | CTSD | 14 | 4 | CATHEPSIN D PRECURSOR. |
| 79 | IPI00011250; IPI00647149 | UCHL3 | 24 | 4 | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L3; 17 KDA PROTEIN. |
| 93 | IPI00013723 | PIN1 | 30 | 3 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1. |
| 97 | IPI00013894; IPI00479946 | STIP1 | 17 | 10 | STRESS-INDUCED-PHOSPHOPROTEIN 1; STIP1 PROTEIN. |
| 127 | IPI00018246; IPI00220663; IPI00220665; IPI00220667 | HK1 | 22 | 20 | ISOFORM 1 OF HEXOKINASE-1; ISOFORM 2 OF HEXOKINASE-1; ISOFORM 3 OF HEXOKINASE-1; ISOFORM 4 OF HEXOKINASE-1. |
| 129 | IPI00018398 | PSMC3 | 10 | 3 | 26S PROTEASE REGULATORY SUBUNIT 6A. |
| 132 | IPI00018740 | IDI2 | 30 | 5 | ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE 2. |
| 142 | IPI00020210 | DYSF | 5 | 10 | DYSFERLIN_V1. |
| 160 | IPI00021841 | APOA1 | 42 | 12 | APOLIPOPROTEIN A-I PRECURSOR. |
| 161 | IPI00021890; IPI00747151 | HSD17B8 | 16 | 3 | ESTRADIOL 17-BETA-DEHYDROGENASE 8; 24 KDA PROTEIN. |
| 171 | IPI00022463; IPI00798430 | TF | 20 | 14 | SEROTRANSFERRIN PRECURSOR.; TRANSFERRIN VARIANT (FRAGMENT). |
| 179 | IPI00024046 | CDH13 | 8 | 4 | CADHERIN-13 PRECURSOR. |
| 182 | IPI00024664; IPI00375145 | USP5 | 15 | 9 | ISOFORM LONG OF UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 5; ISOFORM SHORT OF UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 5. |
| 202 | IPI00025874 | RPN1 | 9 | 4 | DOLICHYL-DIPHOSPHOOLIGOSACCHARIDE-PROTEIN GLYCOSYLTRANSFERASE 67 KDASUBUNIT PRECURSOR. |
| 275 | IPI00100980 | EHD2 | 18 | 9 | EH DOMAIN-CONTAINING PROTEIN 2. |
| 283 | IPI00164623; IPI00783987 | C3 | 21 | 24 | 187 KDA PROTEIN.; COMPLEMENT C3 PRECURSOR (FRAGMENT). |
| 295 | IPI00176903; IPI00513773 | PTRF | 18 | 5 | ISOFORM 1 OF POLYMERASE I AND TRANSCRIPT RELEASE FACTOR; ISOFORM 2 OF POLYMERASE I AND TRANSCRIPT RELEASE FACTOR. |
| 303 | IPI00215715; IPI00745082 | CAMK2A | 13 | 4 | ISOFORM A OF CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II ALPHACHAIN; CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE IIA ISOFORM 2. |
| 310 | IPI00215983 | CA1 | 56 | 10 | CARBONIC ANHYDRASE 1. |
| 314 | IPI00216138 | TAGLN | 39 | 8 | TRANSGELIN. |
| 325 | IPI00217458 | GPT | 17 | 6 | ALANINE AMINOTRANSFERASE 1. |
| 326 | IPI00217466 | HIST1H1D | 21 | 5 | HISTONE H1.3. |
| 328 | IPI00217871; IPI00647328 | ALDH4A1 | 20 | 8 | DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, MITOCHONDRIALPRECURSOR; ALDEHYDE DEHYDROGENASE 4 FAMILY, MEMBER A1. |
| 366 | IPI00219772 | NDUFB7 | 12 | 3 | NADH DEHYDROGENASE [UBIQUINONE] 1 BETA SUBCOMPLEX SUBUNIT 7. |
| 387 | IPI00221127 | MYLK2 | 18 | 9 | MYOSIN LIGHT CHAIN KINASE 2, SKELETAL/CARDIAC MUSCLE. |
| 407 | IPI00293867; IPI00472043 | DDT | 34 | 4 | D-DOPACHROME DECARBOXYLASE; SIMILAR TO D-DOPACHROME TAUTOMERASE. |
| 409 | IPI00294073 | COQ7 | 27 | 5 | UBIQUINONE BIOSYNTHESIS PROTEIN COQ7 HOMOLOGUE. |
| 415 | IPI00295386 | CBR1 | 21 | 5 | CARBONYL REDUCTASE [NADPH] 1. |
| 435 | IPI00301028 | TRIM72* | 38 | 14 | ISOFORM 1 OF TRIPARTITE MOTIF-CONTAINING PROTEIN 72. |
| 438 | IPI00302592; IPI00333541; IPI00644576 | FLNA | 7 | 10 | FILAMIN A, ALPHA; FILAMIN-A; FILAMIN A, ALPHA. |
| 443 | IPI00303882 | M6PRBP1 | 28 | 8 | ISOFORM B OF MANNOSE-6-PHOSPHATE RECEPTOR-BINDING PROTEIN 1. |
| 452 | IPI00307755 | PRKAA2 | 8 | 4 | 5′-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2. |
| 463 | IPI00220906; IPI00333838 | ACOT1; ACOT2 | 15 | 5 | ISOFORM 1 OF ACYL-COENZYME A THIOESTERASE 2; ACYL-COENZYME A THIOESTERASE 1. |
| 470 | IPI00376005; IPI00411704 | EIF5A | 26 | 3 | ISOFORM 2 OF EUKARYOTIC TRANSLATION INITIATION FACTOR 5A-1.; ISOFORM 1 OF EUKARYOTIC TRANSLATION INITIATION FACTOR 5A-1. |
| 476 | IPI00011454; IPI00383581; IPI00441414; IPI00472068 | GANAB | 9 | 5 | ISOFORM 2 OF NEUTRAL ALPHA-GLUCOSIDASE AB PRECURSOR; ISOFORM 1 OF NEUTRAL ALPHA-GLUCOSIDASE AB PRECURSOR; ISOFORM 3 OF NEUTRAL ALPHA-GLUCOSIDASE AB PRECURSOR; 107 KDA PROTEIN. |
| 477 | IPI00384280 | PCYOX1 | 8 | 4 | PRENYLCYSTEINE OXIDASE PRECURSOR. |
| 502 | IPI00420049; IPI00739670 | C1orf170 | 14 | 5 | SIMILAR TO ZK84.1; SIMILAR TO ZK84.1. |
| 522 | IPI00470535 | CACNA2D1 | 8 | 7 | DIHYDROPYRIDINE RECEPTOR ALPHA 2 SUBUNIT |
| 541 | IPI00554737 | PPP2R1A | 24 | 10 | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A 65 KDA REGULATORY SUBUNIT AALPHA ISOFORM. |
| 554 | IPI00745872 | ALB | 46 | 33 | ISOFORM 1 OF SERUM ALBUMIN PRECURSOR. |
| 555 | IPI00216256; IPI00746165 | WDR1 | 20 | 6 | ISOFORM 2 OF WD REPEAT PROTEIN 1; ISOFORM 1 OF WD REPEAT PROTEIN 1. |
| 564 | IPI00792715 | ENO2 | 7 | 2 | 37 KDA PROTEIN. |
| 565 | IPI00793379 | 35 | 44 | 165 KDA PROTEIN. | |
| 593 | IPI00008964 | RAB1B | 37 | 5 | RAS-RELATED PROTEIN RAB-1B. |
| 605 | IPI00012272 | MYBPH | 14 | 4 | MYOSIN-BINDING PROTEIN H. |
| 635 | IPI00022432; IPI00646384 | TTR | 33 | 3 | TRANSTHYRETIN PRECURSOR.; 13 KDA PROTEIN. |
| 636 | IPI00022488 | HPX | 10 | 4 | HEMOPEXIN PRECURSOR. |
| 637 | IPI00022977 | CKB | 16 | 4 | CREATINE KINASE B-TYPE. |
| 639 | IPI00024107; IPI00218467; IPI00788661 | SNCA | 45 | 4 | ISOFORM 1 OF ALPHA-SYNUCLEIN.; ISOFORM 2–4 OF ALPHA-SYNUCLEIN.; 12 KDA PROTEIN. |
| 654 | IPI00030154; IPI00479722; IPI00748256 | PSME1 | 33 | 8 | PROTEASOME ACTIVATOR COMPLEX SUBUNIT 1; 29 KDA PROTEIN.; PROTEASOME ACTIVATOR SUBUNIT 1 ISOFORM 2. |
| 656 | IPI00032633 | COQ6 | 7 | 3 | UBIQUINONE BIOSYNTHESIS MONOOXYGENASE COQ6. |
| 665 | IPI00168620 | ADPRHL1 | 14 | 4 | ISOFORM 1 OF [PROTEIN ADP-RIBOSYLARGININE] HYDROLASE-LIKE PROTEIN 1. |
| 675 | IPI00217960; IPI00396630 | LOC730418; PRKACA | 21 | 6 | ISOFORM 2 OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT; ISOFORM 1 OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT. |
| 708 | IPI00329646 | PCBD2 | 36 | 4 | PTERIN-4 ALPHA-CARBINOLAMINE DEHYDRATASE 2. |
| 712 | IPI00394809 | APOOL | 27 | 6 | APOLIPOPROTEIN O-LIKE PRECURSOR. |
| 787 | IPI00032313 | S100A4 | 28 | 3 | PROTEIN S100-A4 |
Molecular Weight Distributions
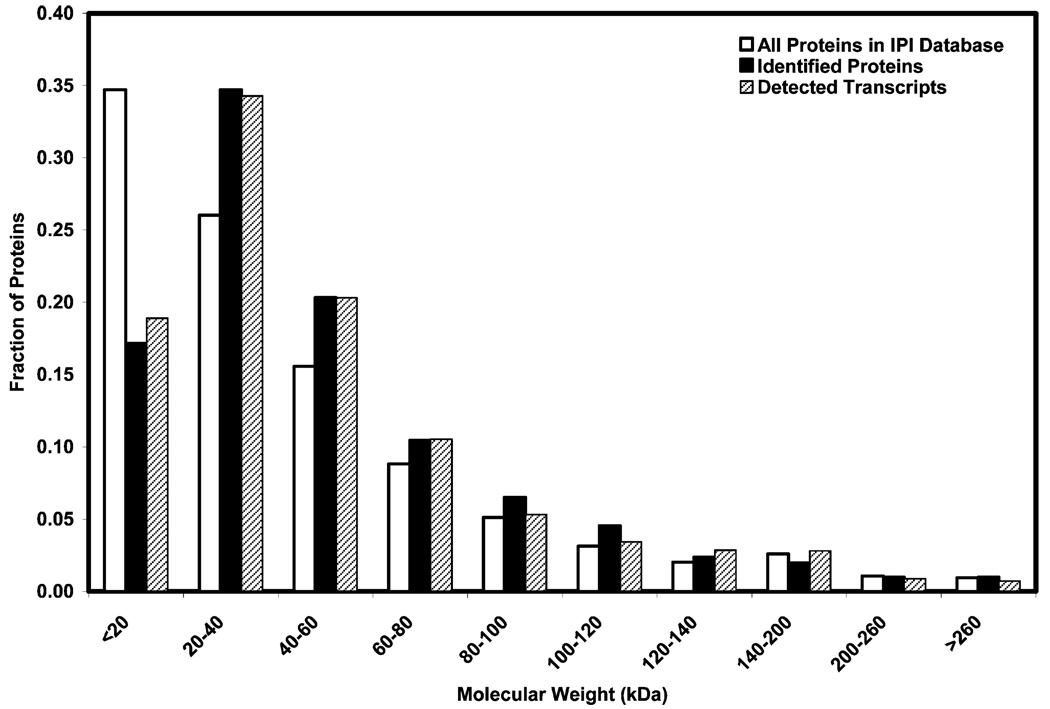
The MW distributions of all 507 identified proteins and corresponding proteins of 5193 present transcripts as well as all proteins listed in the IPI human database are shown in Figure 2. The MW distributions of all 507 identified proteins and corresponding proteins for the 5193 present transcripts are similar. Compared to the expected values based on the human proteome cataloged in the IPI human database, we identified a lower than expected proportion of proteins under 20 kDa. This may be an artifact of the 1Dgel from which very small proteins could be lost preferentially.
Figure 2.
Molecular weight distribution of identified proteins and transcripts in human skeletal muscle.
Subcellular Localization
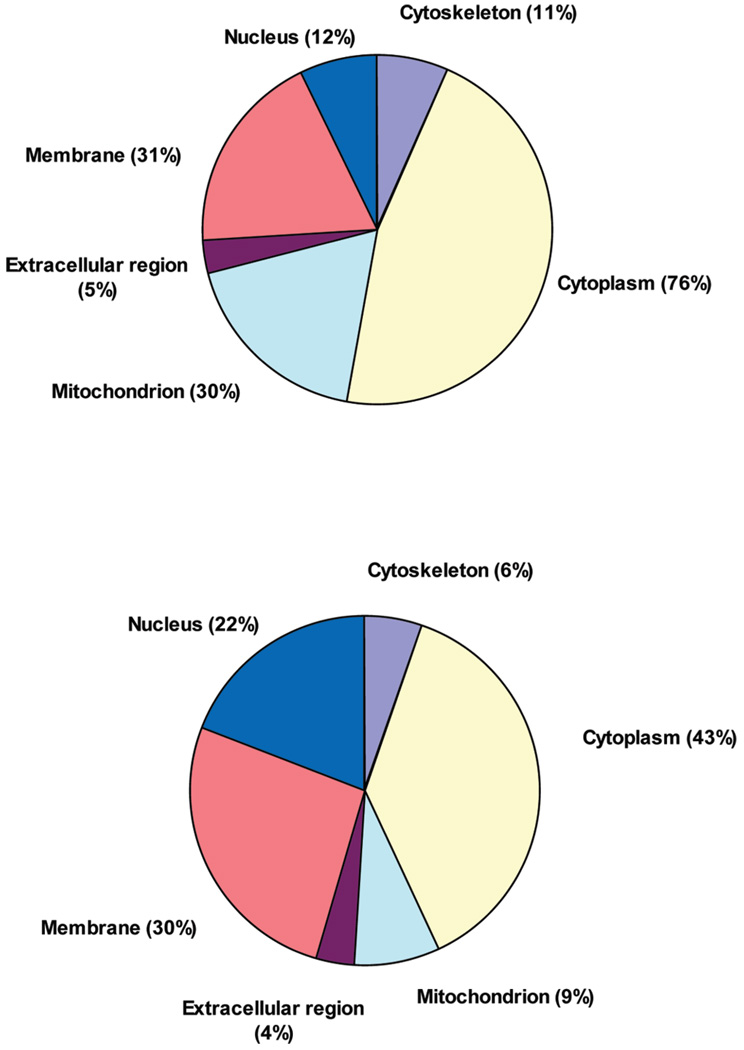
It was possible to assign the subcellular location of 438 out of the 507 proteins identified by mass spectrometry from at least four out of six subjects. Sixty-nine remained unassigned because they have no GO annotation in the GOA database (Figure 3, top panel). Of the proteins with GO annotations, 76% could be assigned to cytoplasm, representing the predominant subcellular location of all identified proteins in this study. Membrane proteins compromised the next highest (31%) subcellular location of identified proteins. Mitochondrial proteins represented 30% of all identified proteins. Nuclear proteins represented the fourth largest group, with 12% of all identified proteins. Cytoskeletal proteins represented 11% of all identified proteins and extra-cellular proteins constituted 5% of all identified proteins. Some proteins could be assigned to multiple subcellular locations. The subcellular locations of the 59 proteins, detected by mass spectrometry with no corresponding transcripts detected by microarray, were all assigned. Of these, 32%, 22%, 19%, 19%, 12% and 3% could be assigned to cytoplasm, mitochondrion, membrane, nucleus, extracellular, and cytoskeleton, respectively.
Figure 3.
Subcellular location of identified proteins (top panel) and transcripts (bottom panel) in human skeletal muscle.
The subcellular location of 3185 out of the 5193 transcripts identified by microarray from at least four out of six subjects could be assigned, while 2008 remained unassigned (Figure 3, bottom panel). Of the identified transcripts, 43% could be assigned to cytoplasm, representing the largest subcellular location of all identified transcripts in this study. Membrane transcripts (30%) were the second dominant subcellular location of identified transcripts. Nuclear transcripts represented the third largest group, with 22% of all identified transcripts and 485 of the 5193 transcripts detected (9%) could be attributed to the mitochondrion. Cytoskeletal transcripts represented the fifth largest group, with 6% of all identified transcripts. As was the case for proteins, some transcripts were also assigned to multiple subcellular locations.
It is noted that both transcripts and proteins for the mitochondrion are overrepresented in skeletal muscle. It has been estimated that mitochondrial proteins comprise 4.8% of the total human proteome.49 In skeletal muscle, 485 of the 5193 transcripts (9%) detected by microarray could be attributed to the mitochondrion, and in contrast, 150 of the 507 proteins (30%) detected by mass spectrometry were attributed to the mitochondrion. The overrepresentation of mitochondrial proteins reflects the importance of energy supply for the primary function of skeletal muscle, that is, contraction for movement. The percentages for cytoplasm and cytoskeleton proteins also were higher than those for cytoplasm and cytoskeleton transcripts. In contrast, the percentage of identified nucleus transcripts was higher than that for identified nucleus proteins.
Global Relationship between Human Muscle Proteome and Transcriptome
We employed bootstrap analysis, a widely used statistic tool,45,46 to study the relationship between human muscle proteome and transcriptome based on their GO information as described in the Methods section. In total, there were 1109 GO terms associated with the identified proteins. Proteins assigned to a child GO term were also assigned to its parent term. For example, there are 13 proteins were assigned to mitochondrial matrix and 68 to mitochondrial envelope. All of them were also assigned to their parent term, mitochondrion. Another example is ion binding that has 3 direct child terms: anion binding, cation binding and metal ion binding. While no protein was assigned to anion binding, 94 proteins were assigned to cation binding and 114 were assigned to metal ion binding. All of them were also assigned to ion binding.
We built the confidence level of relationships of the detection of proteins and transcripts using bootstrap. As an example, consider the situation where 23 proteins were assigned to the extracellular region, and the transcripts for 16 of these also were detected. In this analysis, we randomly selected 23 proteins from all 507 proteins and determined how many of these have their corresponding transcripts detected, and the fraction was recorded. This process was repeated 10 000 times to obtain 10 000 such fractions. The fifth and 95th percentiles of the 10 000 fractions, in this case, were 78% and 87%, respectively. The observed fraction in this case (16/23 or 70%) was less than the fifth percentile (78%) of the empirically derived distribution of fractions. This indicates that proteins assigned to the extracellular region have fewer corresponding transcripts detected than would be expected by chance. Since many proteins can be transported into muscle from blood, this relationship for extracellular region proteins is not surprising. GO terms with assigned proteins having fewer or more transcripts detected than would be expected by chance are listed in Table 2.
Table 2.
GO Terms with Assigned Proteins Have Fewer or More Transcripts Detected than Would Be Expected by Chance
| GO name | GO category a | GO number | no. of proteins found | no. of proteins with transcripts present | experimental fraction b | fifth percentile by bootstrap | 95th percentile by bootstrap |
|---|---|---|---|---|---|---|---|
| cholesterol metabolic process | P | 8203 | 5 | 1 | 20% | 60% | 100% |
| cholesterol biosynthetic process | P | 6695 | 4 | 1 | 25% | 50% | 100% |
| steroid biosynthetic process | P | 6694 | 6 | 2 | 33% | 67% | 100% |
| antigen binding | F | 3823 | 3 | 1 | 33% | 67% | 100% |
| hormone metabolic process | P | 42445 | 3 | 1 | 33% | 67% | 100% |
| ubiquitin thiolesterase activity | F | 4221 | 3 | 1 | 33% | 67% | 100% |
| steroid metabolic process | P | 8202 | 8 | 3 | 38% | 63% | 100% |
| vacuole | C | 5773 | 5 | 2 | 40% | 60% | 100% |
| lysosome | C | 5764 | 5 | 2 | 40% | 60% | 100% |
| sterol biosynthetic process | P | 16126 | 5 | 2 | 40% | 60% | 100% |
| phosphotransferase activity, alcohol group as acceptor | F | 16773 | 10 | 5 | 50% | 70% | 100% |
| Golgi apparatus | C | 5794 | 8 | 4 | 50% | 63% | 100% |
| protein serine/threonine kinase activity | F | 4674 | 8 | 4 | 50% | 63% | 100% |
| protein amino acid phosphorylation | P | 6468 | 12 | 7 | 58% | 75% | 100% |
| kinase activity | F | 16301 | 18 | 12 | 67% | 72% | 100% |
| extracellular region | C | 5576 | 23 | 16 | 70% | 78% | 100% |
| lyase activity | F | 16829 | 20 | 14 | 70% | 75% | 100% |
| protein modification process | P | 6464 | 33 | 24 | 73% | 79% | 97% |
| alcohol metabolic process | P | 6066 | 34 | 26 | 76% | 79% | 97% |
| mitochondrial envelope | C | 5740 | 68 | 65 | 96% | 82% | 94% |
| integral to membrane | C | 16021 | 55 | 53 | 96% | 80% | 95% |
| electron transport | P | 6118 | 58 | 56 | 97% | 81% | 95% |
| mitochondrial inner membrane | C | 5743 | 61 | 59 | 97% | 82% | 95% |
| mitochondrial membrane | C | 31966 | 65 | 63 | 97% | 82% | 94% |
| structural molecule activity | F | 5198 | 43 | 42 | 98% | 79% | 95% |
| generation of precursor metabolites and energy | P | 6091 | 95 | 93 | 98% | 83% | 94% |
| monovalent inorganic cation transmembrane transporter activity | F | 15077 | 25 | 25 | 100% | 76% | 96% |
| energy derivation by oxidation of organic compounds | P | 15980 | 31 | 31 | 100% | 77% | 97% |
C, cellular component; F, molecular function; and P, biological process
Fraction of proteins with transcript present
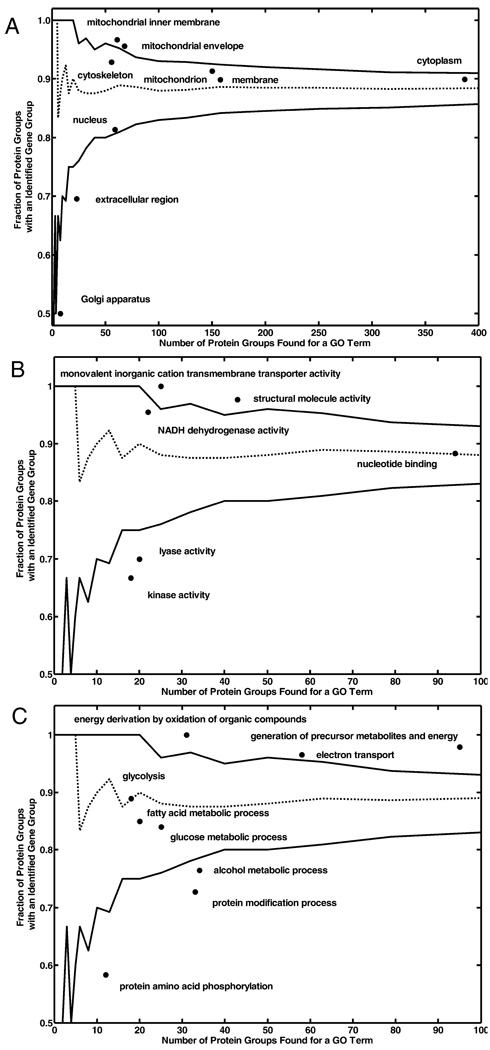
Figure 4A shows the protein–transcript relationships of 6 major cellular components: mitochondrion, cytoplasm, cytoskeleton, nucleus, membrane, and the extracellular region. In the mitochondrion category, 91% of 150 identified proteins had their corresponding transcripts detected, which is within the fifth and 95th percentile of the empirically derived distribution of fractions. In addition, 93% of identified cytoskeleton proteins and 81% of identified nucleus proteins, 90% cytoplasmic proteins and 90% membrane proteins identified by HPLC-ESI-MS/MS had their corresponding transcripts detected by microarray. In contrast, extracellular region proteins had fewer corresponding transcripts detected than would be expected by chance, as described above. Golgi apparatus proteins also had fewer corresponding transcripts detected. Although mitochondrial proteins had about the same number of transcripts detected as would be expected by chance, mitochondrial inner membrane and envelope proteins had more transcripts detected than would be expected by chance. In total, 97% and 96% of detected mitochondrial inner membrane and envelope proteins had their respective transcripts detected by microarray.
Figure 4.
Protein–transcript relationships. (A) Protein–transcript relationships of 6 major cellular components: mitochondrion, cytoplasm, cytoskeleton, nucleus, membrane, and extracellular region. (B) Protein–transcript relationships of some molecular functions. (C) Protein–transcript relationships of some biological processes. Dash line indicates the median of the empirically derived distribution of the 10 000 fractions. Solid lines above and below the dash line depict upper and lower 95% confidence intervals of the 10 000 fractions, respectively.
Figure 4B shows protein–transcript relationships of some molecular functions of interest. Proteins assigned to monovalent inorganic cation transmembrane transporter activity (GO ID 15077) and structural molecule activity (GO ID 5198) had more transcripts detected than would be expected by chance, while those for lyase activity (GO ID 16829) and kinase activity (GO ID 16301) had fewer transcripts detected than would be expected by chance. In addition, protein assigned to NADH dehydrogenase activity (GO ID 3954) and nucleotide binding (GO ID 166) had about the same number of transcripts detected as would be expected by chance.
Figure 4C shows protein–transcript relationships of some biological processes. Proteins assigned to electron transport (GO ID 6118), energy derivation by oxidation of organic compounds (GO ID 15980) and generation of precursor metabolites and energy (GO ID 6091) had more transcripts detected than would be expected by chance, while proteins assigned to alcohol metabolic process (GO ID 6066) and protein modification process (GO ID 6464) and protein amino acid phosphorylation (GO ID 6468) had fewer transcripts detected than just by chance. The proteins involving in fatty acid metabolic process (GO ID 6631), glycolysis (GO ID 6096), and glucose metabolic process (GO ID 6006) had about the same number of transcripts detected as would be expected by chance.
Comparison of Muscle Proteome and Transcriptome from the Same Subject
To determine whether the data obtained from two separate groups of subjects is an accurate reflection of such data obtained from a single subject, we performed proteomic and transcriptomic studies from a single muscle biopsy from one healthy volunteer. In this subject, 635 unique protein groups were identified, which is similar to the average number of identified proteins, 594, from the 6 subjects described above. The majority of these proteins, 533 (84%), had corresponding transcripts detected by microarray analysis. As mentioned earlier, there were 507 protein groups identified in four out of six subjects, and 448 (88%) of them had corresponding transcripts detected by microarray analysis.
For the single muscle biopsy, 5978 transcripts were called “present”, and this is similar to the 5193 transcripts called present from 4 out of 6 subjects. Among the 5978 present transcripts, 519 had their corresponding proteins detected by mass spectrometry. There were 103 out of 635 proteins (16%) identified by mass spectrometry that did not have their corresponding transcripts detected by microarray analysis. The transcripts for 10 of these 103 proteins were not present as probes in the microarray chips. Thus, 93 of the 635 identified proteins did not have corresponding transcripts detected even though the transcripts were present as probes on the chips.
Molecular Weight Distributions
The MW distributions of all 635 identified proteins and corresponding proteins of 5978 present transcripts as well as all proteins listed in the IPI human database are shown in Supplemental Figure 1. The MW distributions of all 635 identified proteins and corresponding proteins for the 5978 present transcripts are similar to each other and also similar to the MW distributions of the proteome and transcriptome from the two groups of subjects shown in Figure 2.
Subcellular Localization
The subcellular location pie-chart for the proteome and transcriptome from the same subject is shown in Supplemental Figure 2. For the proteome, 73%, 28%, 24%, 12%, 11% and 5% of identified proteins could be assigned to cytoplasm, membrane, mitochondrion, nucleus, cytoskeleton and extracellular region, respectively. There is no significant difference by paired t-test between the subcellular location for the proteome from this subject and the proteome from the group of six subjects (detected from four out of six subjects 4 out of 6 subjects). For the transcriptome, 38%, 27%, 8%, 19%, 6% and 4% of identified transcripts could be assigned to cytoplasm, membrane, mitochondrion, nucleus, cytoskeleton and extracellular region, respectively. Again, there is no significant difference by paired t-test between the subcellular location for the transcriptome from this subject and the transcriptome from the group of six subjects (detected from four out of six subjects 4 out of 6 subjects).
Conclusions
Mechanisms underlying changes in muscle structure and metabolism during different physiological and pathophysiological conditions have traditionally been studied by focusing on a small number of genes or proteins. Transcriptional profiling has been applied to define the molecular signature of denervation, immobilization, exercise-training, age-related sarcopenia, insulin resistance and muscular dystrophy.6–13,50 However, global profiling of temporal changes in metabolic enzymes and structural proteins and their post-translational modifications in skeletal muscle in vivo has been limited by the lack of appropriate proteomic technology. Thus, global comparison of the proteome and transcriptome has not been feasible. The present study shows that it is possible to obtain a global comparison of the proteome and transcriptome by combing HPLC-ESI-MS/MS with microarray technology.
It must be noted that most of the muscle biopsies used for proteomic analysis and microarray analysis were from different subjects. This is mitigated by the fact that all subjects were healthy and nonobese with similar characteristics. In addition, the muscle biopsies were obtained under identical conditions. Nevertheless, the concordance between the proteome and transcriptome is striking, considering the fact that different groups of subjects were used for these analyses. Had the same subjects been used for both analyses, these relationships likely would have been even more marked. The one attempt on the proteomic analysis and microarray analysis on the same healthy subject revealed dramatic similarity between the results obtained from the same subject and from the two groups of subjects as regard to number of identified protein/transcripts, molecular distribution, and subcellular localization. This indicates that the variations between the transcriptome and the proteome are not due to variations in the individual test subjects. It is worth noting that, even in the one individual subject’s proteome–transcriptome comparison, mitochondrial proteins were also overrepresented compared to their identified transcripts.
In conclusion, this study represents the first detailed, large-scale comparison of the human skeletal muscle proteome and transcriptome. These data demonstrate the utility of combining these two methods of analysis for small, human tissue samples, making this approach a potentially valuable tool in elucidating changes in the proteome/transcriptome associated with human disease.
Supplementary Material
Acknowledgment
This work was supported by NIH grants R01DK47936 and R01DK66483 (L.J.M.).
Abbreviations
- FA
formic acid
- ACN
acetonitrile
- CID
collision-induced dissociation
- OXPHOS
oxidative phosphorylation
- GO
Gene Ontology annotation
- MW
molecular weight
- MS/MS
tandem mass spectrometry
Footnotes
Supporting Information Available: Figures of molecular weight distribution subcellular location of identified proteins and transcripts in human skeletal muscle; table of proteins identified in human muscle biopsies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hojlund K, Beck-Nielsen H. Impaired glycogen synthase activity and mitochondrial dysfunction in skeletal muscle. Markers or mediators in type 2 diabetes. Curr. Diabetes Rev. 2006;2:375–395. doi: 10.2174/1573399810602040375. [DOI] [PubMed] [Google Scholar]
- 2.Krebs M, Roden M. Molecular mechanisms of lipid-induced insulin resistance in muscle, liver and vasculature. Diabetes Obes. Metab. 2005;7(6):621–632. doi: 10.1111/j.1463-1326.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- 3.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 4.Pirola L, Johnston AM, Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47(2):170–184. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 5.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49(5):677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 6.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol. Genomics. 2005;21(2):253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 7.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U.S.A. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 9.Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51(6):1913–1920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 10.Richardson DK, Kashyap S, Bajaj M, Cusi K, Mandarino SJ, Finlayson J, DeFronzo RA, Jenkinson CP, Mandarino LJ. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J. Biol. Chem. 2005;280(11):10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 11.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12(22):2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 12.Batt J, Bain J, Goncalves J, Michalski B, Plant P, Fahnestock M, Woodgett J. Differential gene expression profiling of short and long term denervated muscle. FASEB J. 2006;20(1):115–117. doi: 10.1096/fj.04-3640fje. [DOI] [PubMed] [Google Scholar]
- 13.Teran-Garcia M, Rankinen T, Koza RA, Rao DC, Bouchard C. Endurance training-induced changes in insulin sensitivity and gene expression. Am. J. Physiol. Endocrinol. Metab. 2005;288(6):E1168–E1178. doi: 10.1152/ajpendo.00467.2004. [DOI] [PubMed] [Google Scholar]
- 14.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18(3–4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 15.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta -subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003;278(12):10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 17.Hittel DS, Hathout Y, Hoffman EP, Houmard JA. Proteome analysis of skeletal muscle from obese and morbidly obese women. Diabetes. 2005;54(5):1283–1288. doi: 10.2337/diabetes.54.5.1283. [DOI] [PubMed] [Google Scholar]
- 18.Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P. The human muscle proteome in aging. J. Proteome Res. 2006;5(6):1344–1353. doi: 10.1021/pr050414x. [DOI] [PubMed] [Google Scholar]
- 19.Capitanio D, Vigano A, Ricci E, Cerretelli P, Wait R, Gelfi C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics. 2005;5(10):2577–2586. doi: 10.1002/pmic.200401183. [DOI] [PubMed] [Google Scholar]
- 20.Gelfi C, De Palma S, Cerretelli P, Begum S, Wait R. Two dimensional protein map of human vastus lateralis muscle. Electrophoresis. 2003;24(1–2):286–295. doi: 10.1002/elps.200390025. [DOI] [PubMed] [Google Scholar]
- 21.Gelfi C, De Palma S, Ripamonti M, Eberini I, Wait R, Bajracharya A, Marconi C, Schneider A, Hoppeler H, Cerretelli P. New aspects of altitude adaptation in Tibetans: a proteomic approach. FASEB J. 2004;18(3):612–614. doi: 10.1096/fj.03-1077fje. [DOI] [PubMed] [Google Scholar]
- 22.Adachi J, Kumar C, Zhang Y, Mann M. In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol. Cell. Proteomics. 2007;6(7):1257–1273. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Brockmann R, Beyer A, Heinisch JJ, Wilhelm T. Posttranscriptional expression regulation: what determines translation rates. PLoS Comput. Biol. 2007;3(3):e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YR, Juan HF, Huang HC, Huang HH, Lee YJ, Liao MY, Tseng CW, Lin LL, Chen JY, Wang MJ, Chen JH, Chen YJ. Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J. Proteome Res. 2006;5(10):2727–2742. doi: 10.1021/pr060212g. [DOI] [PubMed] [Google Scholar]
- 25.Greenbaum D, Jansen R, Gerstein M. Analysis of mRNA expression and protein abundance data: an approach for the comparison of the enrichment of features in the cellular population of proteins and transcripts. Bioinformatics. 2002;18(4):585–596. doi: 10.1093/bioinformatics/18.4.585. [DOI] [PubMed] [Google Scholar]
- 26.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 27.Nissom PM, Sanny A, Kok YJ, Hiang YT, Chuah SH, Shing TK, Lee YY, Wong KT, Hu WS, Sim MY, Philp R. Transcriptome and proteome profiling to understanding the biology of high productivity CHO cells. Mol. Biotechnol. 2006;34(2):125–140. doi: 10.1385/mb:34:2:125. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt MW, Houseman A, Ivanov AR, Wolf DA. Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol. Syst. Biol. 2007;3:79. doi: 10.1038/msb4100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unwin RD, Whetton AD. Systematic proteome and transcriptome analysis of stem cell populations. Cell Cycle. 2006;5(15):1587–1591. doi: 10.4161/cc.5.15.3101. [DOI] [PubMed] [Google Scholar]
- 30.Yin L, Tao Y, Zhao K, Shao J, Li X, Liu G, Liu S, Zhu L. Proteomic and transcriptomic analysis of rice mature seed-derived callus differentiation. Proteomics. 2007;7(5):755–768. doi: 10.1002/pmic.200600611. [DOI] [PubMed] [Google Scholar]
- 31.Joosen R, Cordewener J, Supena ED, Vorst O, Lammers M, Maliepaard C, Zeilmaker T, Miki B, America T, Custers J, Boutilier K. Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Physiol. 2007;144(1):155–12. doi: 10.1104/pp.107.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox B, Kislinger T, Emili A. Integrating gene and protein expression data: pattern analysis and profile mining. Methods. 2005;35(3):303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 33.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–440. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 34.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol. Cell. Proteomics. 2004;3(10):960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.McNicoll F, Drummelsmith J, Muller M, Madore E, Boilard N, Ouellette M, Papadopoulou B. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics. 2006;6(12):3567–3581. doi: 10.1002/pmic.200500853. [DOI] [PubMed] [Google Scholar]
- 36.Xun Z, Sowell RA, Kaufman TC, Clemmer DE. Protein expression in a Drosophila model of Parkinson's disease. J. Proteome Res. 2007;6(1):348–357. doi: 10.1021/pr060488o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP, Auer G, Ried T. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes, Chromosomes Cancer. 2007;46(1):10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- 38.Lorenz P, Ruschpler P, Koczan D, Stiehl P, Thiesen HJ. From transcriptome to proteome: differentially expressed proteins identified in synovial tissue of patients suffering from rheumatoid arthritis and osteoarthritis by an initial screen with a panel of 791 antibodies. Proteomics. 2003;3(6):991–1002. doi: 10.1002/pmic.200300412. [DOI] [PubMed] [Google Scholar]
- 39.Ruse CI, Tan FL, Kinter M, Bond M. Intregrated analysis of the human cardiac transcriptome, proteome and phosphoproteome. Proteomics. 2004;4(5):1505–1516. doi: 10.1002/pmic.200300682. [DOI] [PubMed] [Google Scholar]
- 40.Hojlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, Langlais P, Weintraub ST, Mandarino LJ. Characterization of the human skeletal muscle proteome by one-dimensional Gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell. Proteomics. 2008;7(2):257–267. doi: 10.1074/mcp.M700304-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 42.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J. Proteome Res. 2006;5(1):64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 45.Davison AC, Hinkley DV, Schechtman E. Efficient Bootstrap Simulation. Biometrika. 1986;73(3):555–566. [Google Scholar]
- 46.Davison AC, Hinkley DV, Young GA. Recent developments in bootstrap methodology. Statist. Sci. 2003;18(2):141–157. [Google Scholar]
- 47.Cavelier C, Lorenzi I, Rohrer L, von Eckardstein A. Lipid efflux by the ATP-binding cassette transporters ABCA1 and ABCG1. Biochim. Biophys. Acta. 2006;1761(7):655–666. doi: 10.1016/j.bbalip.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Dullens SP, Plat J, Mensink RP. Increasing apoA-I production as a target for CHD risk reduction. Nutr. Metab. Cardiovasc. Dis. 2007;17(8):616–528. doi: 10.1016/j.numecd.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20(11):1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 50.Urso ML, Scrimgeour AG, Chen YW, Thompson PD, Clarkson PM. Analysis of human skeletal muscle after 48 h immobilization reveals alterations in mRNA and protein for extracellular matrix components. J. Appl. Physiol. 2006;101(4):1136–1148. doi: 10.1152/japplphysiol.00180.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.