Advances in magnetic resonance imaging (MRI) have extended the application of this technique from routine clinical diagnosis to in vivo cellular imaging.1–3 Gd(III) complexes are commonly used in MR imaging to enhance local contrast. Gd(III) ions reduce the longitudinal relaxation time (T1) of surrounding water protons, rendering an increase in signal intensity in an appropriately weighted MR image.4,5
The inherent lack of sensitivity of Gd(III) contrast agents limits the application of MR for cellular imaging when compared to agents used for positron emissive tomography (PET)6 and light microscopy.7 A number of Gd(III) complexes (low and high molecular weights) have been reported for in vitro and in vivo applications.8–12 However, the majority of these agents have not been optimized with regard to ease of synthesis, biocompatibility, cellular translocation, and molecular rotational correlation time (τR), lifetime (τm), and the number of coordinated water molecules (q).
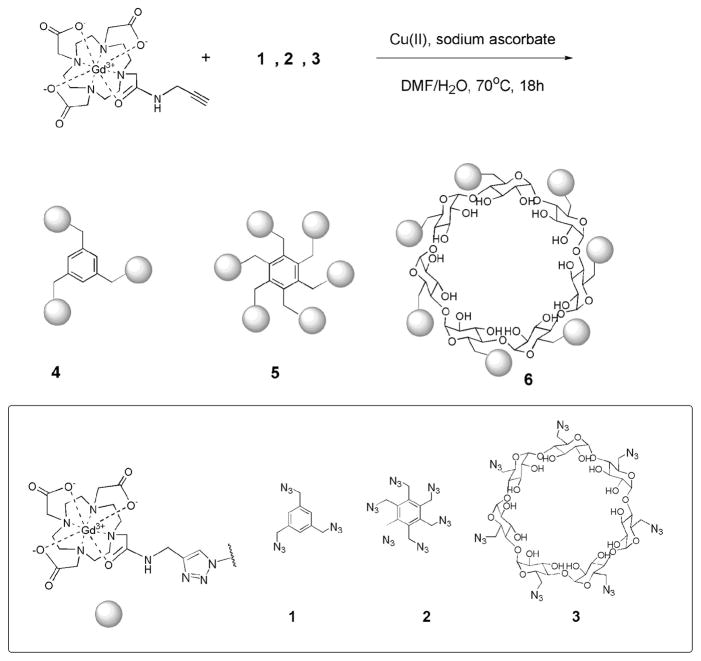
Here, we describe a facile and inexpensive approach to prepare three new low molecular weight, monodispersed MR contrast agents with multiple Gd(III) complexes. Click chemistry is employed to generate three pretemplated macromolecular architectures.13 According to the Solomon–Bloembergen–Morgan theory, elongation of τR will result in an increase in relaxivity.4 Moreover, the robust reaction between the azide substrate and the DOTA-Gd-alkyne derivative forms a triazole ring linkage (Figure 1). The rigid nature of the triazole linker hinders the local rotation of the Gd(III) complexes to further enhance the relaxivity.14,15 The complexes possess from three to seven Gd(III) chelates/molecule and exhibit excellent biocompatibility and high molecular relaxivities (Table 1).
Figure 1.
Synthetic scheme of multimeric MR contrast agents via click chemistry.
Table 1.
Gd(III) Ionic and Molecular Relaxivities of the New MRI Contrast Agents 4, 5, and 6 at 60 MHz, 37 °C
| compounds | MW (g/mol) | Gd ionic r1 (mM−1 s−1) | no. of Gd | molecular r1 (mM−1 s−1) |
|---|---|---|---|---|
| Gd-DOTA-alkyne derivative | 595.70 | 3.21 | 1 | 3.21 |
| 4 | 2030.34 | 5.90 ±0.15 | 3 | 17.7 ± 0.47 |
| 5 | 3982.57 | 10.97 ±0.13 | 6 | 65.8 ± 0.76 |
| 6 | 5480.01 | 12.20 ±0.54 | 7 | 85.4 ±3.74 |
β-Cyclodextrin (β-CD) has been shown to associate with DOTA-Gd and DTPA-Gd derivatives in a noncovalent fashion and form inclusion complexes.16 Each cyclodextrin molecule can host only one Gd(III) complex with an association constant between 103 and 104M.17 Covalent attachment of Gd(III) complexes to β-CD ensures the stability of the assembly, which is crucial in conducting long-term cell tracking.
The DOTA-Gd-alkyne derivative was synthesized by modification of existing procedures.18,19 Metalation of the DOTA-alkyne ligand was performed before the cycloaddition reaction because DOTA has a high affinity for Cu(II).20 The trismethylazido benzene, 1, hexakis(azidomethyl) benzene, 2, and heptakis-6-azido-6-deoxy- β-cyclodextrin, 3, were synthesized from corresponding commercially available halides in one step. These azide compounds are known to be stable at room temperature; however, they should be handled with extreme care due to the explosive nature of organic azides.21 The click chemistry was carried out in a water/dimethylformamide mixture with heating or under microwave irradiation (Figure 1). HPLC and Sephadex size exclusion chromatography were used to purify these complexes. High resolution MS and MALDI-MS characterization of complexes 4, 5, and 6 confirmed the formation of the products (see Table 1 and Supporting Information).
To evaluate the in vitro efficiency of these agents, T1 relaxivity was measured on a Bruker Minispec 60 MHz relaxometer at 37 °C in Millipore water using a standard inversion–recovery pulse sequence. The relaxivity of the Gd-DOTA-alkyne derivative was measured to be 3.21 mM−1 s−1 at 60 MHz, which is comparable to the reported value of DOTA-Gd (Dotarem, r1 = 2.7–3.1 mM−1 s−1 at 60 MHz, 37 °C).22
After conjugation with 1, 2, and 3, the measured relaxivities of the complexes increased dramatically and are summarized in Table 1. With an increase of the molecular weight and restricted rotation of the Gd(III) chelates, the relaxivity per Gd(III) increased 2–4-fold. The molecular relaxivities of the three new agents are significantly higher than those of commercially available agents.
MR phantoms were imaged using a 4.7 T Bruker Biospec MR Imager with increasing molecular concentrations of DOTA-Gd, 4, 5, and 6 in water as indicated in Figure 2A. T1 weighted spin–echo images were obtained.
Figure 2.
(A) T1 weighted spin–echo MR phantom images of DOTA-Gd, 4, 5, and 6 in Millipore water at equal molar concentration of each contrast agent. Images were acquired at 4.7 T at 25 °C (TR/TE = 300/18.4 ms). Concentrations (from left to right): 62, 51, 37, 21, 7, 0 μM. (B) T1 weighted spin–echo images of pelleted NIH-3T3 cells incubated with DOTA-Gd, 5, and 6 at approximately equal molar concentrations of Gd(III) (0, 7.60, 7.6, and 8.3 mM Gd(III), respectively). Images were acquired at 4.7 T at 25 °C (TR/TE = 500/12.5). Each sample diameter is 1 mm.
Cell labeling studies were carried out using 5 and 6 due to their high molecular relaxivities. NIH-3T3 cells were incubated with DOTA-Gd, 5, and 6 at approximately equal molar concentration of Gd(III). Normalizing the incubation concentration of the agents according to the Gd(III) ion was done to accurately demonstrate the efficacy of our new agents as cellular labels. Cells were incubated for 4 h at 37 °C and 5% CO2 in complete media containing DOTA-Gd, 5, or 6. The cells were rinsed three times in phosphate buffered saline and pelleted in 1 mm capillary tubes. Cell phantom images were acquired on the 4.7 T system described above with an unlabeled cell pellet as a control. The images clearly show that cells labeled with 5 and 6 display a brighter image than with DOTA-Gd at approximately the same incubation concentration of Gd(III) (Figure 2B).
A number of factors influence the observed signal intensity in cell imaging, including relaxivity, intracellular concentration, and subcellular localization.23 To evaluate the uptake of 5 and 6 in NIH-3T3 cells, MR images of cell pellets were acquired. The Gd(III) content in these cells was quantified by ICP-MS. Cells were incubated with increasing concentrations of Gd(III) using each contrast agent in complete media for 4 h at 37 °C and 5% CO2 (see Supporting Information). A linear concentration-dependent uptake profile was observed for all three agents. The concentrations of Gd(III) in cells labeled with each agent were slightly different but of the same order of magnitude. The uptake mechanism and the subcellular localization of the agents are under investigation.
Furthermore, the biocompatibility of DOTA-Gd, 5, and 6 was examined. Cellular viability assays indicated that a very high percentage of NIH-3T3 cells survived after incubation with DOTA-Gd, 5, or 6 (see Supporting Information). Future investigation will be carried out to determine the longevity of the label and any long-term effects on cellular function.
In summary, we have prepared three new MR contrast agents with varying numbers of Gd(III) complexes via click chemistry. This inexpensive and facile synthesis results in contrast agents with increased relaxivity and good biocompatibility. The agents are monodispersed and have relatively low molecular weights. We have demonstrated that these agents do not affect cellular viability in culture and are able to effectively label cells as observed in acquired MR images.
Supplementary Material
Acknowledgments
This research was supported by Center of Excellence in Translational Human Stem Cell Research, 1P50 NS054287-01 and NIH grant 1RO1 EB005866-01. MR Imaging was performed on instruments supported by NIH/NCRR Grant S10 RR15685-01.
Footnotes
Supporting Information Available: Experimental details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Voisin P, Ribot EJ, Miraux S, Bouzier-Sore AK, Lahitte JF, Bouchaud V, Mornet S, Thiaudiere E, Franconi JM, Raison L, Labrugere C, Delville MH. Bioconjugate Chem. 2007;18:1053–1063. doi: 10.1021/bc060269t. [DOI] [PubMed] [Google Scholar]
- 2.Frullano L, Meade TJ. J Biol Inorg Chem. 2007;12:939–949. doi: 10.1007/s00775-007-0265-3. [DOI] [PubMed] [Google Scholar]
- 3.Modo MM, Bulte JWW, editors. Molecular and Cellular MR Imaging. CRC Press; Boca Raton, FL: 2007. [Google Scholar]
- 4.Merbach AE, Toth E, editors. The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. 1. Wiley; New York: 2001. [Google Scholar]
- 5.Recent reports cite NSF toxicity exclusively in patients with renal failure with one class of DTPA Gd(III) chelate and has prompted studies validating the in vivo stability and safety of Gd(III) agents: Kurtkoti J, Snow T, Hiremagalur B. Nephrology. 2008;13:235–241. doi: 10.1111/j.1440-1797.2007.00912.x.Dillman JR, Ellis JH, Cohan RH, Strouse PJ, Jan SC. Am J Roentgenol. 2007;189:1533–1538. doi: 10.2214/AJR.07.2554.
- 6.Sharma V, Luker Gary D, Piwnica-Worms D. J Magn Reson Imaging. 2002;16:336–351. doi: 10.1002/jmri.10182. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 8.Crich SG, Biancone L, Cantaluppi V, Duo D, Esposito G, Russo S, Camusi G, Aime S. Magn Reson Med. 2004;51:938–944. doi: 10.1002/mrm.20072. [DOI] [PubMed] [Google Scholar]
- 9.Brekke C, Morgan SC, Lowe AS, Meade TJ, Price J, Williams SCR, Modo M. NMR Biomed. 2007;20:77–89. doi: 10.1002/nbm.1077. [DOI] [PubMed] [Google Scholar]
- 10.Anderson EA, Isaacman S, Peabody DS, Wang EY, Canary JW, Kirshenbaum K. Nano Lett. 2006;6:1160–1164. doi: 10.1021/nl060378g. [DOI] [PubMed] [Google Scholar]
- 11.Hooker JM, Datta A, Botta M, Raymond KN, Francis MB. Nano Lett. 2007;7:2207–2210. doi: 10.1021/nl070512c. [DOI] [PubMed] [Google Scholar]
- 12.Hueber MM, Staubli AB, Kustedjo K, Gray MHB, Shih J, Fraser SE, Jacobs RE, Meade TJ. Bioconjugate Chem. 1998;9:242–249. doi: 10.1021/bc970153k. [DOI] [PubMed] [Google Scholar]
- 13.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Greenfield MT, Spiller M, Mcmurry TJ, Lauffer RB, Caravan P. Angew Chem, Int Ed. 2005;44:6766–6769. doi: 10.1002/anie.200502245. [DOI] [PubMed] [Google Scholar]
- 15.Nicolle GM, Toth E, Schmitt-Willich H, Raduchel B, Merbach AE. Chem—Eur J. 2002;8:1040–1048. doi: 10.1002/1521-3765(20020301)8:5<1040::aid-chem1040>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 16.Barge A, Cravotto G, Robaldo B, Gianolio E, Aime S. J Inclusion Phenom Macrocycl Chem. 2007;57:489–495. [Google Scholar]
- 17.Aime S, Gianolio E, Terreno E, Menegotto I, Bracco C, Milone L, Cravotto G. Magn Reson Chem. 2003;41:800–805. [Google Scholar]
- 18.Prasuhn DE, Jr, Yeh RM, Obenaus A, Manchester M, Finn MG. Chem Commun. 2007;126:9–71. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 19.Viguier RFH, Hulme AN. J Am Chem Soc. 2006;128:11370–11371. doi: 10.1021/ja064232v. [DOI] [PubMed] [Google Scholar]
- 20.Smith SV. J Inorg Biochem. 2004;98:1874–1901. doi: 10.1016/j.jinorgbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert EE, Voreck WE. Propellants, Explos, Pyrotech. 1989;14:19–23. [Google Scholar]
- 22.Rohrer M, Bauer H, Mintorovitch J, Requardt M, Weinmann HJ. Invest Radiol. 2005;40:715–724. doi: 10.1097/01.rli.0000184756.66360.d3. [DOI] [PubMed] [Google Scholar]
- 23.Terreno E, Crich SG, Belfiore S, Biancone L, Cabella C, Esposito G, Manazza AD, Aime S. Magn Reson Med. 2006;55:491–497. doi: 10.1002/mrm.20793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.