Abstract
The N terminal region of hepatitis delta antigen (HDAg), referred to here as NdAg, has a nucleic acid chaperone activity that modulates the ribozyme activity of hepatitis delta virus (HDV) RNA and stimulates hammerhead ribozyme catalysis. We characterized the nucleic acid binding properties of NdAg, identified the structural and sequence domains important for nucleic acid binding, and studied the correlation between the nucleic acid binding ability and the nucleic acid chaperone activity. NdAg does not recognize the catalytic core of HDV ribozyme specifically. Instead, NdAg interacts with a variety of nucleic acids and has higher affinities to longer nucleic acids. The studies with RNA homopolymers reveal that the binding site size of NdAg is around nine nucleotides long. The extreme N terminal portion of NdAg, the following coiled-coil domain and the basic amino acid clusters in these regions are important for nucleic acid binding. The nucleic acid–NdAg complex is stabilized largely by electrostatic interactions. The formation of RNA–protein complex appears to be a prerequisite for facilitating hammerhead ribozyme catalysis of NdAg and its derivatives. Mutations that reduce the RNA binding activity or high ionic strength that destabilizes the RNA–protein complex, reduce the nucleic acid chaperone activity of NdAg.
INTRODUCTION
Hepatitis delta virus (HDV) contains a single-stranded circular RNA genome of 1.7 kb (1–3, 4–6 for reviews). Hepatitis delta antigen (HDAg), the only protein encoded by HDV, is essential for viral replication and virion assembly (7). HDAg is a basic protein, and binds in vitro to several regions of HDV RNA, which possesses a rod-like structure (8,9). This RNA binding property is required for various biological functions of HDAg, such as the nuclear cytoplasm shuttling of HDV RNA (10,11), the trans-activation activity on viral replication (12) and viral RNA packaging (13). HDV RNAs of genomic and antigenomic senses individually contain a ribozyme domain that requires adopting a pseudoknot-like structure to undergo self-cleavage (14–18). The ribozyme activity of each sense HDV RNA is essential for generating the 1.7 kb RNA during viral replication (19). Nevertheless, the active ribozyme structure exists only transiently during viral replication because of the highly self-complementary nature of the HDV RNA sequence. HDAg can enhance, though is not required for, the processing of multiple-unit-length HDV RNA in vivo. Conceivably, HDAg per se or together with other cellular factor modulates HDV ribozyme activity by affecting HDV RNA folding (20).
RNA chaperones refer to proteins that aid in the process of RNA folding by preventing misfolding or by resolving misfolded species (21). The RNA chaperone activities of a number of proteins that bind nucleic acid with broad specificity, such as the nucleocapsid protein of HIV, hnRNP A1 protein, Escherichia coli protein StpA and several ribosomal proteins, have been identified (22–25). In the test tube, HDAg has been shown to have an RNA chaperone activity (26,27). Several HDAg peptides are able to modulate the cis-cleaving activity of HDV RNA genome subfragments by altering RNA structure as well as stimulate the catalysis of hammerhead trans-acting ribozymes by promoting the annealing of ribozymes to the cognate substrates and by accelerating ribozyme turnover. The functional domain of HDAg resides in the N terminal region (26), which is rich in basic amino acids and contains the cryptic RNA binding domain (residues 2–27) (28), the coiled-coil domain (residues 12–60) (29), and the nuclear localization signal (residues 69–88) (30) of HDAg. The major RNA binding domain of HDAg (residues 97–146) (8,12), nevertheless, does not possess and is not required for the RNA chaperone activity in vitro (26). Moreover, synthetic complementary RNAs were also used to study the molecular basis of the stimulatory effect of the N terminal domain of HDAg on RNA structural rearrangement. The results demonstrate that the N terminal domain of HDAg could accelerate the annealing of complementary sequences in a selective fashion and promote strand exchange for the formation of a more extended duplex (27).
In this paper, we describe assays for studying the correlation between the nucleic acid binding ability and the RNA chaperone activity of HDAg peptides with HDV genome subfragments and a hammerhead trans-acting ribozyme and cognate substrate as the experimental materials. We investigate the role of the previously identified structural/functional domains of the N terminal region of HDAg on the nucleic acid–protein interaction. We also use less complicated DNA or RNA homopolymers to characterize the nucleic acid binding properties of the N terminal domain of HDAg in detail.
MATERIALS AND METHODS
Nucleic acids
HDV 726–770 and HJ12L [nt 683–770 (31)] are HDV genome fragments. mH2, ΔH1 and ΔH2 are mutants of HJ12L. mH2 is a mutant of HJ12L that contains three mismatched base pairs in the extended H2. ΔH1 and ΔH2 are deletion mutants, ΔH1 had the cleavage site and the 5′ strand of H1 deleted and ΔH2 had the 3′ strand of H2 deleted. KSR4 contains four copies of hammerhead ribozyme HH16, and PRP19S contains a copy of the target of HH16 as described by Huang and Wu (26).
Most of the RNAs in this study were synthesized by phage polymerases (Promega) using synthetic DNA (for HH16), a polymerase chain reaction amplified DNA (for ΔH1, ΔH2, mH2 and HDV 726–770), or a linearized plasmid DNA as the template (for HJ12L, KSR4 and PRP19S). The in vitro transcribed RNAs were labeled by [α-32P]CTP (3000 Ci/mmol, NEN). Poly rU and poly rA of heterogeneous sizes were purchased from Amersham Pharmacia Biotech. The RNA fragment S, dT homopolymers of various sizes and dA36 were made by solid phase chemical synthesis. The RNA fragment S and nucleic acid homopolymers were 5′-end labeled by T4 polynucleotide kinase (Promega) and [γ-32P]ATP (6000 Ci/mmol, NEN). The 32P-labeled nucleic acids were purified by denaturing polyacrylamide gel electrophoresis, resuspended in TE buffer [10 mM Tris–HCl (pH 8.0) and 0.1 mM EDTA], and stored at –20°C. The concentration of the 32P-labeled nucleic acid was calculated from its radioactivity and the specific activity of the labeled nucleotide. Prior to being used in ribozyme cleavage reactions, mobility-shift analyses and binding assays, each purified nucleic acid was denatured at 95–100°C for 2–3 min and cooled to room temperature for at least 5 min before use.
HDAg peptides
The cDNAs of HDAg peptides were amplified by polymerase chain reactions with synthetic oligonucleotides as primers. The polymerase chain reaction products were cloned to the NdeI and BamHI sites of vector pET15b (Novagen). The insertion mutant and the substitution mutants of NdAg were constructed by site-directed mutagenesis. The sequence of each recombinant clone was determined by DNA sequencing. The HDAg peptides were expressed in E.coli BL21 (DE3) cells. The proteins were purified by nickel column chromatography or by phosphocellulose column chromatography as previously described (26). Fractions containing the HDAg peptide were snap frozen in liquid nitrogen and stored at –70°C. The concentration of each protein was determined by ninhydrin assay. Prior to being used in cleavage reactions, binding assays or mobility shift assays, the purified protein was diluted with protein dilution buffer [50 mM HEPES–NaOH (pH 7.9), 1 M NaCl, 1 mM EDTA and 20% (v/v) glycerol] to the desired concentrations and used as 10× stock.
Mobility-shift analyses
To analyze the formation of RNA–HDAg peptide complexes, cis-cleavage reactions of HJ12L in the presence of different HDAg peptides in a total volume of 10 µl were carried out at 37°C for 60 min. After the addition of 2 µl of 25% Ficoll, the resulting complexes were resolved at 4°C on a native 10% polyacrylamide (30:1 acrylamide–bisacrylamide)–1× TBE (89 mM Tris–borate, 1 mM EDTA) gel and were visualized by autoradiogram. Alternatively, the purified HJ12L or the 3′-cleavage product of HJ12L was incubated with different HDAg peptides in 40 mM Tris–HCl (pH 7.5), 0.1 M NaCl, 0.02 mM EDTA and 2% glycerol in a total volume of 10 µl at 37°C for 60 min before gel electrophoresis.
Filter binding assay
The affinity of HDAg peptides to various nucleic acids was examined by the double-filter binding assay (32,33). The RNP complexes were generated by incubating a fixed concentration of 32P-labeled nucleic acid with increasing concentrations of HDAg peptide (0–10 µM) for 30 min at 37°C in the standard binding buffer [40 mM Tris–HCl (pH 7.5), 0.1 M NaCl, 0.02 mM EDTA, 2% glycerol and 40 µg/ml bovine serum albumin] in a total volume of 50 µl. The reaction was filtered through a BA 85 nitrocellulose membrane (Schleicher and Schuell) overlaid on a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) in a 60-well slot blot apparatus (Schleicher and Schuell). The filters were pre-soaked in Tris–NaCl buffer [40 mM Tris–HCl (pH 7.5) and 0.1 M NaCl] for at least 5 min, and the slots were washed with 100 µl of Tris–NaCl buffer before and after the reaction was filtered. The RNP complexes were retained on a nitrocellulose membrane and the unbound RNA was trapped by the Hybond N+ nylon membrane. Removal of the bovine serum albumin from the standard binding buffer significantly increased the background binding of the labeled RNA and caused variable retention efficiencies in the plateau region of the binding curve (data not shown), but the binding curve was not altered by increasing the bovine serum albumin concentration from 40 to 80 µg/ml (data not shown). The radioactivity on both filters was quantitated using a PhosphorImager (Molecular Dynamics), and the fraction of input RNA bound was calculated. A binding curve was plotted as the fraction of input RNA bound as a function of HDAg peptide concentration. Typically, background binding in the absence of HDAg peptide is less than 2%, and the binding curve reaches a plateau at which near 100% of the input RNA is bound. The total HDAg peptide at the mid-point of the binding curve was defined as [HDAg peptide]1/2.
Ribozyme reactions
All cleavage reactions were carried out in TMN buffer [40 mM Tris–HCl (pH 7.5), 12 mM MgCl2, 0.1 M NaCl, 0.02 mM EDTA and 2% glycerol] in which NaCl, EDTA and glycerol were from the HDAg peptide stock, and MgCl2 was used to initiate the cleavage reaction. The purified HDV ribozymes and HDV RNA fragments were pre-incubated for 30 min at 37°C with or without an HDAg peptide in a total volume of 9 µl. Following pre-incubation, 1 µl of MgCl2 was added to a final concentration of 12 mM to initiate the cis-cleavage reaction and incubation was continued for various time periods at 37°C. After terminating the cis-cleavage reaction by addition of 10 µl of stop solution [50 mM EDTA (pH 8.0), 7 M urea, 0.005% xylene cyanol and 0.005% Bromophenol Blue], the precursor RNA and the cleavage products were resolved on a denaturing polyacrylamide gel and visualized by autoradiography. The radioactivity of each RNA was quantitated using a PhosphorImager (Molecular Dynamics), and the molar ratio of the uncleaved precursor RNA at each time point was calculated.
For the single turnover trans-cleavage reaction of hammerhead ribozyme, 0.5 nM of PRP19S and 5 nM of KSR4 were pre-incubated with or without an HDAg peptide for <0.5 min at 25°C (for S/HH16) or for 20 min at 37°C (for PRP19S/KSR4). The trans-cleavage reaction was initiated by the addition of MgCl2 and incubation was continued for various time periods at 37°C. After the reaction was terminated, the substrate, ribozyme and cleavage products in the reaction mixture were resolved on denaturing polyacrylamide gels (25% for S/HH16 and 3.5% for PRP19S/KSR4). The radioactivity of each RNA was determined, and the fraction of uncleaved substrate at each time point was calculated.
Circular dichroism
Circular dichroism (CD) spectra were measured on a Jasco J-720 spectropolarimeter equipped with a water bath (Neslab RTE-211) for controlling the temperature of the cell. Constant N2 flushing was employed. Ellipticity is reported as mean residue ellipticity ([θ]) in deg cm2 dmol–1 and calculated from the equation [θ] = [θ]obs (mrw)/10lc, where [θ]obs is the ellipticity measured in degrees, mrw is the mean residue weight, c is the peptide concentration (g ml–1) and l is the optical path length of the cell (cm). CD spectra were obtained by collecting data at 0.5 nm intervals from 245 to 195 nm using a quartz cuvette with 0.1 cm path length at 37°C. Ten scans were obtained to average each spectrum. Buffer blank was run for each set of CD spectra, the data were averaged and smoothed for baseline correction using JEES program (Jasco).
RESULTS
Interaction of HDAg truncated proteins and HDV ribozyme HJ12L
We investigated the interaction between HDAg truncated proteins and an HDV ribozyme. HDAg truncated proteins were overexpressed in and were purified from E.coli as His-tagged fusion proteins as described (26). SdAg contains the full-length small delta antigen; NMdAg contains the first 143 amino acids, NdAg contains the N terminal domain (aa 1–88), MdAg contains the middle one-third (aa 89–143) and CdAg contains the C terminal domain (aa 89–195) of HDAg; and NdAg-tag contained the first 88 amino acids as that of NdAg but without the His-tag (Fig. 1A and B). HJ12L contains the ribozyme domain of the HDV genome (nt 683–770) (Fig. 2A). HJ12L could adopt the catalytically active pseudoknot-like structure and underwent cis-cleavage in the absence of protein factor in vitro (data not shown). The pre-mixing of HJ12L with SdAg, NMdAg, NdAg or NdAg-tag prior to the initiation of cis-cleavage by the addition of MgCl2 caused the attenuation of the cis-cleaving activity of HJ12L, while the cis-cleavage reaction of HJ12L was not affected at all by the same or even higher concentration of MdAg or CdAg (data not shown).
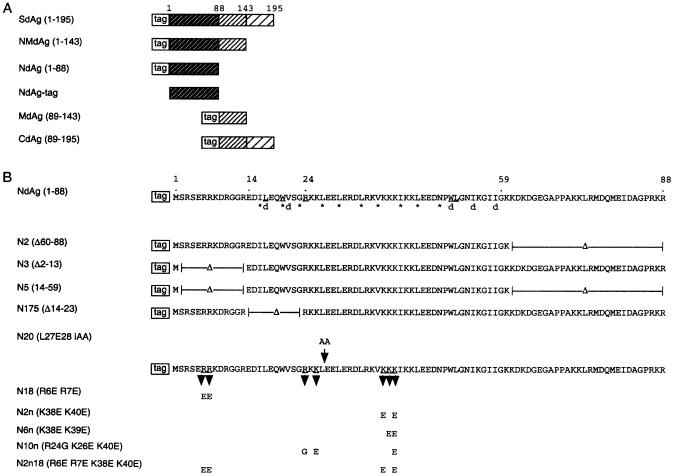
Figure 1.
(A) Schematic diagram of the HDAg truncated proteins. 1, 88, 143 and 195 correspond to the number of amino acids of HDAg. The tag has the sequence of MGSSH HHHHH SSGLV PRGSH. (B) NdAg and its mutants. Residues marked with an asterisk make up the a and d positions in the heptad repeat in the coiled-coil domain. The residues marked with ‘d’ are involved in the dimer–dimer interface in the crystal structure of peptide δ12–60 (Y) that contains residues 12–60 of HDAg (29). The underlined residues participate in the hydrophobic interaction in the dimer. N2, N3 and N5 are truncated mutants; N175 is an internal deletion mutant; N20 has two alanine residues inserted to Leu27 and Glu28; and N18, N2n, N6n, N10n and N2n18 have combinations of basic amino acids substituted.
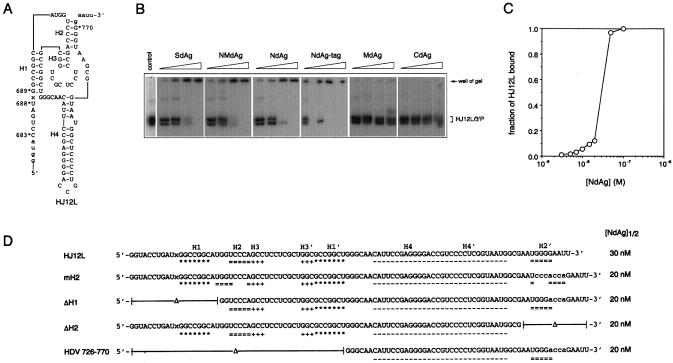
Figure 2.
Interaction of HDV ribozymes and HDAg truncated proteins. (A) The sequence and proposed secondary structure of HDV ribozyme HJ12L. HJ12L contains nt 683–770 of the HDV genome. H1–H4 are the four helical regions of HDV ribozyme, and ‘x’ is the cleaving point. The sequence in the lower case letters was derived from the cloning vector during in vitro transcription. (B) The mobility shift assay: 3.5 nM of HJ12L was incubated without any protein (control), or with 0.375, 0.75, 1.5 or 3 ng/µl of each HDAg truncated protein for 30 min before the cis-cleavage reaction was initiated. The cis-cleavage reaction was carried out at 37°C for 60 min, and the reaction mixtures were separated on a non-denaturing 10% polyacrylamide gel. The free HJ12L and its 3′-cleavage product are indicated on the right. (C) The filter binding assay. The NdAg binding curve of 3.5 nM HJ12L in standard binding buffer at 37°C is shown. (D) HJ12L mutants and their interaction with NdAg. Nucleotides corresponding to the four helical regions of each ribozyme molecule, i.e. H1–H4 and H1′–H4′, are marked; and ‘x’ is the cleaving point. The sequences in lower case letters are the substituted residues. The filter binding assay was carried out with 3.5 nM RNA at 37°C and the total NdAg concentration at the midpoint of the binding curve of each RNA defined as [NdAg]1/2 is summarized.
We analyzed the binding of each HDAg truncated protein to the RNAs in the cis-cleavage reaction by electrophoresis mobility shift assay. In the absence of protein, ∼50% of the HJ12L RNA underwent cis-cleavage (data not shown), and the HJ12L RNA and its 3′ cleavage product (HJ12L3′P) migrate as two distinct bands on the native polyacrylamide gel (Fig. 2B, lane 1), while each band contained both RNA species when analyzed by a denaturing polyacrylamide gel (data not shown). Each HDAg truncated protein that inhibited the ribozyme activity of HJ12L formed a complex with the RNAs in the cis-cleavage reaction, and the RNA–protein complex that did not migrate from the well of the gel appears to be very large. However, neither MdAg nor CdAg that did not interfere with the cis-cleavage of HJ12L shifted HJ12L or HJ12L3′P under the same conditions (Fig. 2B).
Furthermore, we used a filter-binding assay to examine the binding of each HDAg truncated protein to HJ12L. The standard buffer for the filter-binding assay did not contain any divalent cation, therefore HJ12L could not undergo cis-cleavage. The NdAg binding isotherm of HJ12L shows that the formation of RNA–protein complex for 3.5 nM HJ12L was detected at ∼5 nM NdAg, had a midpoint at ∼30 nM NdAg, and reached a plateau at 50 nM NdAg (Fig. 2C). There was little difference between the HJ12L binding curves of NdAg and NdAg-tag, while the formation of the HJ12L–protein complexes was not detectable with up to 1 µM MdAg or CdAg under the same conditions (data not shown). Purified HJ12L3′P interacted with these HDAg truncated proteins in a manner indistinguishable from that of HJ12L (data not shown). The findings derived from the filter-binding assay were quite similar to those of the mobility shift assay. However, probably due to the dissociation of the RNA–protein complexes during gel electrophoresis, the complexes could be detected at lower protein concentrations using the filter-binding assay.
These results show that the N terminal region of HDAg could interact with the HDV ribozyme and attenuate its cis-cleaving activity. In contrast, neither the His-tag of the fusion protein nor the previously identified HDV RNA binding domain that resides in aa 89–143 of HDAg are required for these activities.
NdAg interacts with HDV ribozyme specifically?
The ability of NdAg to interact with HJ12L and inhibit HJ12L cis-cleavage could suggest that either NdAg specifically binds to the catalytic core of the HDV ribozyme and blocks cis-cleavage or, alternatively, that NdAg alters the overall structure of the HDV ribozyme and attenuates the cis-cleaving activity. Assuming that NdAg can specifically recognize the catalytic core of HDV ribozyme, active and defective ribozyme mutants may bind NdAg with distinct affinities because HDV ribozyme requires a specific structure for autocatalytic cis-cleavage. To test this possibility, a variety of HDV ribozyme mutants were made (Fig. 2D) and their NdAg binding affinities were determined by the filter-binding assay. mH2, ΔH1, ΔH2 and HDV 726–770 had at least one of the essential structural/sequence elements of the HDV ribozyme disrupted or deleted, and none of them possessed cis-cleaving activity (data not shown). The binding studies show that each of these RNAs bound NdAg in a manner similar to that of HJ12L (Fig. 2D) and NdAg did not activate any of the defective mutants (data not shown). These results show that NdAg interacts with HDV RNA of various length, sequence and structure. Therefore, NdAg is not likely to recognize the catalytic core of the HDV ribozyme specifically.
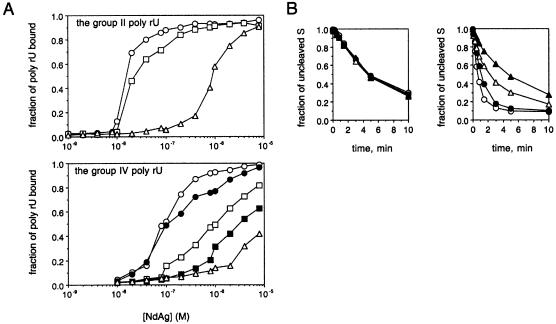
Interaction of NdAg with hammerhead ribozyme and cognate substrate
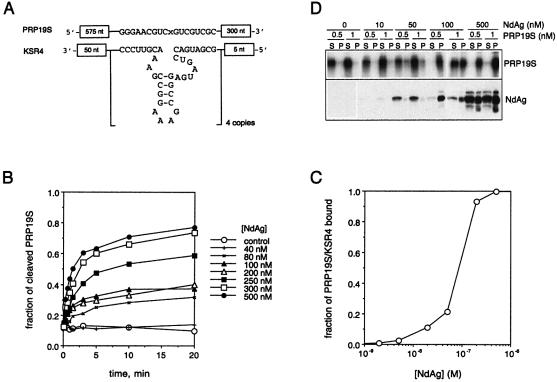
NdAg is able to facilitate the trans-cleavage reactions between hammerhead ribozymes and cognate substrates in vitro by acting as an RNA chaperone (26,27), indicating NdAg interacts with non-HDV RNA as well. Here we used the KSR4/PRP19S hammerhead ribozyme system to investigate the correlation between the stimulatory activity of NdAg on the cleavage of PRP19S (the substrate) by KSR4 (the ribozyme) and the ability of NdAg to bind the ribozyme and substrate RNAs. KSR4 contains four copies of hammerhead ribozyme HH16 and PRP19S contains a copy of the target of HH16 (Fig. 3A). The cleavage of 0.5 nM PRP19S by 5 nM KSR4 was promoted by 80 nM and higher concentrations of NdAg: after pre-incubating with sufficiently high concentrations of NdAg, PRP19S was cleaved rapidly once the trans-cleavage reaction was initiated, and the extent of cleavage approached a plateau value when the trans-cleavage reaction was allowed to continue for more than 5 min (Fig. 3B). However, only a fraction of the PRP19S molecules was cleaved; the value of the extent of cleavage at plateau increased as the NdAg concentration increased, and the extent of cleavage reached the maximum value at 300–500 nM NdAg. To obtain information on the binding of NdAg to the RNAs in the trans-cleavage reaction, we performed a filter-binding assay. As illustrated in Figure 3C, the formation of RNA–NdAg complex(es) was detected at ∼10 nM NdAg, had a midpoint at ∼100 nM NdAg, and reached a plateau at ≥300 nM NdAg, which correlates well with the stimulatory effect of different concentrations of NdAg on the trans-cleavage reaction. These results imply that the formation of RNA–NdAg complex(es) is a prerequisite for activating ribozyme catalysis although not all of the RNAs in the RNP–complex(es) participate in the trans-cleavage reaction, and that there is no turnover in the stimulatory activity of NdAg probably due to the strength of the RNA–protein interaction. Furthermore, we found that the PRP19S RNA as well as other RNAs, which are soluble at concentrations of 0.5 and 1 nM, could be induced to precipitate by 100 and 500 nM NdAg although ∼50% of NdAg aggregated at these concentrations (Fig. 3D and data not shown). Therefore, NdAg and RNA can form a coaggregate.
Figure 3.
Interaction of NdAg with hammerhead ribozyme KSR4 and cognate substrate PRP19S. (A) Schematic diagrams of KSR4 and PRP19S RNAs. KSR4 contains four copies of the hammerhead ribozyme HH16, and PRP19S contains a copy of the HH16 target S. Only the sequences of HH16 and S are shown, and ‘x’ is the cleaving point. (B) The cleavage of PRP19S (0.5 nM) by KSR4 (5 nM) stimulated by NdAg. Two RNAs were incubated with the indicated concentration of NdAg at 37°C for 30 min before the trans-cleavage reaction was initiated by the addition of MgCl2. The fraction of PRP19S that underwent cleavage is plotted against the reaction time. (C) The NdAg binding curve for PRP19S (0.5 nM) and KSR4 (5 nM) in standard binding buffer at 37°C. (D) The coaggregation of RNA with NdAg. A 25 µl reaction mixture in standard binding buffer containing 0.5 or 1 nM of PRP19S and the indicated concentration of NdAg was incubated at 37°C for 10 min in a 1.5 ml tube. The samples were centrifuged at 14 000 r.p.m. for 2 min, and the supernatant and the pellet were separated. The RNA in each fraction was resolved on a 3.5% polyacrylamide–7 M urea gel and analyzed by autoradiography. The protein in each fraction was resolved by 15% SDS–PAGE, and NdAg was detected by western blotting with an antiserum raised against NdAg.
Analysis of NdAg mutants
NdAg is rich in basic amino acids, and it contains the cryptic RNA binding domain, the coiled-coil domain, and the nuclear localization signal of HDAg. Here we investigated the importance of the individual structural/functional domain and certain basic amino acid clusters of NdAg to the RNA binding activity and to the stimulatory effect on hammerhead ribozyme catalysis. Various NdAg mutants (Fig. 1B) were expressed as His-tag fusion proteins and each protein was purified to >95% homogeneous for this study.
Mutant N2 had residues 60–88 deleted from NdAg, which contains the nuclear localization signal; mutant N3 had residues 2–13 deleted from NdAg, which contains the cryptic RNA binding domain; and mutant N5 contained residues 14–59 of NdAg only, which contain the coiled-coil domain. All three truncated mutants could adopt the α-helical structure as that of NdAg (Fig. 4A, and the data of N3 was not shown). N2 bound RNA with an affinity similar to that of NdAg, whereas N3 and N5 bound RNA less tightly than NdAg (Fig. 4B). These results suggest that the coiled-coil domain of NdAg by itself is not sufficient for the high affinity of RNA binding, and that although the cryptic RNA binding domain and the nuclear localization signal of NdAg are both rich in basic amino acid, the former rather than the latter may directly participate in RNA binding.
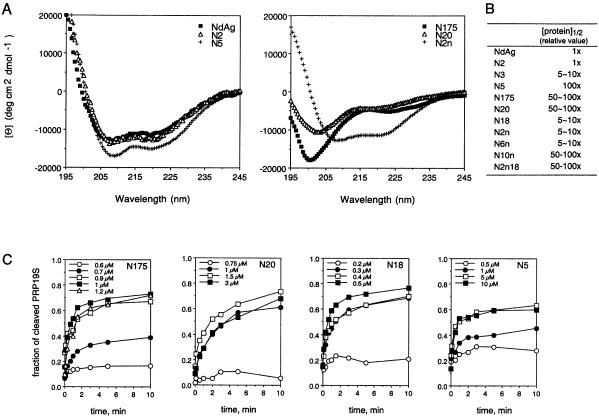
Figure 4.
The analysis of NdAg mutants. (A) Circular dichroism (CD) spectrum. Spectrum of each protein was taken with 0.6 µM of protein in 10 mM potassium phosphate (pH 7.6) and 0.1 M KF. The spectra of NdAg, N2, N5 and N2n exhibited double minima at 208 and 222 nm indicating both proteins have α-helical structures. (B) The RNA binding affinity. The filter binding assay was carried out in the presence of 0.5 nM PRP19S, 5 nM KSR4 and 0–10 µM of protein in standard binding buffer at 37°C for 30 min. The concentration of the input protein at the midpoint of the binding curve defined as [protein]1/2 was determined. The [protein]1/2 value was used to present the relative RNA binding affinity of each protein, and the smaller the value, the higher the RNA binding affinity. The [protein]1/2 value of NdAg that was determined in the experiment shown in (C) was standardized to 1. (C) The stimulatory effect on the cleavage of PRP19S (0.5 nM) by KSR4 (5 nM). Two RNAs were incubated with the indicated concentration of NdAg mutant at 37°C for 30 min before the trans-cleavage reaction was initiated by the addition of MgCl2. The fraction of PRP19S that underwent cleavage is plotted against the reaction time.
Mutant N20 had two alanine residues inserted between residues 27 and 28 of NdAg that reside in the coiled-coil domain, and mutant N175 had the residues 14–23 of NdAg deleted (Fig. 1B), which are important for stabilizing the α-helical structure and for the formation of a coiled-coil dimer of HDAg peptide δ12–60 (29,34). The alteration in each variant indeed caused the loss of the α-helical structure (Fig. 4A). In addition, both proteins bound RNA with an affinity significantly lower than that of NdAg (Fig. 4B). Since N20 and N175 contain as many basic residues as that of NdAg, their low RNA binding affinity indicates that certain structure in addition to the high density-positive charges is important for the high affinity of RNA binding.
To further analyze the importance of the basic amino acid clusters of the cryptic RNA binding domain and the coiled-coil domain of NdAg to the RNA binding activity, we prepared mutants of NdAg in which combinations of two to four of the arginine and lysine residues were replaced by non-basic amino acids (mutants N18, N2n, N6n, N10n and N2n18, Fig. 1B). We found that all substitution mutants could adopt the α-helical structure, and each mutant bound RNA with an apparent affinity lower than that of NdAg (Fig. 4B). Thus, the basic amino acids near the N terminus and in the coiled-coil domain are important for the RNA–NdAg interaction.
Furthermore, we analyzed the stimulatory effect of each NdAg variant on the trans-cleavage of PRP19S by KSR4. We found that each mutant protein stimulated the trans-cleavage reaction although higher concentrations of the weaker RNA binders were required to obtain the maximum stimulatory effect on the trans-cleavage reaction (some of the data are shown in Fig. 4C). These findings are consistent with the speculation that the RNA binding ability and the stimulatory activity of the HDAg peptide are correlated.
NdAg binds different nucleic acids with various affinities
The highly basic nature of NdAg suggests it may bind nucleic acids with broad specificity, and our studies indeed illustrate that NdAg can interact with RNAs of various sequences and structures. Here, we performed competition experiments to examine the interaction of NdAg with other nucleic acids. About 0.05 ng/µl (or 0.5 nM) labeled KSR4 (∼220 nt long) and a near saturating concentration of NdAg (10 nM) were mixed with various unlabeled competitor nucleic acids at concentrations up to 500 ng/µl; after equilibration, the reactions were filtered. This method of competition using unlabeled nucleic acids results in a reduction in the amount of KSR4 retained on the filter as the concentration of the competitor is increased. The amount of KSR4–NdAg complexes was reduced by ∼50% at 0.05 ng/µl of COS-1 cell RNA or E.coli cell RNA. However, more than 1 ng/µl of plasmid DNA or salmon sperm DNA, and more than 10 ng/µl of yeast tRNA, were required to reduce the amount of KSR4–NdAg complexes to the same level (Table 1; the data for E.coli cell RNA and salmon sperm DNA are not shown). Competition assays with 0.05 ng/µl of HDV 654–965 RNA (∼300 nt long) or PRP19S (∼900 nt long) as the labeled nucleic acid gave similar results (data not shown). These findings confirm that NdAg binds a variety of nucleic acids, and show that the affinity of NdAg to different nucleic acids follows the order of long RNA > long double strand DNA > the highly structured tRNA.
Table 1. Summary of the competition experiment.
| Competitor (ng/µl) | θ (Fc/Fo) | ||
|---|---|---|---|
| COS-1 cell RNA | Plasmid DNA | Yeast tRNA | |
| 0 | 100% | 100% | 100% |
| 0.01 | 90–95% | 90–95% | n.d. |
| 0.05 | 40–45% | 90–95% | n.d. |
| 0.1 | 10–15% | 80–85% | n.d. |
| 1 | <5% | 80–85% | 100% |
| 10 | <5% | 25–30% | 100% |
| 50 | <5% | 5–10% | 30–40% |
| 100 | <5% | <5% | 25–30% |
| 500 | n.d. | n.d. | <5% |
The competition experiment was carried out in standard binding buffer at 37°C for 30 min with 10 nM NdAg, 0.5 nM labeled KSR4 (∼0.05 ng/µl), and 0–500 ng/µl of various nucleic acids as the competitor. θ is the ratio of Fc and Fo which represent the fraction of labeled KSR4 retained on nitrocellulose filter in the presence of a competitor and in the absence of a competitor, respectively. The value of Fo ranged from 0.5 to 0.8 among different experiments. n.d., not determined.
Interaction of NdAg with less complicated nucleic acids
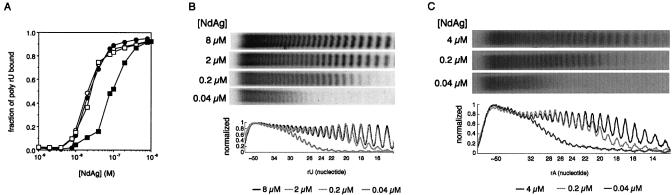
We then used less complicated nucleic acid molecules to characterize the nucleic acid binding property of NdAg. We first studied the interaction between NdAg and rU or rA homopolymer of various sizes. Poly rU or poly rA of heterogeneous sizes were 5′ end labeled, and the rU or rA homopolymers were purified as groups according to size. Three groups of poly rU greater than 90 nt long bound NdAg with similar affinities, and all or no binding occurred within a narrow protein concentration range suggesting NdAg binding is a cooperative process (groups I–III, Fig. 5A). The group of 55∼15 nt-poly rU had lower affinity to NdAg, and the titration curve was broader than that of the longer poly rUs (group IV, Fig. 5A). The group IV poly rU–NdAg complex was eluted from the filter, the NdAg was removed and the RNA was isolated and resolved on a high resolution denaturing polyacrylamide gel. We analyzed the composition of the rU homopolymers bound at different NdAg concentrations and quantified the amount of poly rU with different sizes (Fig. 5B). We found that at saturated NdAg concentrations (2–10 µM), different sizes of poly rU were bound almost equally. However, at the sub-saturating NdAg concentrations (0.02– 1 µM), long poly rUs were bound more efficiently than short poly rUs. In the case of 0.04 µM NdAg, the >32 nt-poly rUs were completely bound; the binding efficiency of the 32∼ 24 nt-poly rUs was reduced progressively as the size of poly rU decreased; and the 23∼15 nt-poly rUs were not bound at all. In the case of 0.2 µM NdAg, the >23 nt-poly rUs were completely bound; and the 23∼15 nt-poly rUs were bound with progressively lower efficiencies as the RNA became shorter. These results show that NdAg binds poly rU cooperatively and the apparent binding affinity increases dramatically as the size of rU homopolymer increases by ∼9 nt, which suggests the binding site size of NdAg is around 9 nt with poly rU as the template. These binding properties of NdAg are not unique to poly rU since the studies with rA homopolymer of 55∼15 nt long gave similar results (Fig. 5C).
Figure 5.
Interaction of NdAg with rU homopolymer of various sizes. (A) The NdAg binding curves for four groups of poly rU. The binding curve was obtained with 0.01 nM of poly rU in standard binding buffer at 37°C. The sizes of groups I (open circle), II (filled circle) and III poly rU (open square) were all greater than 90 nt long, and the size range of the group IV poly rU (filled square) was 55∼15 nt. (B) The analysis of the composition of the rU homopolymer of group IV bound at various NdAg concentrations. The bound poly rU was isolated from the filter and resolved on a 20% polyacrylamide–7 M urea gel, and dT homopolymers of various sizes were the size markers. The intensity of each rU homopolymer on the X-ray film was quantitated with Imagegate software. (C) The analysis of the composition of the rA homopolymer of 55∼15 nt long bound at various NdAg concentrations.
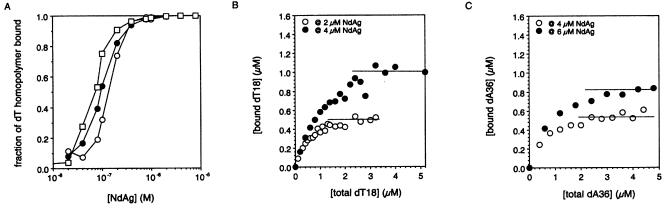
DNA homopolymers of defined size were used for the NdAg binding study as well. Unexpected high background binding to the nitrocellulose filter was observed for dT homopolymers greater than 20 nt long (data not shown), while shorter dT homopolymers did not have this problem. The titration curves of dT18, dT16 and dT14 shown in Figure 6A indicate that the NdAg binding affinity decreased as the dT homopolymer became shorter. Consistently, NdAg bound dT12, dT10 and dT8 with progressively lower affinities, and NdAg did not bind dT6 at all at 5 µM under the same conditions (data not shown).
Figure 6.
Interaction of NdAg to DNA homopolymers. (A) The NdAg binding curves for dT14 (open circle), dT16 (filled circle) and dT18 (open square) (0.01 nM of each) in standard binding buffer at 37°C. (B) Stoichiometry of NdAg binding to dT18; 2 or 4 µM of NdAg was incubated with varying concentrations of dT18 in standard binding buffer at 37°C for 30 min. The bound dT18 plotted against the total dT18 of one experiment is shown here, and the dashed line indicates the level off point of bound dT18 for two NdAg concentrations. The data obtained from 10 independent experiments gave the mean level off values of 0.94 (±0.14) µM of bound dT18 for 4 µM NdAg and 0.45 (±0.05) µM of bound dT18 for 2 µM of NdAg. (C) Stoichiometry of NdAg binding to dA36; 4 or 6 µM of NdAg was incubated with varying concentrations of dA36 in standard binding buffer at 37°C for 30 min. The bound dA36 plotted against the total dA36 is shown and the level off value of bound dA36 was 0.51 (±0.06) µM and 0.81 (±0.04) µM for 4 and 6 µM of NdAg, respectively. The dashed line indicates the level off point of bound dA36.
We tried to determine the stoichiometry of nucleic acid–NdAg complex by titrating excess NdAg, i.e. greater than the apparent Kd, with increasing amounts of nucleic acid homopolymer. We used dT18 and dA36 for this study because they both bound NdAg with sufficiently high affinities and gave reliable stoichiometric data. For both DNA homopolymers, the bound DNA homopolymer increased until a titration point was achieved and leveled off thereafter (Fig. 6B and C). The mean concentration of bound dT18 at level off points was 0.94 (±0.14) µM for 4 µM NdAg and 0.45 (±0.05) µM for 2 µM NdAg. The mean concentration of bound dA36 at level off points was 0.81 (±0.04) µM for 6 µM NdAg and 0.51 (±0.06) µM for 4 µM NdAg. This corresponds to an approximate 1:4 ratio of dT18 to NdAg and an approximate 1:8 ratio of dA36 to NdAg if all DNA homopolymers and NdAg are active in binding. Assuming that the binding site size of NdAg is 9 nt and NdAg binds nucleic acid as coiled-coil dimer, NdAg may bind dT18 as two dimers and bind dA36 as four dimers.
The formation and the dissociation of nucleic acid–NdAg complex
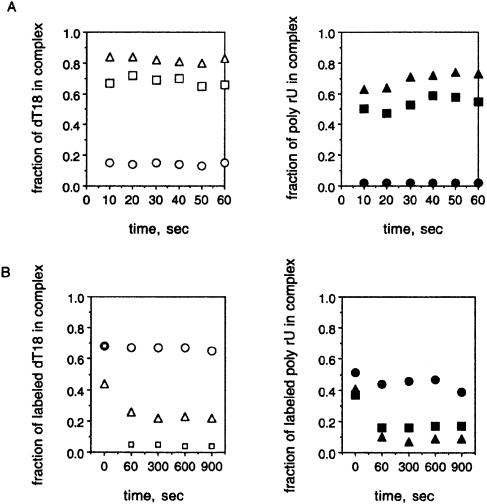
To assess the kinetics of nucleic acid–NdAg complexes formation, we mixed labeled nucleic acid with different concentrations of NdAg, and withdrew and filtered aliquots through nitrocellulose filters after very short intervals. Results obtained with dT18 as well as with poly rU (the group II) show that the maximum binding was obtained by the first time point for a sub-saturating concentration of NdAg (Fig. 7A). Hence, the formation of nucleic acid–NdAg complex is a rapid process.
Figure 7.
The kinetics of the formation and the dissociation of the nucleic acid–NdAg complex. (A) Association kinetics: 1 nM dT18 was incubated with 0 nM (open circle), 50 nM (open square) or 100 nM NdAg (open triangle); or 0.01 nM poly rU (group II) was incubated with 0 nM (filled circle), 5 nM (filled square) or 10 nM NdAg (filled triangle) in 500 µl standard binding buffer at 37°C. At the indicated times, 50-µl aliquots were withdrawn and filtered. The fraction of labeled nucleic acid retained on the filter was plotted against the reaction time. (B) Dissociation kinetics: 1 nM labeled dT18 was incubated with 100 nM NdAg, or 0.01 nM labeled poly rU (the group II) was incubated with 10 nM NdAg in 500 µl standard binding buffer at 37°C. After 30 min of incubation, 50 µl of the reaction mixture was withdrawn and filtered as the time 0 control, and the radioactivity retained on the nitrocellulose filter represented the amount of the labeled dT18–NdAg complex or the labeled poly rU–NdAg complex. Then 5 µl of water, or 5 µl of concentrated but unlabeled dT18 (1 µM oligo or 18 µM nucleotide) or poly rU (1 µM nucleotide) was added to the reaction mixture. At the indicated time points, 50-µl aliquots were withdrawn and filtered. The fraction of the remaining labeled dT18–NdAg or poly rU–NdAg complex was plotted against the reaction time. The reaction of labeled dT18 with water (open circle), dT18 (open square) or poly rU (open triangle) as the trap is shown in the left panel and the reaction of labeled poly rU with water (filled circle), dT18 (filled square) or poly rU (filled triangle) as the trap is shown in the right panel.
To obtain information on the dissociation kinetics of the nucleic acid–NdAg complex, we formed complex using labeled dT18 or poly rU (the group II) and a sub-saturating concentration of NdAg. A sufficient amount of unlabeled dT18 or poly rU (with an average size of >150 nt) that can compete with the labeled nucleic acid for NdAg binding was then added, and the amount of the remaining labeled nucleic acid–NdAg complex over time was monitored. The dissociated labeled nucleic acid would compete with the unlabeled nucleic acid to rebind to NdAg, resulting in a reduction in the amount of labeled complex over time after the addition of the unlabeled nucleic acid. In the case of the labeled dT18–NdAg complex, the amount of the labeled complex decreased rapidly upon addition of unlabeled dT18 (∼1000-fold excess) or poly rUs (∼50-fold excess), while a control experiment in which water was added at zero time showed almost no decrease in the amount of the labeled complex over time (Fig. 7B). In the case of the labeled poly rU–NdAg complex, the labeled complex decreased rapidly upon the addition of unlabeled dT18 (∼12 000-fold excess) or poly rUs (∼650-fold excess) as well (Fig. 7B). Since a lower amount of poly rU is required than dT18 to detect the dissociation of each labeled complex, NdAg has a higher affinity to longer polynucleotide.
The effect of salt on nucleic acid–NdAg interaction
To examine the influence of salt on poly rU–NdAg interaction, we changed the NaCl concentration of the standard binding buffer. We found that the NdAg concentration required to achieve the mid-point of the binding curve increased when the NaCl concentration of the binding buffer was raised from 0.1 M. The results obtained from the study of a group of poly rU >90 nt long (group II) and the 55∼15 nt poly rU (group IV) are shown in Figure 8A. The apparent binding affinity decreases with increasing NaCl concentration. Thus, electrostatic contacts contribute largely to the nucleic acid–NdAg interaction.
Figure 8.
(A) The effect of salt on binding affinity. The NdAg binding curves of the group II poly rU that had an average size of 155 nt and the group IV poly rU obtained at 37°C in standard binding buffer (open circle), or with NaCl added to 0.2 M (filled circle), 0.3 M (open square), 0.4 M (filled square) or 0.5 M (open triangle). (B) Salt concentration dependence of the facilitation effect of NdAg on HH16 catalysis. The trans-cleavage reaction of 0.5 nM S and 5 nM HH16 was carried out in the absence of NdAg (left panel) or in the presence of 120 nM NdAg (right panel) at 25°C in TMN buffer with NaCl added to 0.1 M (open circle), 0.15 M (filled circle), 0.2 M (open triangle) or 0.3 M (filled triangle). The fraction of uncleaved S was plotted against the reaction time.
We also investigated the effect of salt on the stimulatory activity of NdAg on hammerhead ribozyme catalysis. Hammerhead ribozyme HH16 catalyzed the cleavage of the 17-nucleotide S in trans. With 0.5 nM S and 5 nM HH16, the substrate RNA S was cleaved at a constant rate at 0.1–0.3 M NaCl (Fig. 8B, left panel), indicating the assembly of the hammerhead catalytic domain and the chemical steps of the cleavage process are not perturbed by up to 0.3 M NaCl. The S RNA was cleaved at an elevated rate when the trans-cleavage reaction was carried out in the presence of 120 nM NdAg at 0.1 M NaCl. However, the stimulatory effect of NdAg was reduced at 0.2 M NaCl, and was eliminated at 0.3 M NaCl (Fig. 8B, right panel). The reduction in the nucleic acid binding activity of NdAg at high ionic strengths may account for the attenuation of the stimulatory activity of NdAg on HH16 catalysis.
DISCUSSION
We observed previously and in this study that the N terminal domain of HDAg, referred to here as NdAg, can modulate the cis-cleaving activity of an HDV genome subfragment and stimulate the trans-cleavage reaction between a hammerhead ribozyme and cognate substrate in vitro (26,27). In the present study, NdAg, which contains the first 88 amino acids of HDAg, was expressed as a His-tagged fusion protein, and we established assays to characterize the nucleic acid binding properties of NdAg. The highly basic nature of NdAg suggests that the protein would bind nucleic acid with a broad specificity, and that the nucleic acid–NdAg complexes are mainly stabilized by electrostatic interaction. The binding studies in this report confirm these speculations. The studies with NdAg truncated mutants reveal that the extreme N terminal portion of NdAg (residues 2–13) contributes to the nucleic acid binding affinity, whereas the His-tag and the residues 60–88 of NdAg could individually be deleted without affecting the nucleic acid binding ability. Hence, the nucleic acid binding domain resides in residues 1–59 of HDAg. Mutagenesis analysis of NdAg shows that the substitution of combinations of basic residues near the N terminus and in the coiled-coil domain, and the insertion or deletion mutation in the coiled-coil domain that disrupts the heptad repeat but does not reduce the total number of basic residues, individually caused the attenuation of the nucleic acid binding activity. Thus, certain structural features in addition to a high density of positive charges of NdAg are required for the high affinity nucleic acid binding. It is likely that the adoption of α-helical structure or/and the formation of coiled-coil dimer assists the line-up of the basic sidechains and other functional groups of NdAg for nucleic acid binding.
We studied the interaction between NdAg and nucleic acid homopolymers to gain some insight into the nucleic acid binding properties of NdAg and the characteristics of the nucleic acid–NdAg complex. The binding studies with RNA homopolymers under protein excess conditions reveal that NdAg binds nucleic acid cooperatively, and the cooperativity contributes significantly to the apparent binding affinity. In addition, the NdAg binding affinity increases dramatically as the size of the rU or rA homopolymer increases by ∼9 nt, suggesting the binding site size of NdAg is around 9 nt long. The stoichiometry studies with shorter DNA homopolymers dT18 and dA36 reveal that on average one dT18 molecule binds four NdAg molecules, and one dA36 binds eight NdAg molecules. Assuming that NdAg forms a coiled-coil dimer under the experimental conditions and that NdAg binds DNA homopolymer in a manner similar to that of rU or rA homopolymer, i.e. with a binding site size of about 9 nt, each dT18 and dA36 may bind two and four NdAg dimers, respectively. The NdAg dimers may bind in tandem on the polynucleotide and nucleic acid–NdAg complexes may interact with one another to form higher order complex(es).
The crystal structure of the δ12–60(Y) peptide, corresponding to residues 12–60 of HDAg, has been solved. In the crystal, the peptide monomer is composed of a long helix, a bend at Pro49, and a short helix; peptide monomers form antiparallel coiled-coil dimers; and each dimer associates with three other dimers to form a doughnut-like octamer with a hole 50 Å in diameter (29). The central hole of the octamer is large enough to accommodate one nucleic acid helix. The basic sidechains on the sides of the coiled-coil dimer and in the hole of the octamer may interact with the phosphate backbone of the nucleic acid molecule. The corresponding region in NdAg likely has a structure similar to the δ12–60(Y) peptide. However, the extreme N terminal portion of NdAg that participates in nucleic acid binding is missing in the crystal structure. The structure of the nucleic acid–NdAg complex may be very complicated and the exact arrangement of the nucleic acid–NdAg complex remains to be characterized.
NdAg and all of its mutants made in this report stimulate the hammerhead ribozyme catalysis. Several lines of evidence strongly suggest that the stimulatory activity of NdAg or its derivative is dependent upon the nucleic acid binding ability of the protein: (i) the extent of the trans-cleavage reaction of PRP19S and KSR4 correlates with the amount of the PRP19S/KSR4–NdAg complex; (ii) NdAg mutants that bind nucleic acid less tightly stimulate the trans-cleavage reaction of PRP19S and KSR4 less efficiently; and (iii) high ionic strength that weakens the nucleic acid binding affinity attenuates the stimulatory activity on the trans-cleavage reaction of S and HH16. Hence, the formation of the RNP complex seems to be a prerequisite for the stimulatory effect on hammerhead ribozyme catalysis, and NdAg or its mutant likely acts as a nucleic acid chaperone that elevates the mutual accessibility between the hammerhead ribozyme and cognate substrate. Consequently, there is no turnover in the stimulatory activity on the assembly of the hammerhead catalytic domain. One can speculate that the high RNA binding activity of NdAg and some of its mutants allows them to enhance the hammerhead ribozyme catalysis more effectively than some other highly basic peptides, such as K7 (26), YK9A and Rev 34–50 (24), and cationic detergent cetyltrimethylammonium bromide (CTAB) (24), since a much lower concentration of the effector is required.
Previous studies have shown that some proteins that bind nucleic acids with broad specificity, including bacteriophage T4 gene 32 protein (22) and influenza A virus nucleoprotein NP (35), do not possess a nucleic acid chaperone activity. Therefore, nucleic acid binding activity is not equivalent to nucleic acid chaperone activity; i.e. certain unidentified features are also required for facilitating nucleic acid structural rearrangement. We suggest that the stimulatory effect of NdAg and its mutants on hammerhead ribozyme trans-cleavage reaction may arise from the following mechanisms: (i) The highly basic protein facilitates the opening-up of the intramolecular structure upon binding, resulting in a relatively extended RNA structure that maximizes the probability of intermolecular duplex formation. (ii) The positive charges of the peptide cause a shielding of the negative charges of the phosphate backbone which bridges together the annealing strands to increase their effective concentration. (iii) The selective strand annealing activity and selective strand exchange activity of the protein promote the assembly of the hammerhead catalytic domain between the substrate and ribozyme RNAs. Additional details regarding the complex molecular mechanisms that underlie the nucleic acid chaperone activity of NdAg remain under study.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr M. S. Lin for English editing. This work was supported by Academia Sinica, Republic of China, and by grants from the National Science Council, Republic of China (NSC 89-2311-B-001–077; NSC 90-2311-B-001–023; and NSC 91-2311-B-001–013).
REFERENCES
- 1.Kos A., Dijkema,R., Arnberg,A.C., van der Merde,P.H. and Schellekens,H. (1986) The hepatitis delta virus possesses a circular RNA. Nature, 323, 558–560. [DOI] [PubMed] [Google Scholar]
- 2.Wang K.S., Choo,Q.L., Weiner,A.J., Ou,J.H., Najarian,R.C., Thayer,R.M., Mullenbach,G.T., Denniston,K.J., Gerin,J.L. and Houghton,M. (1986) Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature, 323, 508–514. [DOI] [PubMed] [Google Scholar]
- 3.Makino S., Chang,M.F., Shieh,C.K., Kamahora,T., Vannier,D.M., Govindarajan,S. and Lai,M.M.C. (1987) Molecular cloning and sequencing of a human hepatitis delta virus RNA. Nature, 329, 343–346. [DOI] [PubMed] [Google Scholar]
- 4.Lai M.M.C. (1995) The molecular biology of hepatitis delta virus. Annu. Rev. Biochem., 64, 259–286. [DOI] [PubMed] [Google Scholar]
- 5.Gerin J.L., Casey,J.L. and Purcell,R.H. (2001) Hepatitis delta virus. In Knipe,D.M. and Howley,P.M. (eds), Fields’ Virology. Lippincott Williams and Wilkins, pp. 3037–3050. [Google Scholar]
- 6.Taylor J.M. (2003) Replication of human hepatitis delta virus: recent developments. Trends Microbiol., 11, 185–190. [DOI] [PubMed] [Google Scholar]
- 7.Chao M., Hsieh,S.Y. and Taylor,J.M. (1990) Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol., 64, 5066–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J.H., Chang,M.F., Baker,S.C., Govindarajan,S. and Lai,M.M.C. (1990) Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J. Virol., 64, 4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao M., Hsieh,S.Y. and Taylor,J.M. (1991) The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J. Virol., 65, 4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou H.C., Hsieh,T.Y., Sheu,G.T and Lai,M.M.C. (1998) Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol., 72, 3684–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavanez J.P., Cunha,C., Silva,M.C.A., David,E., Monjardino,J. and Carmo-Fonseca,M. (2002) Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA, 8, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C.Z., Lin,J.H., Choa,M., McKnight,K. and Lai,M.M.C. (1993) RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol., 67, 2221–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H.W., Chen,P.J., Lee,C.Z., Wu,H.L. and Chen,D.S. (1994) Packaging of hepatitis delta virus RNA via the RNA-binding domain of hepatitis delta antigens: different roles for the small and large antigens. J. Virol., 68, 6363–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo M.Y.P., Sharmeen,L., Dinter-Gottlieb,G. and Taylor,J.M. (1988) Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol., 62, 4439–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrotta A.T. and Been,M.D. (1991) A pseudoknot structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature, 350, 434–436. [DOI] [PubMed] [Google Scholar]
- 16.Jeng K.S., Daniel,A. and Lai,M.M.C. (1996) A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J. Virol., 70, 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih I.H. and Been,M.D. (2002) Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem., 71, 887–917. [DOI] [PubMed] [Google Scholar]
- 18.Ferre-D'Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 19.Macnaughton T.B., Wang,Y.J. and Lai,M.M.C. (1993) Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J. Virol., 67, 2228–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeng K.S., Su,P.Y. and Lai,M.M.C. (1996) Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J. Virol., 70, 4205–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand E.L. and Rossi,J. (1994) Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J., 13, 2904–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coetzee T., Herschlag,D. and Belford,M. (1994) Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev., 8, 1575–1588. [DOI] [PubMed] [Google Scholar]
- 24.Herschlag D., Khosla,M., Tsuchihashi,Z. and Karpel,R.L. (1994) An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J., 13, 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang A., Derbyshire,V., Galloway Salvo,J.L. and Belford,M. (1995) Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA, 1, 783–793. [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Z.S. and Wu,H.N. (1998) Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem., 273, 26455–26461. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z.S., Su,W.H., Wang,J.L. and Wu,H.N. (2003) Selective strand annealing and selective strand exchange promoted by the N-terminal domain of hepatitis delta antigen. J. Biol. Chem., 278, 5685–5693. [DOI] [PubMed] [Google Scholar]
- 28.Poisson F., Roingeard,P., Baillou,A., Dubois,F., Bonelli,F., Calogero,R.A. and Gouderau,A. (1993) Characterization of RNA-binding domains of hepatitis delta antigen. J. Gen. Virol., 74, 2473–2477. [DOI] [PubMed] [Google Scholar]
- 29.Zuccola H.J., Rozzelle,J.E., Lemon,S.M., Erickson,B.W. and Hogle,J.M. (1998) Structural basis of the oligomerization of hepatitis delta antigen. Structure, 6, 821–830. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y.P., Yeh,C.T., Ou,J.H. and Lai,M.M.C. (1992) Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol., 66, 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H.N. and Lai,M.M.C. (1990) RNA conformational requirements of self-cleavage of hepatitis delta virus RNA. Mol. Cell. Biol., 10, 5575–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong I. and Lohman,T.M. (1993) A double-filter method for nitrocellulose-filter binding: application to protein–nucleic acid interactions. Proc. Natl Acad. Sci. USA, 90, 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirupati H.K., Shaw,L.C. and Lewin,A.S. (1999) An RNA binding motif in the Cbp2 protein required for protein-stimulated RNA catalysis. J. Biol. Chem., 274, 30393–30404. [DOI] [PubMed] [Google Scholar]
- 34.Rozzelle J.E. Jr, Wang,J.G., Wanger,D.S., Erickson,B.W. and Lemon,S.M. (1995) Self-association of a synthetic peptide from the N terminus if the hepatitis delta virus protein into an immunoreactive alpha-helical multimer. Proc. Natl Acad. Sci. USA, 92, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clodi E., Semrad,K. and Schroeder,R. (1999) Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J., 18, 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]