Summary
We develop a new class of models, dynamic conditionally linear mixed models, for longitudinal data by decomposing the within-subject covariance matrix using a special Cholesky decomposition. Here ‘dynamic’ means using past responses as covariates and ‘conditional linearity’ means that parameters entering the model linearly may be random, but nonlinear parameters are nonrandom. This setup offers several advantages and is surprisingly similar to models obtained from the first-order linearization method applied to nonlinear mixed models. First, it allows for flexible and computationally tractable models that include a wide array of covariance structures; these structures may depend on covariates and hence may differ across subjects. This class of models includes, e.g., all standard linear mixed models, antedependence models, and Vonesh–Carter models. Second, it guarantees the fitted marginal covariance matrix of the data is positive definite. We develop methods for Bayesian inference and motivate the usefulness of these models using a series of longitudinal depression studies for which the features of these new models are well suited.
Keywords: Covariance matrix, Heterogeneity, Hierarchical models, Markov chain Monte Carlo, Missing data, Unconstrained parameterization
1. Introduction
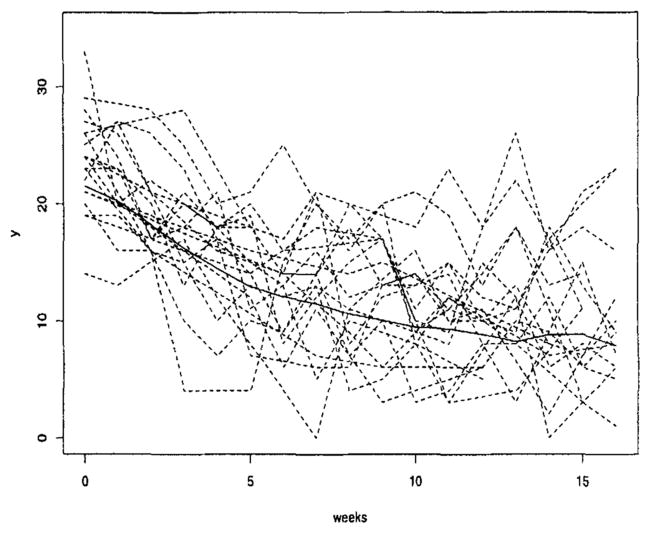
In clinical trials and observational studies, repeated measures on the same subject over time are correlated. In this article, we explore the dependence structure of longitudinal data from a series of five depression studies conducted in Pittsburgh from 1982 to 1992 (Thase et al., 1997). Patients were assigned active treatment and measured weekly for 16 weeks. Earlier work (Thase et al., 1997) explored the time to recovery from depression. Here we will examine the rate of improvement and dependence in weekly depression scores over this 16-week period for the 549 subjects with no missing baseline covariates (Figure 1). Three main questions of interest for this analysis were as follows: (1) Is the combination drug/psychotherapy treatment more effective than the psychotherapy-only treatment in improving patients’ depression? (2) Is initial severity an important predictor of patient improvement? (3) Do treatment and initial severity interact in their impact on the rate of improvement? This last question relates to practice guide-lines for treatment of major depression, which emphasizes the importance of symptom severity in determining the need for antidepressant drugs (Thase et al., 1997).
Figure 1.
Trend in depression scores over the 16-week treatment period. The solid line is the line through the observed mean at each week. The dashed lines are the observed curves for a random sample of 20 subjects.
For these 549 patients, about 30% (2840) of the possible measurements were missing mostly intermittently. There are several reasons for the missingness. Several of the studies measured depression biweekly for part or all of the active phase of treatment, so we have some observations missing by design. Some subjects dropped out (about 16%). Of the dropouts, some were missing completely at random (MCAR) while some were related to treatment, such as side effects or being so depressed they would need an alternative treatment (Patricia Houck, personal communication). Previous analyses with these data and analyses of similar data (Gibbons et al., 1993) have assumed the missingness was at random (MAR). Within our modeling framework, discussed below, the MAR assumption implies that dropout is explained by measured covariates in the model and/or observed responses prior to dropout (e.g., poor performance early in the study). Future work will examine the sensitivity of inferences to this assumption using pattern mixture models (e.g., Hogan and Laird, 1997), where we use the class of models developed here for each missing data pattern; however, this is beyond the scope of the current article.
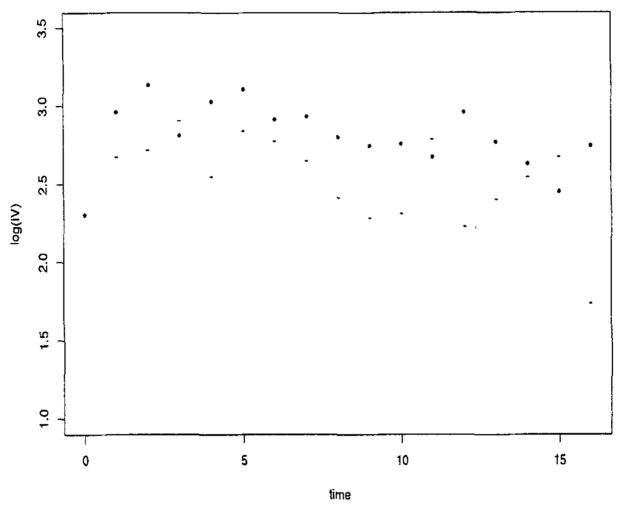
We develop a class of models with the following features: (1) flexibility in modeling the covariance structure parsimoniously and (2) ease in allowing the covariance structure to vary across subjects. The first feature is motivated by the fact that a 17 × 17 covariance matrix has 153 parameters and one needs to reduce the number of parameters to be estimated. The second feature is motivated by preliminary exploration of components of the covariance matrix, which suggests that the covariance structure may differ by the initial severity of depression in the patient. In particular, the variances for the severely depressed patients (defined at baseline) are higher than those not severely depressed (see Figure 2). Our class of models includes random effects models with residual auto-correlation, which has been used previously in the analysis of depression data (Gibbons et al., 1993).
Figure 2.
Logarithm of innovation variances versus time for low (−) and high (*) severity.
Having motivated the features of this class of models for analyzing the depression dataset, we review some models for longitudinal data in the current literature and, in the process, introduce our model in more detail. Many methods for handling dependence are introduced as an afterthought in conjunction with the desire to model accurately the marginal means of the responses within the framework of generalized linear models (McCullagh and Nelder, 1989, Chapter 9; Diggle, Liang, and Zeger, 1994). A notable example is the linear mixed models (Laird and Ware, 1982),
| (1) |
where yi is an ni × 1 vector of responses measured on the ith subject, β is the vector of unknown fixed effects parameters, bi is a q × 1 vector of unknown random effects parameters, and Xi and Zi are known ni × p and ni × q design matrices, respectively. The roles of the random term Zibi in modeling the individual effects (conditional means) and covariances simultaneously carry over to our more general treatment of dependence if one views Zibi as a stochastically weighted sum of the columns of the design matrix Zi. Lately, there is a growing tendency to view the repeated measures on a subject over time as a curve and model it using certain extensions of latent curve analysis (Rao, 1958) and multivariate growth curve models (Potthoff and Roy, 1964). The key mathematical tool in this framework is the spectral decomposition (Karhunen-Loève expansion) of a covariance matrix Σ (stochastic process), with its eigenspace taking the place of the column space of Zi (Rice and Silverman, 1991; Ramsay and Silverman, 1997; Brumback and Rice, 1998; Wang, 1998; Scott and Handcock, 1999; Verbyla et al., 1999). The close relationships between the linear mixed models and its predecessor, the multivariate growth curve models and its more recent reincarnation as functional data analysis (Ramsay and Silverman, 1997), are established by Mikulich et al. (1999) and Scott and Handcock (1999), respectively. These developments reveal the salient feature of the linear mixed models as a source for flexible and powerful models for capturing dependence in the repeated measure data; for its limitations, however, see Lindsey (1999, p. 119).
An extension of (1) known as conditionally linear mixed models (Vonesh–Carter models) is given by
| (2) |
with every component as in (1) except that now f is a known nonlinear function of its arguments (Vonesh and Carter, 1992). This model, which is linear in the random effects and nonlinear in the fixed effects, subsumes many earlier models and is closely related to the Sheiner, Rosenberg, and Melmon (1972) and Beal and Sheiner’s (1982) models.
Our focus in this article is on the accurate modeling of the covariance matrix of the responses. We show that another generalization of (2) called dynamic conditionally linear mixed models is universal for modeling dependence in the sense that it can handle essentially any dependence structure. The key idea is that the covariance matrix Σ of a mean-zero random vector y = (y1,…,yn) can be diagonalized by a lower triangular matrix constructed from the regression coefficients when yt is regressed on its predecessors y1,…,yt−1. More precisely, for t = 2,…,n, we have
| (3) |
where T and D are unique matrices, T is a unit lower triangular having ones on its diagonal and −ϕtj at its (t, j) th position, j < t, and D is diagonal with as its diagonal entries (Pourahmadi, 1999, 2000). We show that, after accounting for the means, (3) leads to representing the vector yi of repeated measurements on the ith subject with covariance Σi as a model similar to (2) with two additional design matrices, both random, depending on the response yi and, in addition, depending nonlinearly on the fixed effect parameters β and linearly on the random mean parameters bi. The resulting marginal covariance matrices are guaranteed to be positive definite and can differ across individuals through either explained (covariates) or unexplained (random effects) heterogeneity.
The outline of the article is as follows. Section 2 reviews dynamic conditionally linear mixed models. These models allow for a wide selection of covariance structures within the frame-work of a simple, conditionally linear mixed model. Section 3 discusses Bayesian inference, including computational details and model comparisons. Section 4 offers a complete analysis of the depression dataset and, in the process, introduces some of the types of covariance structures that can be modeled in this framework. For these data, model 5 in Table 1 stands out as a clear indication of the degree of flexibility one has in modeling covariances using the factorization (3).
Table 1.
The DIC and design matrices for several models fitted to the depression data. The DIC of the best fitting model is in bold type. Design matrices: Zi: * = (polyt(2)); Ai: + = (It−j=1 × severityi, It−j=2), + + = (It−j=1, It−j=2, It−j=3, I t−j=4), + + + = (polyt(1) × I t−j=1, polyt−j(4) × It−j > l), + + + + = (polyt(1) × It−j=1 × drugi, polyt−j(4) × It−j>l × drugi); Ui: * = (It−j = l); Li: + = (severityi × It=0, It>0 × Iseverityi=1, It>0 × Iseverityi=0 × polyt(3)).
| Model | Zi | Ai | Ui | Li | DIC* | DIC | D̄ | D(θ̄) | pD |
|---|---|---|---|---|---|---|---|---|---|
| ID | — | — | — | 28,677 | 28,542 | 28,512 | 28,481 | 31 | |
| UN | — | — | — | — | 25,536 | 25,045 | 24,931 | 24,817 | 114 |
| 1 | * | — | — | — | 25,408 | 25,249 | 25,212 | 25,175 | 37 |
| 2 | * | + | — | + | 25,024 | 24,934 | 24,913 | 24,892 | 21 |
| 3 | — | ++ | — | + | 25,069 | 24,956 | 24,930 | 24,905 | 26 |
| 4 | — | +++ | * | + | 25,174 | 24,910 | 24,850 | 24,789 | 61 |
| 5 | — | ++++ | — | + | 25,025 | 24,886 | 24,855 | 24,823 | 32 |
2. Dynamic Mixed Models for Longitudinal Data
In this section, we introduce a dynamic extension of (2) that offers considerable conceptual and computational flexibility in modeling the covariance of correlated data. Consider a longitudinal study where m subjects enter the study. Let yi = (yi1,…,yi,ni)′ stand for the vector of repeated measurements on subject i, yit be the response measured at (not necessarily equidistant) times indexed by t = 1,…,ni (i = 1,…,m), and xit = (x1it,…,xpit)′ be the covariates. Often it is of interest to assess the effects of the covariates on the response of interest using regression models while accounting for the dependence among the repeated measurements.
Let Σi = cov(yi) be the covariance matrix of the measurements on the ith subject and be its Cholesky decomposition as in (3). Following Pourahmadi (1999) and Daniels and Pourahmadi (2001), the nonredundant entries of Ti and Di, denoted by ϕi,tj, which will be called the generalized autoregressive parameters (GARP), and , which will be called the innovation variances (IV), can be modeled using time and/or subject-specific covariate vectors ai,tj and li,t by setting
| (4) |
where γ and λ are q1 × 1 and q2 × 1 vectors of unknown dependence and variance parameters, respectively. Then the dynamic model (3) for the ith subject takes the form . Upon substituting for ϕi,tj from (4), it reduces to
| (5) |
where is a stochastically weighted sum of the covariates ai,tj with yij’s as the weights, reminiscent of the random term Zibi in (1).
If the levels, variations, and shapes of subject-specific trajectories vary considerably across subjects (Figure 1), then one may replace γ by γ + gi, which is random and subject dependent, and add the effect of the covariates xit to (5), as in (2). To this end, we introduce two new design matrices and parameter vectors, (Zi, bi) and (Ui, gi), and additionally allow the flexibility for the marginal mean to be nonlinear in the fixed parameters β through f (·) by considering the model
| (6) |
where εi ~ N(0, Di), bi ~ N(0, Gb), gi ~ N(0, Gg), and Ai = (Ai1,…,Aini) and Ui = (Ui1,…,Uini) are dynamic design matrices constructed from the ai,tj and ui,tj (subset of ai,tj associated with the random components of γ, gi), respectively, as in (5), but with yit replaced by yit − f (xit, β) − zitbi. Such centering of the design matrix is needed for the identification of the mean parameters (β, bi) and the covariance parameters (γ, gi) (see (7) below).
We call (6) a dynamic conditionally linear mixed model. Special cases of this model include all nonlinear mixed effects models in Vonesh and Carter (1992), where gi and γ equal zero; random coefficient autoregressive models (Rahiala, 1999), where bi equals zero, f(·) is the identity function, and Ai and Ui are not centered; and hence the random coefficient polynomial and growth curve models. The model in (6) allows the flexibility of fitting many complex covariance structures using the conceptual and computational tools often used to fit the standard linear mixed models.
Alternatively, for computational convenience, we rewrite (6) as
| (7) |
where and . This formulation also makes apparent how computations can be done very simply using the Gibbs sampler and induces a marginal covariance structure on yi of the familiar form . This model allows the covariance matrix to vary across subjects using either subject-specific covariates, ai,tj and li,t, and/or random effects, gi. While we have arrived at this extension of (2) via a purely algebraic factorization of a covariance matrix, historically, (2) had appeared first in the literature of pharmacokinetics (Sheiner et al., 1972; Beal and Sheiner, 1982) using linearization methods to approximate nonlinear mixed models with a form additive in the random effects and individual errors. A major motivation for such approximations was the desire to adapt the existing estimation and inferential procedure and software packages for the linear mixed models. For a detailed review of the linearization methods and the relevant software, see Davidian and Giltinan (1995, Chapter 6).
A referee has pointed out that, in models (2) and (6), the fixed and random effects appear on different scales. Though these models have some limitations, they are useful in approximating more general nonlinear mixed models, and in some case studies, they are motivated by the substantive knowledge and are supported by the data; see Heitjan (1991).
3. Bayesian Inference
In this section, we discuss prior specifications and model fitting using the Gibbs sampler. In addition, we discuss model comparison using the output from the Gibbs sampler.
3.1 Priors and Computations
We propose independent, noninformative normal priors on β γ, and λ, with large variances and inverse Wishart priors on Gb and Gg, with degrees of freedom equal to dim(Gb) and dim(Gg), respectively. To sample from the posterior distribution of the parameters, we develop a simple Gibbs sampler. The full conditional distributions of all parameters have known forms, either normals, inverse Wisharts, or inverse gammas, except for λ. Specifically, when sampling from the full conditional distribution of Gb, β, bi, and the missing data, we use formulation (7); when sampling γ, gi, and D(λ), we use formulation (6). To sample λ, we implement a Metropolis–Hastings algorithm, using a normal approximation to the full conditional distribution of λ as the candidate distribution. For the models with f (·) not the identity function, nonlinear models in the fixed mean parameters, the full conditional distribution of β will not be a known form. Here we also implement a similar Metropolis–Hastings algorithm. More details of the Bayesian implementation can be found in Davidian and Giltinan (1995, Chapter 8) and Daniels and Pourahmadi (2001).
We use data augmentation (Tanner and Wong, 1987) to integrate over the missing data. Operationally, this involves imputing the missing responses using equation (7) and assumes the missing responses are MAR. More specifically, the complete set of 17 observations for subject i are divided into its observed components (yobs) and missing components (ymis), so that yi = (yobs, ymis). Given that yi conditional on all the mean and covariance parameters is multivariate normal, the conditional distribution of ymis | yobs will also be multivariate normal. This distribution is the full conditional of the missing data for subject i given the observed data.
3.2 Model Comparison
The complexity of these models make standard model comparisons using the AIC and BIC difficult since both the sample size and the dimension of the parameter space are not obvious. Consequently, to compare models with or without random effects in the mean or covariance parameters, we will use the recent deviance information criterion (DIG) (Spiegelhalter et al., 2001). The structure of this statistic allows for automatic computation of the dimension of the parameter space and has a form similar to the Akaike information criterion (AIC): a goodness-of-fit term, the deviance evaluated at the posterior mean of the parameters, and a penalty term, two times the effective number of parameters, computed as the mean deviance minus the deviance evaluated at the posterior mean. Thus,
| (8) |
where θ̄ is the posterior mean of θ and , where is the posterior mean of the deviance.
However, we point out that the DIC has a problem similar to the AIC; when the sample size gets large, it favors models with too many parameters. As a result, we might alter the DIC to a form similar to the BIC,
| (9) |
where m is the number of subjects. We will use both DIC and DIC* for model comparison in Section 4.
4. Longitudinal Depression Data
4.1 Model Building and Selection
The Hamilton Rating Scale for Depression (HRSD), measured weekly, was used as the measure of depression for these studies. Similar to Thase et al. (1997), we pooled the data across studies. We did not include indicators of study in our model, which was motivated by the fact that they were confounded with age and gender so we adjusted for study by including these covariates in the model. For our analysis, we modeled the rate of improvement of HRSD over the 17 weeks (baseline + 16 weeks of treatment). The main questions of interest were addressed in the Introduction.
We restricted ourselves to the dynamic conditionally linear mixed models (6) with f (·) the identity function. As discussed in Section 2, this class of models allows the covariance structure to vary across subjects, either through random parameters of the covariance and/or covariates, i.e., through the design matrices Ai and Ui. These models were fit using Gibbs sampling algorithm described in Section 3.1. To compare the different covariance models, we used the DIC and DIC*. Here we defined the deviance as
| (10) |
where as in (7), yobs is the observed data, and the other parameters and missing data have been integrated out. We also mention that pD depends on how we parameterize θ Fortunately, in our analysis, similar conclusions were drawn when we set θ = (β, gi, γ λ Gg). This explains, e.g., why pD for the unstructured model in Table 1 is not 167, the number of parameters in the unstructured covariance matrix plus the 14 fixed mean parameters (β).
4.2 Exploratory Analysis
Our exploratory work on these data is presented next. A quadratic trend in weekly Hamilton scores (see Figure 1) fit well. We considered interactions of this trend with the four possible combinations of the two binary covariates: initial severity of depression in the patient, with two levels, high versus low; and treatment (drug), with two levels, drug and psychotherapy versus only psychotherapy. Estimation and testing of these interactions addresses one of the questions of this analysis—how treatment and severity affect the rate of improvement. To account for the differences between studies, we also included age and gender of the patient. Thus, the design vector xit for the ith patient at time t is 14 × 1, xit = (polyt(2), drugi × polyt(2), severityi × polyt(2), drugi × severityi × polyt(2), genderi, agei), where polya (k) is a kth-order orthogonal polynomial in a.
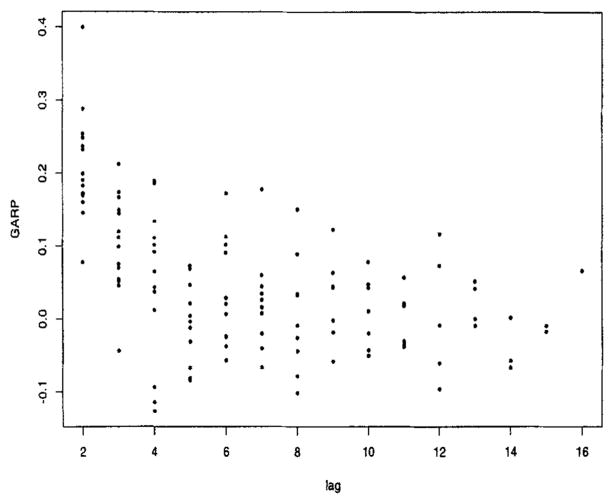
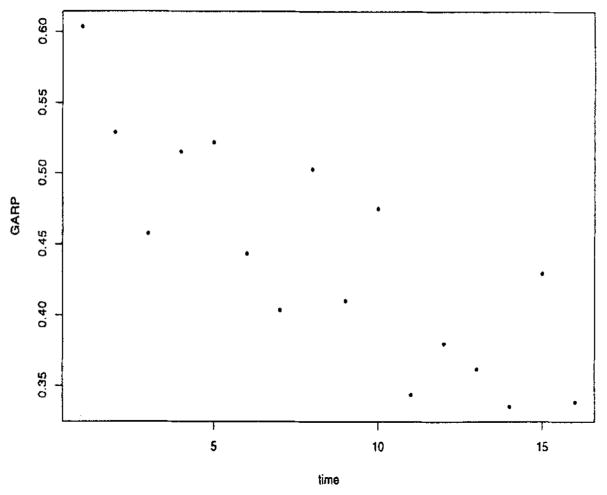
We then explored the covariance structure. First, we examined the plot of versus time (Figure 2). The variability at baseline, , was much lower than at the other time points, and the variability for the low severity patients was consistently lower than the high severity patients. Based on this plot and examination of credible intervals of coefficients and the DIC, we fit a separate intercept at baseline for each category of severity and a flat line for high severity from weeks 1 to 16 and a cubic for low severity over the same time period. Conditional on this model for the log , we then explored models for the generalized autoregressive parameters (GARP). Empirical regressograms (Pourahmadi, 1999) of the GARP appear in Figures 3 and 4, namely the plot of the estimated ϕt,,t−j in (3) versus j and the plot of ϕt,t−1 versus t, respectively. Figure 4 suggests the lag-1 GARP ϕt,t−1 has a roughly linear relationship with time. No apparent patterns were obvious for the other ϕt,t−j over time. Figure 3 suggested that lag-1, -2, -3, and -4 GARPs were relatively large and important and that a fourth-order polynomial, polyt−j, (4), fit the data well. These observations were used to suggest parametric models for ϕt,,j’s giving rise to structured (parametric) models for GARP. In addition, there was an important interaction between the GARP models and treatment (drug) (not shown in figures). These exploratory plots along with examination of 95% credible intervals and the DIC were used to guide our analysis.
Figure 3.
GARP versus lag.
Figure 4.
Lag-1 GARP versus time.
As a comparison with more standard approaches to modeling the covariance structure, we also fit an independence model, an unstructured covariance matrix model, a random effects model, i.e., with random quadratic curves (Zi, = polyt(2)), and a random effects model with residual auto-correlation and a structured model on (four parameters corresponding to the two levels of severity and baseline versus weeks 1–16).
4.3 Results
The DICs for a subset of models we fit, including the best fitting ones, appear in Table 1. Clearly, the independence (ID) model fits poorly and the unstructured (UN) model fits better.
Model 5 fit best overall. For the GARP, this model included a fourth-order polynomial in t − j, j ≥ 2, for ϕt,t−j first-order polynomial in t for ϕt,t−j, with both sets of coefficients depending on drug. For the innovation variances, the model included a third-order polynomial in t for log , t = 1,…, 16, and a separate parameter for for low-severity patients and two intercepts for t = 0 and t = 1,…,16, respectively, for high-severity patients. Model 4, which allows for unexplained heterogeneity, with a random intercept in ϕt,t−1, was less competitive. Model 2, the random effects model with residual autocorrelation and a structured model for , with both the dependence and variance parameters depending on severity, fit about as well as the best model without a random quadratic curve (model 5). In general, the best fitting models support the covariance structure differing by either severity or drug or both.
Table 2 shows the estimates of the mean parameters for the best fitting model, model 5. Using this table, we address the three main questions of interest for this analysis: (1) There was not a significant effect of drug and psychotherapy versus psychotherapy only in terms of rate of improvement, although the sign of the coefficients suggested those on the combination therapy did better. (2) Initial severity was an important predictor of rate of improvement. Those patients who were severely depressed at the start of the studies improved more quickly than those not so severely depressed (95% credible interval for the severity by linear interaction excluded zero). In addition, the magnitude of effects were larger for initial severity than for treatment. (3) There were no significant interactions between treatment and severity on rate of improvement. However, the signs of the coefficients suggested the rate of improvement was most rapid for those with high initial severity and on the combination drug/psychotherapy treatment. Age and gender were not statistically significant.
Table 2.
Posterior means and 95% credible intervals for the coefficients in model 5 from Table 1
| Covariate | Estimate |
|---|---|
| Intercept | 11.0 (10.3, 11.8) |
| Linear | −0.59 (−0.67, −0.52) |
| Quadratic | 0.014 (0.009, 0.019) |
| Severity | 3.35 (2.30, 4.37) |
| Severity × linear | −0.20 (−0.33, −0.08) |
| Severity × quadratic | 0.0062 (−0.0014, 0.014) |
| Drug | −0.74 (−1.77, 0.26) |
| Drug × linear | −0.084 (−0.19, 0.022) |
| Drug × quadratic | 0.0013 (−0.0058, 0.0081) |
| Severity × drug | −0.92 (−2.29, 0.48) |
| Severity × drug × linear | −0.12 (−0.27, 0.035) |
| Severity × drug × quadratic | 0.0068 (−0.0028, 0.016) |
| Age | 0.013 (−0.0016, 0.028) |
| Gender | −0.017 (−0.46, 0.42) |
5. Discussion
We have proposed a class of models and a unified method to fit these, which allows the user to fit a wide variety of potentially parsimonious covariance structures to unbalanced longitudinal data including models in which there is heterogeneity in the covariance structure across subjects, either explained or unexplained by covariates. In addition, we have suggested a simple approach to compare these models within the Bayesian paradigm using the DIC; however, we recommend using the DIC only as a guide to finding better fitting covariance structures. In the depression example, we found initial severity to be an important determinant of both the improvement rate and the covariance structure in depressed patients and drug to be less important. For these data, model 5 stands out as a clear indication of the degree of flexibility one has in modeling covariances using the factorization (3) and the ensuing dynamic conditionally linear model (6).
Future work on these models will address the impact of heterogeneity in the covariance structure on estimation of β; Daniels and Kass (2001) discuss gains in mean squared error for estimating β when the covariance structure is properly modeled for nonheterogeneous situations. In terms of the depression data, models that allow for nonignorable missingness (dropout) will be explored as detailed in the Introduction. In addition, the sensitivity of the overall inferences to individual studies might be addressed by using a cross-validation approach (Thase et al., 1997). Finally, we are currently exploring using the modified Cholesky decomposition to smoothly incorporate covariates into a random effects matrix.
Résumé
Nous développons une nouvelle classe de modèles, les modèles mixtes dynamiques conditionnellement linéaires, pour des données longitudinales, en déomposant la matrice de covariance intra-sujet par une décomposition de Cholesky particulière. Dans notre approche, dynamique se réfère à l’utilisation des réponses passées comme covariables, et la linéarité conditionnelle signifie que les paramètres linéaire du modèle peuvent être aléatoires tandis que les paramètres non-linéaires sont non aléatoires. Ce dispositif offre plusieurs avantages, et est étonnamment similaire aux modèles obtenus par une méthode de linéarisation au premier ordre appliquée aux modèles mixtes non linéaires. Tout d’abord il s’applique à des modèles flexibles, accessibles aux calculs et incluant une large gamme de structures de covariance; ces structures peuvent dépendre des covariables et par là-même varier entre sujets. Cette classe de modèles inclut par exemple tous les modèles linéaires mixtes standard, les modèles d’antédépendance, et les modèles de Vonesh-Carter. Ensuite, il garantit que la matrice de covariance marginale ajustée soit définie positive. Nous développons des méthodes pour l’inférence bayésienne, et nous justifions l’intérêt de ces modèles en utilisant une série d’études longitudinales sur la dépression pour lesquelles les caractéristiques de ces nouveaux modèles sont particulièrement bien adaptées.
Acknowledgments
The work of the second author was supported by NIH grant CA85295-01A1. The authors would like to thank Professor Joel Greenhouse for providing the data and clarifying several issues. The authors would also like to thank Patricia Houck, M.S.H, for clarifying issues regarding dropout in the studies and Dr Edward Vonesh for helpful discussions on the nature of scales in the nonlinear mixed models.
Contributor Information
M. Pourahmadi, Division of Statistics, Northern Illinois University, DeKalb, Illinois 60115, U.S.A. pourahm@math.niu.edu
M. J. Daniels, Department of Statistics, Iowa State University, Ames, Iowa 50011, U.S.A. mdaniels@iastate.edu
References
- Beal SL, Sheiner LB. Estimating population kinetics. CRC Critical Reviews in Biomedical Engineering. 1982;8:195–222. [PubMed] [Google Scholar]
- Brumback BA, Rice JA. Smoothing spline models for the analysis of nested and crossed samples of curves. Journal of the American Statistical Association. 1998;93:961–976. [Google Scholar]
- Daniels MJ, Kass RE. Shrinkage estimators for covariance matrices. Biometrics. 2001;57:1173–1184. doi: 10.1111/j.0006-341x.2001.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Pourahmadi M. Bayesian analysis of covariance matrices and dynamic models for longitudinal data. Department of Statistics, Iowa State University; Ames: 2001. Preprint 2001–03. [Google Scholar]
- Davidian M, Giltinan DM. Nonlinear Models for Repeated Measurements. New York: Chapman and Hall; 1995. [Google Scholar]
- Diggle PJ, Liang KY, Zeger SL. Analysis of Longitudinal Data. Oxford: Oxford University Press; 1994. [Google Scholar]
- Gibbons RD, Hedeker D, Elkin I, Waternaux C, Kraemer HC, Greenhouse JB, Shea MT, Imber SD, Sotsky SM, Watkins JT. Some conceptual and statistical issues in the analysis of longitudinal psychiatric data. Archives of General Psychiatry. 1993;50:739–750. doi: 10.1001/archpsyc.1993.01820210073009. [DOI] [PubMed] [Google Scholar]
- Heitjan DF. Nonlinear modeling of serial immunologic data: A case study. Journal of the American Statistical Association. 1991;86:891–898. [Google Scholar]
- Hogan JW, Laird NM. Mixture models for the joint distribution of repeated measures and event times. Statistics in Medicine. 1997;16:239–257. doi: 10.1002/(sici)1097-0258(19970215)16:3<239::aid-sim483>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Laird NM, Ware JJ. Random-effects models for longitudinal data. Biometrics. 1982;38:973–979. [PubMed] [Google Scholar]
- Lindsey JK. Models for Repeated Measurements. 2. Oxford: Clarendon Press; 1999. [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2. Cambridge: Chapman and Hall; 1989. [Google Scholar]
- Mikulich SK, Zerbe GO, Jones RH, Crowley TJ. Relating the classical covariance adjustment techniques of multivariate growth curve models to modern univariate mixed effects models. Biometrics. 1999;55:957–964. doi: 10.1111/j.0006-341x.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- Pothoff RF, Roy SN. A generalized multivariate analysis of variance model useful especially for growth curve problems. Biometrika. 1964;52:313–326. [Google Scholar]
- Pourahmadi M. Joint mean-covariance models with applications to longitudinal data: Unconstrained parameterisation. Biometrika. 1999;86:677–690. [Google Scholar]
- Pourahmadi M. Maximum likelihood estimation of generalized linear models for multivariate normal covariance matrix. Biometrika. 2000;87:425–435. [Google Scholar]
- Rahiala M. Random coefficient autoregressive models for longitudinal data. Biometrika. 1999;86:718–722. [Google Scholar]
- Ramsay JO, Silverman BW. Functional Data Analysis. New York: Springer; 1997. [Google Scholar]
- Rao CR. Some statistical methods for comparison of growth curves. Biometrics. 1958;14:1–17. [Google Scholar]
- Rice JA, Silverman BW. Estimating the mean and covariance structure nonparametrically when the data are curves. Journal of the Royal Statistical Society, Series B. 1991;53:233–243. [Google Scholar]
- Scott MA, Handcock MS. IEE Working Paper 13. Institute of Education and the Economy, Columbia University; 1999. Latent curve covariance models for longitudinal data. [Google Scholar]
- Sheiner LB, Rosenberg B, Melmon KL. Modelling of individual pharmacokinetics for computer-aided drug dosing. Computers and Biomedical Research. 1972;5:441–459. doi: 10.1016/0010-4809(72)90051-1. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, Van der Linde A. Bayesian measures of model complexity and fit. Division of Biostatistics, University of Minnesota; Minneapolis: 2001. Report 2001–013. [Google Scholar]
- Tanner MA, Wong WH. The calculation of posterior distributions by data augmentation. Journal of the American Statistical Association. 1987;82:528–540. [Google Scholar]
- Thase ME, Greenhouse JB, Frank E, Reynolds CF, III, Pilkonis PA, Hurley K, Grochocinski V, Kupfer DJ. Treatment of major depression with psychotherapy or psychotherapy–pharmacotherapy combinations. Archives of General Psychiatry. 1997;54:1009–1015. doi: 10.1001/archpsyc.1997.01830230043006. [DOI] [PubMed] [Google Scholar]
- Verbyla AP, Cullis BR, Kenward MG, Welham SJ. The analysis of designed experiments and longitudinal data by using smoothing splines (with discussion) Applied Statistics. 1999;48:269–311. [Google Scholar]
- Vonesh EF, Carter RL. Mixed effects non-linear regression for unbalanced measures. Biometrics. 1992;48:1–18. [PubMed] [Google Scholar]
- Wang Y. Smoothing spline models with correlated random errors. Journal of the American Statistical Association. 1998;93:341–348. [Google Scholar]