Abstract
The gene that codes for dopamine receptor D2 (DRD2 on chromosome 11q23) has long been a prime functional and positional candidate risk gene for schizophrenia. Collectively, prior case–control studies found a reliable effect of the Ser311Cys DRD2 polymorphism (rs1801028) on risk for schizophrenia, but few other polymorphisms in the gene had ever been evaluated and no adequately powered family-based association study has been performed to date. Our objective was to test 21 haplotype-tagging and all three known nonsynonymous single-nucleotide polymorphisms (SNPs) in DRD2 for association with schizophrenia in a family-based study of 2408 Han Chinese, including 1214 affected individuals from 616 families. We did not find a significant effect of rs1801028, but we did find significant evidence for association of schizophrenia with two multi-marker haplotypes spanning blocks of strong linkage disequilibrium (LD) and nine individual SNPs (Ps < 0.05). Importantly, two SNPs (rs1079727 and rs2283265) and both multi-marker haplotypes spanning entire LD blocks (including one that contained rs1801028) remained significant after correcting for multiple testing. These results further add to the body of data implicating DRD2 as a schizophrenia risk gene; however, a causal variant(s) in DRD2 remains to be elucidated by further fine mapping of the gene, with particular attention given to the area surrounding the third through fifth exons.
Keywords: allele, dopamine, haplotype, linkage disequilibrium, PBAT
Introduction
In the study of schizophrenia, few facts are as widely accepted as these: (1) genes influence risk for the disorder; and (2) its symptoms are effectively ameliorated by drugs that antagonize D2 dopamine receptors. It is therefore not surprising that the gene that codes for the D2 dopamine receptor (DRD2) was among the first evaluated for allelic association with the illness. Over a decade ago, Arinami and co-workers1 found a trend and then significant evidence2 for an effect of the Cys allele of the Ser311Cys single-nucleotide polymorphism (SNP) of DRD2 on risk for schizophrenia. However, 21 of 22 subsequent replication attempts failed, after which the result obtained by Arinami et al.2 became widely regarded as a type-I inferential error and the possibility of an association was considered remote.3 Yet, as we previously documented,4 this pattern of nonreplication was most likely attributable to low statistical power among the replication studies which had, on average, a 55.7% chance of detecting an effect as large as that observed by Arinami et al.2 (odds ratio (OR) of 3.1), but only a 7.5% chance of detecting what we identified by meta-analysis5 as the ‘best estimate’ of the magnitude of this polymorphism’s effect on risk for schizophrenia (OR = 1.3). In addition to the statistical significance of the OR determined from our meta-analysis, the reliability of this association was bolstered by several facts including (1) there was no evidence that the effect was attributable to publication bias, where only positive reports might be accepted into the literature and contribute to the meta-analysis; (2) there was no evidence of heterogeneity among the studies, suggesting that the effect of DRD2 was consistent across samples; and perhaps most importantly, (3) the significance of the overall effect observed by meta-analysis persisted even when the large and influential study by Arinami et al.2 was removed from the analysis. These observations suggested that the effect of this DRD2 polymorphism on schizophrenia risk is reliable and uniform across populations, although the magnitude of its effect is small.5
Even though the collective body of evidence gleaned from these case–control studies is sufficient to re-establish DRD2 as a prime candidate gene for schizophrenia,6 it must also be interpreted in light of its limitations. Case–control studies (and thus meta-analyses of such studies) are subject to various biases that can induce false-positive associations. Foremost among them is population stratification, whereby differences in allele frequencies observed between cases and controls reflect natural (that is, not illness-associated) variations between the different ancestral groups or cohorts from which each sample is drawn. Thus, before DRD2 can be established firmly as a risk gene for the disorder, it is crucial to amass further support for this association using family-based association methods that are robust to population stratification and associated biases.
To date, only one family-based association study of DRD2 in schizophrenia has been published,3 and this study clearly indicated a trend toward association that was similar in direction and magnitude (OR = 1.7) to the effect observed in case–control studies; however, statistical significance was not attained, likely due to the small sample size employed (n=64 families). Therefore, the first aim of the present study was to determine if an effect of the Ser311Cys polymorphism (rs1801028) on risk for schizophrenia could be detected in a much larger (and possibly genetically loaded) sample of 616 families affected with multiple cases of the disorder. In addition, although the Ser311Cys polymorphism is a functional polymorphism affecting the receptor’s affinity for dopamine,7 it is also possible that another polymorphism in the gene is the true causal variant, and that the Ser311Cys SNP shows association with the disorder either independent of such a polymorphism or simply through strong linkage disequilibrium (LD) with an as-yet-unrecognized variant. Thus, the second aim of our study was to establish the degree of LD between the Ser311Cys variant and the 23 other SNPs that collectively represent all known nonsynonymous mutations and much of the haplotype diversity of DRD2 in the Han Chinese population. The third study objective was to more precisely specify the locus within DRD2 that influences risk for schizophrenia by examining patterns of association with all 24 SNPs and haplotypes of these SNPs. To our knowledge, this is the first family-based association study of schizophrenia that tested both haplotype-tagging and all nonsynonymous SNPs in DRD2, and thus provides the most comprehensive examination yet of the role of DRD2 in influencing risk for schizophrenia.
Materials and methods
Ascertainment
A detailed description of ascertainment and clinical assessment methods for this study is given by Hwu et al.8 Briefly, probands were recruited from six data-collection field-research centers throughout Taiwan. To be included in the study, the family was required to be of Han Chinese ancestry and to meet the entry criteria adopted by the US National Institute of Mental Health’s Schizophrenia Genetics Initiative; that is, the family had to have two siblings with schizophrenia or schizoaffective disorder, depressed type (which family studies suggest may be an alternate expression of schizophrenia susceptibility genes9). When diagnosing schizoaffective disorder, the fourth edition of the American Psychiatric Association’s Diagnostic and Statistical Manual of Mental Disorders10 requires that ‘symptoms that meet criteria for a mood episode (be) present for a substantial portion of the total duration of active and residual periods.’ We operationalized ‘substantial portion’ as one-third or more, with the mood symptoms contributing to the functional impairments of the patient. Of the 1214 affected individuals included in the present study, only eight were diagnosed with schizoaffective disorder, depressed subtype.
Clinical assessment
Each proband underwent a diagnostic screen by a research psychiatrist using medical records and a semi-structured interview that was based on Diagnostic and Statistical Manual of Mental Disorders-IV. Following this screen, we administered the Mandarin Chinese version11 of the Diagnostic Interview for Genetic Studies.12 These data were also supplemented by a semi-structured itemized assessment of psychopathology in family members using the Family Interview for Genetic Studies.13 The interviewers who administered the Diagnostic Interview for Genetic Studies and Family Interview for Genetic Studies were college graduates specializing in psychology or psychiatric nursing and had 2–3 years of clinical experience with psychiatric patients as well as extensive training on each instrument.
Best-estimate final diagnoses were made independently by two board-certified research psychiatrists based on all the clinical information that was collected. When these psychiatrists disagreed, a third diagnostician (H-GH) resolved the disagreement by reviewing all data schedules and medical records, and if necessary, by conferring with the field psychiatrist who cared for the patient. Informed consent was obtained from all participants and all procedures involving human subjects were approved by the Institutional Review Boards of all project sites.
DNA sample acquisition and distribution
Approximately 10 ml of blood was drawn from each subject and immediately shipped to the National Institute of Mental Health Center for Collaborative Studies of Mental Disorders at the Rutgers University Cell and DNA Repository, where cells were immortalized via transformation with Epstein–Barr virus. High-quality DNA was successfully extracted from 2448 of these cell lines and sent to the principal investigator (MTT) at UCSD for storage and analysis. An aliquot taken from each of these samples was also sent to the Harvard Partners Genotyping Facility at the Harvard Medical School-Partners Healthcare Center for Genetics and Genomics (HPCGG) for genotyping.
Marker selection
Markers were selected for analysis based on two sets of criteria. First, we searched for all known nonsynonymous polymorphisms in DRD2 according to dbSNP, build 126;14 this search resulted in the identification of three SNPs in exon 7, including rs1110977 (an A-to-G transition resulting in substitution of threonine for alanine in codon 351), rs1800496 (a T-to-C transition resulting in substitution of proline for serine in codon 310) and rs1801028 (a G-to-C transversion resulting in the Ser311Cys polymorphism). Second, we searched for all nonredundant (r2= 1.0) haplotype-tagging SNPs of DRD2 with minor allele frequencies ≥ 0.05 in the Han Chinese population by applying the Tagger Pairwise algorithm15 to data from the International HapMap Project, release 21/phase II;16 this search resulted in the identification of an additional 21 SNPs, all of which mapped to intronic regions of the gene.
Genotyping
Polymerase chain reaction assays were designed using SpectroDESIGNER software, version 3.0 0.3 (Sequenom Inc., San Diego, CA, USA) by inputting sequences containing the site of each of 24 SNPs and 100 bp of flanking sequence on either side of each SNP (Supplementary Table 1). Genotypes at 23 of the 24 loci of interest were generated at HPCGG using Sequenom’s iPLEX technology, which is based on multiplexed PCR followed by a minisequencing reaction in a single well. In this process, the size of reaction products is determined directly by matrix-assisted laser desorption/ionization-time of flight mass spectrometry, which yields genotype information.
One marker (rs2734837) amplified poorly and was dropped from the study. The assay for one additional marker (rs1801028) could not be completed using iPlex technology, as nearby flanking SNPs would have inhibited hybridization of the probe sequences to the target sequence. This assay was therefore completed at HPCGG using Sequenom’s Homogeneous MassEXTEND technology that, like iPLEX, utilizes multiplexed PCR and matrix-assisted laser desorption/ionization-time of flight mass spectroscopy to determine genotypes. All procedures were executed per protocol17,18 except for minor changes made to reagent volumes for optimization.
Data cleaning and quality control
Of the 2448 available DNA samples, 38 were from adoptees and two were of such low quality that no usable genotypes were generated. The final data set for the present analyses therefore included genotypes at 23 loci in 2408 individuals (including 1214 affected individuals) from 616 families of various constellations. Descriptive statistics of the sample are presented in Table 1. The desired ascertainment unit for this study consisted of an affected sibling pair and both parents; however, families that were incompletely ascertained were used as appropriate (for example, unaffected individuals (with available DNA) from families where DNA was not available from any affected relatives contributed only to the LD analyses, whereas trios with DNA from only one affected child were used for LD, power calculation of Pedigree Based Association Testing (PBAT) and transmission disequilibrium testing (TDT) analyses).
Table 1.
Descriptive statistics of genotyped subjects
| Statistic | N (% of sample) |
|---|---|
| Individuals | 2408 (100) |
| Affection status | |
| Affected | 1214 (50.4) |
| Unaffected | 932 (38.7) |
| Unknown | 262 (10.9) |
| Sex | |
| Male | 1324 (55.0) |
| Female | 1084 (45.0) |
| Families | 616 (100) |
| Affected per family | |
| 0 | 36 (5.8) |
| 1 | 43 (7.0) |
| 2 | 445 (72.2) |
| 3 | 88 (14.3) |
| 4 | 3 (0.5) |
| 5 | 1 (0.2) |
The accuracy of genotypes was determined by running replicates on 7.15% of all DNA samples. The average discordance rate between replicates was 0.17%, which translates into a 99.83% accuracy rate. Discordant genotypes from replicate samples were excluded from all subsequent analyses. Pedigree inconsistencies (for example, incorrect parental gender or unexpected loops) and Mendelian inconsistencies (which may reflect mis-specified relationships or genotyping errors) were identified using GeneSpring GT software, version 2.0 (Agilent Technologies Inc., Santa Clara, CA, USA). All detected pedigree inconsistencies (n = 5) were identified as data transcription errors and rectified unambiguously by inspecting the original pedigree file. Without additional molecular genetic analyses, it was impossible to determine if the detected Mendelian inconsistencies (n = 30) were due to mis-specified relationships or genotyping error; however, based on the 99.83% accuracy rate of genotyping reported above, one or fewer genotyping errors would be expected among the 46 genotypes generated in each inconsistent parent–child dyad. Thus, in instances in which only one Mendelian inconsistency was detected (n = 11), the stated relationship was assumed to be accurate and genotypes at the inconsistent marker were set to missing. In pedigrees where more than one Mendelian error was detected (n = 19), the inconsistent genotypes were assumed to be accurate and the relationship between the two members of the inconsistent dyad was set to unknown.
As a final quality-control check, each marker was tested for consistency with genotype proportions expected under Hardy–Weinberg equilibrium, and most importantly, no marker showed a significant deviation from expected values among parents (all Ps > 0.11). Among probands, seven markers deviated from expected Hardy–Weinberg equilibrium proportions, but six of these showed significant association with the disease and the seventh approached significance (P = 0.0710); thus, these departures from Hardy–Weinberg equilibrium are almost certainly a product of their preferential overtransmission to affected individuals rather than genotyping error, assortative mating or other potential sources of bias.19
Out of a possible total of 55 384 genotypes (2408 individuals ×23 markers), 55 162 genotypes were generated and retained for analysis after all quality-control checks and data-cleaning procedures were completed, which translates into an overall success rate of 99.60%.
Statistical analyses
The LD block structure of DRD2 was determined using HaploView software, version 3.32.20 For each pair of SNPs, we used Haploview to calculate D′ (which indicates the strength of LD between the two SNPs), r2 (the squared correlation coefficient, which indicates the proportion of variance in one SNP that is accounted for by variance in another) and LOD scores (log of the likelihood OR for LD between the two SNPs, which provides a measure of the significance of the value of D′). Blocks of strong LD were then defined using the parameters established by Gabriel et al.21
On the basis of the results of our prior meta-analysis,5 we hypothesized that DRD2 polymorphisms would exert dominant effects on risk for schizophrenia, so only dominance models of association were evaluated. Our primary family-based association analyses were conducted using the PBAT algorithm22 as implemented in the HelixTree Genetic Analysis Software suite, version 5.20 (GoldenHelix Inc., Bozeman, MT, USA). We chose PBAT instead of the traditional TDT framework due to the flexibility of the former method and ability to extract maximal power from our sample structure. The statistical approach to family-based association analysis implemented in PBAT allows for valid testing of association with any phenotype, sampling structure and pattern of missing marker allele information.22–24 When parental genotypes are missing, PBAT computes a test statistic by conditioning on genotypes of any observed parents and offspring, adjusting for admixture.25 PBAT can also incorporate unaffected offspring (of which there are many in our sample), which often provides a substantial power advantage over the TDT.26,27 All association analyses were restricted to markers (haplotypes or SNPs) for which 10 or more families were informative.
To clarify the haplotypic background of the evaluated SNPs (and to potentially identify significantly associated regions of the gene that might escape detection in single SNP analyses), we tested for association of schizophrenia with haplotypes of markers that spanned the extent of each block of strong LD. The significance of each tested haplotype was evaluated using a simple Bonferroni correction for the number of haplotypes present within each LD block, since these blocks (which are divided by clear recombination hot spots) may be considered to be independent.
In addition to these haplotype analyses, we tested each individual SNP for association with schizophrenia. To control the type-I error rate while testing numerous SNPs, we implemented a two-stage approach combining elements of genomic screening28,29 and spectral decomposition.30 Briefly, we first selected the subset of SNPs that had asymptotic power (1 − β = 1.0, conditional on offspring phenotypes and parental genotypes) relative to the remaining SNPs. The retained markers were generally those with higher minor allele frequencies and heterozygosity rates. The significance of the P-value for each retained marker was then assessed against a critical P-value (α = 0.05), which was corrected for the number of retained markers. Because many of the retained SNPs were in some degree of LD with each other and, thus, not independent, a standard Bonferroni correction would have been too conservative; instead, the adjusted critical P-value that maintained a family-wise error rate of 5% was determined by spectral decomposition of matrices of pairwise LD between SNPs according to the method of Nyholt.30 Finally, to determine the effect size (OR) attributable to each significantly associated SNP, TDTs were conducted on a subset of the sample (all 309 complete parent–child trios) using HaploView software, version 3.32.20
Results
Linkage disequilibrium among DRD2 SNPs
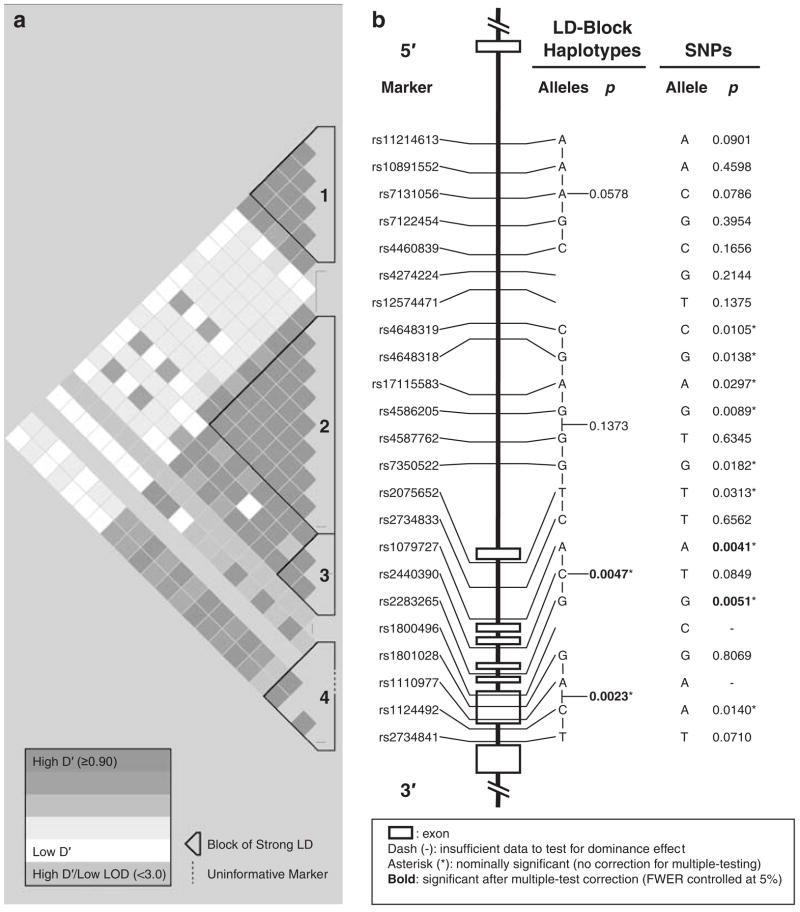
As noted in the Materials and Methods section, one marker (rs2734837) amplified poorly and was dropped from the study. The LD structure of the remaining 23 SNPs is shown in Figure 1a, and various measures of LD between adjacent DRD2 SNPs are provided in Supplementary Table 2. We identified four blocks of strong LD within which all values of D′ between markers met or exceeded 0.90. Block 2 was the largest in terms of the physical distance it spanned (21 kb), the number of total SNPs it contained (n = 8) and the number of SNPs within it that were significantly associated with schizophrenia (n = 6). Block 3 contained the two SNPs and one multi-marker haplotype, which were most reliably associated with schizophrenia (described below).
Figure 1.
Linkage disequilibrium block structure, genomic organization and patterns of association with schizophrenia of dopamine receptor D2 (DRD2) single-nucleotide polymorphisms (SNPs) and haplotypes. (a) The 23 evaluated polymorphisms of DRD2 constituted four blocks (numbered 1–4 from 5′ to 3′) of uninterrupted strong linkage disequilibrium (LD), within which all values of D′ between markers met or exceeded 0.90. Blocks 2–4 also appeared to be subsumed under a larger, higher order block of reasonably strong LD (with interspersed tracks of uninformative markers), which encompassed exons 2–7 and much of the first intron of the gene. (b) Two of four blocks of strong LD were spanned by multi-marker haplotypes that showed nominally significant evidence for association with schizophrenia (*uncorrected Ps < 0.05), and both of these remained significant after corrections for multiple testing were applied to maintain the family-wise type-I error rate (FWER) at 5% (denoted by bold font). Of the 21 SNPs that met the distributional requirements for testing dominance models, nine showed nominally significant evidence for association with schizophrenia in phase I (*Ps < 0.05) and two remained significantly associated with the disorder in phase II after correcting for multiple testing of 12 optimally powered SNPs and maintaining the FWER at 5% using the Nyholt method (Ps < 0.0053, bold).
In addition to these four uninterrupted blocks of strong LD, a larger, higher order block of reasonably strong LD appeared to subsume blocks 2–4 and encompass exons 2–7 and much of the first intron of the gene. However, this large block was interspersed with tracks of uninformative markers having high values of D′ but low LOD scores. The low confidence in D′ suggested by these low LOD scores likely contributed to the fragmentation of this large block into three smaller blocks, each of which had a more uniform pattern of strong LD between markers and higher confidence in their associated D′ values.
The patterns of LD we report here for our Han Chinese sample from Taiwan strongly resemble those observed in the sample of 45 Han Chinese subjects from Beijing evaluated as part of the International HapMap Project. In particular, the same large over-arching block of relatively strong LD is present in both samples, spanning approximately the same distance (32 kb in our sample and 33 kb in the HapMap sample) and demarcated at its 5′ end by the identical marker (rs4648319). Both samples also show evidence of a recombination hot spot in the vicinity of marker rs12574471, which separated the largest LD block in both samples (described above) from the second largest block; this second largest block was also demarcated at its 5′ end by the same marker (rs11214613) in both samples.
Family-based association analyses of haplotypes
Table 2 shows the frequencies of all LD block-spanning haplotypes that were identified in 10 or more informative families. Fourteen distinct five-marker haplotypes were observed in block 1 and, as seen in Figure 1 (‘LD block haplotypes’ column), the haplotype with the strongest evidence for association did not attain even nominal significance (P = 0.0578). In block 2 (which was the largest block of strong LD), 10 eight-marker haplotypes were detected but none attained even a nominal level of significance. In block 3, the most common three-marker haplotype (A-C-G; frequency = 0.544) was significantly associated with schizophrenia (P = 0.0047), and remained reliably associated even after a correction for multiple testing was applied (critical P: 0.05/3 = 0.0177). In block 4 (which contained the most strongly implicated marker from previous work (rs1801028; Ser311Cys)), five distinct haplotypes were detected, and one relatively rare four-marker haplotype (G-A-C-T; frequency = 0.067) was significantly associated with the disorder (P = 0.0023), remaining so even after correcting for multiple testing (critical P 0.05/4 = 0.0125).
Table 2.
DRD2 LD block haplotype frequenciesa
| Block | Haplotypeb | Frequency |
|---|---|---|
| 1 | GTCCT | 0.328 |
| GAAGT | 0.252 | |
| GACGT | 0.185 | |
| AAAGC | 0.113 | |
| GAAGC | 0.041 | |
| GACCT | 0.028 | |
| GTCGT | 0.011 | |
| GTACT | 0.009 | |
| GAACT | 0.006 | |
| AAAGT | 0.005 | |
| GTAGT | 0.003 | |
| AACGC | 0.003 | |
| ATCCC | 0.002 | |
| ATACC | 0.002 | |
| 2 | CGAGGGTC | 0.328 |
| TAGTGTCC | 0.313 | |
| CGGGGGCC | 0.150 | |
| CGAGGGCC | 0.056 | |
| CAGTAGCT | 0.054 | |
| CGGTGTCC | 0.026 | |
| TAGTGTTC | 0.012 | |
| CGGGGGTC | 0.007 | |
| TAATGTTC | 0.003 | |
| TAATGTCC | 0.003 | |
| 3 | ACG | 0.544 |
| GCT | 0.397 | |
| ATG | 0.030 | |
| GCG | 0.013 | |
| ACT | 0.012 | |
| GTT | 0.002 | |
| 4 | GAAT | 0.455 |
| GACG | 0.414 | |
| GACT | 0.067 | |
| CAAT | 0.043 | |
| GAAG | 0.018 |
Abbreviations: DRD2, dopamine receptor D2; LD, linkage disequilibrium.
Frequencies may not total 1.000, as only haplotypes with 10 or more informative families were analyzed.
Markers are in the order from 5′ to 3′, as in Figure 1.
Family-based association analyses of SNPs
Analyses of dominance models were precluded for two of the three nonsynonymous SNPs, including rs1110977, which was totally monomorphic for the A allele, and rs1800496, which was nearly monomorphic for the C allele (only two CT heterozygotes and no TT homozygotes were observed in our sample). The remaining 21 SNPs met the minimum distributional requirements for testing dominance models (that is, a minimum of 10 informative families and all three possible genotypes at each SNP were detected), the results of which are displayed in Figure 1b (‘SNPs’ column). Nine of these 21 SNPs (43%) displayed nominally significant evidence (Ps < 0.05) for association with schizophrenia; however, we did not find a significant effect of rs1801028, which corresponds to the Ser311Cys polymorphism. The nonsignificant effect of this SNP was verified by TDT analysis, in which the Ser allele was transmitted 36 times and the Cys allele was transmitted 34 times from heterozygous parents to affected probands (OR = 1.0, P = 0.8111).
Of the 21 polymorphisms evaluated, 12 SNPs were determined to have optimal power based on observed allele frequencies and rates of heterozygosity, and thus were retained for phase II where the P-values for these SNPs were compared to a critical P-value that was adjusted for the number of optimally powered comparisons performed. On the basis of the patterns of LD among the retained SNPs, the resulting critical P-value for declaring Bonferroni-corrected statistical significance was determined to be 0.0053. It is worth noting that two of the nine nominally significant SNPs (rs2283265 and rs1079727) were judged to be significant after correcting for multiple comparisons. The risk alleles of these two SNPs were also part of the significantly associated three-marker haplotype that spanned block 3 (described above). The effect size associated with the risk alleles of rs2283265 and rs1079727 (determined by TDT analysis) was an OR of 1.1, which was not significant for either marker (Table 3); however, it should be noted that these TDT analyses included far fewer subjects and families than our primary analyses with PBAT and thus had lower power to detect statistically significant evidence for association.
Table 3.
Results of TDT analyses of DRD2 SNPs significantly
| SNP | Allele | T/NTa | χ2 | P | OR |
|---|---|---|---|---|---|
| rs2283265 | G | 282/256 | 1.26 | 0.262 | 1.1 |
| T | 256/282 | ||||
| rs1079727 | A | 281/254 | 1.36 | 0.243 | 1.1 |
| G | 254/281 |
Abbreviations: DRD2, dopamine receptor D2, NT, not transmitted; SNPs, single-nucleotide polymorphisms; T, transmitted; TDT, transmission disequilibrium testing.
Discussion
The objectives of this study were threefold. First, we endeavored to replicate in a family-based study the earlier evidence from case–control studies for an association of schizophrenia with the Ser311Cys polymorphism of DRD2. Second, we sought to establish the degree of LD between the Ser311Cys variant and 23 other SNPs throughout the gene, which captures all known nonsynonymous mutations and the full haplotype diversity of the gene in the Han Chinese population. Third, we aimed to more precisely specify the locus within DRD2 that influences risk for schizophrenia by examining patterns of association with all 24 polymorphisms and haplotypes comprised of pairs of these SNPs. With regard to the first objective, we were unable to directly replicate the finding of association between schizophrenia and the Cys(G) allele of rs1801028. Because of the very low frequency of the Cys allele in our sample (f = 0.04), few families (n = 62) were informative for the analysis of this polymorphism and our power to detect an association of the magnitude previously reported was severely limited (1 − β = 0.05), despite our relatively large overall sample size. However, this marker (and the G allele in particular) was part of a four-marker haplotype that was found to be significantly associated with schizophrenia, a result that persisted even after corrections for multiple testing were applied. In addition (and perhaps of more importance), we successfully accomplished our second and third objectives, and in doing so have observed evidence of an association between schizophrenia and numerous DRD2 variants, some of which are in novel areas of the gene not previously implicated in the disorder and some of which are in the same haplotype block and/or in strong LD with rs1801028 (D′ > 0.90); however, the r2 values for these comparisons were uniformly low (0.02–0.03), suggesting that the implicated SNPs actually accounted for little of the variance in rs1801028.
Although the evidence for association observed in this study is quite strong (with numerous SNPs and haplotypes achieving statistical significance even after corrections for multiple testing were applied), it is well worth noting that the effect sizes attributable to these significantly associated SNPs were quite small, with ORs of only 1.1. This finding provides further support for a complex multifactorial etiology of schizophrenia in which no gene is either necessary or sufficient to cause the illness. The relatively small magnitude of the observed effects may be a common feature of other schizophrenia risk genes as well, underscoring a general need for association analyses of schizophrenia (and other psychiatric and common complex diseases) to be conducted in large samples that have adequate power to detect polymorphisms that increase risk by as little as 10%.
On the basis of the clear biological relevance of the D2 dopamine receptor in schizophrenia, DRD2 was initially proposed as a candidate gene for the disorder based on functional grounds. Yet, DRD2 maps to human chromosome 11q23 (specifically, 105.17cM on the Marshfield map), and therefore might be considered a strong candidate gene on positional grounds as well.31,32 In fact, the evidence for linkage with a putative schizophrenia risk gene at this locus shows an interesting analogy to the body of allelic association evidence for DRD2 itself. Thus, although chromosome 11q23 has not been implicated as a strongly linked locus in most individual genome-wide scans of schizophrenia, the cumulative evidence observed by meta-analysis implicates this locus quite strongly.33 Whether or not DRD2 contributes to the observed linkage with schizophrenia near this region on chromosome 11 remains to be determined, but our data support this possibility.
Despite the strong evidence we have uncovered in support of DRD2 as a risk gene for schizophrenia, the results of this study must be viewed in the context of several potential limitations. First, we did not have the capacity to replicate our findings in an independent sample of families, so the generalizibility of our findings can not yet be established. The ability to generalize our findings may also be hampered by potential clinical (and etiologic) differences between our subjects and the larger population of schizophrenia patients, which may have been introduced by our sampling strategy. Because this was a family-based study, all probands were required to be members of relatively intact families (or at least know how to contact their family members) in order for the pedigree to become enrolled; however, the factors that allow these families to stay intact (for example, high levels of functioning or low levels of paranoia among the patients) may themselves relate to DRD2 polymorphisms, in which case our results might not extend to schizophrenia patients in the general population who present with the alternate clinical profiles. Another possible limitation of this study may relate to our choice of methods for the analysis of family-based data. Thus, although the PBAT algorithm minimizes the influences of admixture and other sources of population stratification on the type-I error rate, it does not completely eradicate them; however, our analyses using the TDT methodology, which more fully neutralizes these potential sources of bias, supported the findings of our analyses using PBAT. A final limitation of our study is that we were unable to draw definitive conclusions regarding several SNPs (including the Ser311Cys polymorphism and both other nonsynonymous SNPs) due to the very low frequencies of their minor alleles and the low levels of inferential power that these afforded; but this limitation may be mitigated to some degree by the wealth of information we were able to derive about these SNPs indirectly by our examination of the LD structure of the gene and our association analyses utilizing haplotypes.
The work reported here (and the limitations recognized above) immediately suggests several avenues for follow-up study. First and foremost, because none of the associated SNPs or haplotypes from our study has been previously investigated in relation to risk for schizophrenia, these results should be replicated, preferably in another large, adequately powered sample of families with multiple members affected with schizophrenia. Such a sample provides the best chance for duplication of our findings, not only due to its likely constitutional similarity to the present sample, but also because such multiply affected families might be enriched for all schizophrenia risk genes, including but not limited to the risk-conferring variants of DRD2 described here. Replication of these findings in a very large case–control study would also be desirable, since this would afford the opportunity to utilize genomic control34 to account for any residual biases in our data that might be attributable to population stratification. Also, because most of the evidence associating DRD2 polymorphisms with schizophrenia (including that reported here and by Arinami et al.2) has been observed in East Asian samples, both family-based and case–control replication studies should sample other major ancestral groups worldwide to determine if the effects of DRD2 on risk for schizophrenia are population-specific or, as suggested by our previous meta-analysis,5 uniform across populations.
Further follow-up work should also investigate the functional ramifications, if any, of the most strongly implicated SNPs. Although some of our most strongly associated haplotypes encompassed exons 3–7, none of our best candidate polymorphisms are ‘functional’ in the traditional sense (that is, none are nonsynonymous or synonymous exonic SNPs, are located at intron/exon boundaries, are located within exonic splicing enhancers or silencers, are in the gene’s promoter or at other transcription factor-binding sites or are recognized microRNA-binding sites). It is still possible that these SNPs somehow regulate the activity of the gene, for example, by creating alternate secondary structures or influencing interactions with transcriptional machinery; however, a more tenable hypothesis is that these SNPs and haplotypes are actually markers for nearby causal SNPs in their respective blocks of strong LD. For example, the synonymous C957T variant (rs6277), which affects mRNA stability and receptor synthesis, resides between two markers (rs1801028 and rs1110977) in LD block 4 and thus may be responsible for the association signal we have detected with the four-marker haplotype that spans this block. The associated region of DRD2 also resides within an extended block of strong LD with neighboring genes (ANKK1 and TTC12) and the previously implicated DRD2/ANKK1 Taq1A variant, which could also contribute to or account for the detected association signals. In contrast, the potentially functional −141C insertion/deletion polymorphism of DRD2 belongs to an extended region of block 1, which was not significantly associated with schizophrenia in our study; however, this does not preclude the possibility that this or any other DRD2 variant not genotyped in our study is a risk-conferring polymorphism for schizophrenia in our sample or in the broader population.
Another possibility is that the responsible SNPs in the vicinity of our associated htSNPs have thus far eluded detection, either because they have not yet been directly evaluated in large enough samples or because they simply await discovery. Therefore, another objective for future studies will be to resequence the relevant portions of the gene flanking the most strongly associated markers (as well as the potentially important promoter, 5′ and 3′ regions of the gene that we have not yet evaluated), with the intention of discovering novel SNPs that might have direct consequences in either the structure or the function of the gene. Once discovered, such polymorphisms should also be directly tested for allelic and genotypic association with the illness, at which point they would be expected to exhibit even stronger and more reliable effects on risk for schizophrenia than the polymorphisms reported on presently. If the associations we have detected are ultimately replicated and validated, DRD2 may begin to fulfill its long-anticipated promise as a genetic marker suitable for risk profiling, early identification and intervention and prevention efforts, or the development of molecular therapeutics for schizophrenia.
Supplementary Material
Acknowledgments
This work was supported in part by a Young Investigator Award from NARSAD, The Mental Health Research Association (SJG), a research project grant (1R01MH059624) from the United States National Institute of Mental Health (MTT) and grants (90-8825PP and 91,92-9113PP) from the National Health Research Institute of Taiwan and the Genomic Medicine Research Program of Psychiatric Disorders of National Taiwan University Hospital. We thank Allison Brown and Maura Regan at the HPCGG for coordinating and performing the genotyping for this project, Sharon Chandler at UCSD for technical assistance, Frank Middleton at SUNY Upstate Medical University for assistance with detecting pedigree inconsistencies and Mendelian errors and Vural Ozdemir at the University of California, Irvine, for critical reading and editing of the manuscript. We further thank our research and clinical collaborators in the Taiwan Schizophrenia Linkage Study Group, including Chih-Min Liu, Wei J Chen, Ming-Ming Tsuang, Shih-Kai Liu, Ming-Hsien Shieh, Tzung-Jeng Hwang, Wen-Chen Ou-Yang, Chun-Ying Chen, Chwen-Cheng Chen, Jin-Jia Lin, Frank Huang-Chih Chou, Ching-Mo Chueh, Wei-Ming Liu, Mei-Hua Hall, Chiao-Chicy Chen, Jia-Jiu Lo, Jia-Fu Lee, Seng Shen, Yung Feng, Shin-Pin Lin, Shi-Chin Guo, Ming-Cheng Kuo, Liang-Jen Chuo, Chih-Pin Lu, Deng-Yi Chen, Huan-Kwang Ferng, Nan-Ying Chiu, Wen-Kun Chen, Tien-Cheng Lee, Hsin-Pei Tang, Yih-Dar Lee, Wu-Shih Wang, For-Wey Long, Tiao-Lai Huang, Jung-Kwang Wen, Cheng-Sheng Chen, Wen-Hsiang Huang, Shu-Yu Yang and Cheng-Hsing Chen. Finally, we thank the hospitals that participated in this study, including National Taiwan University Hospital and Medical College of National Taiwan University, National Taoyuan Psychiatric Center, National Tsaotun Psychiatric Center, National Cheng-Kung University, Kai-Suan Psychiatric Hospital of Kaohsiung City, Yu-Li Veterans Hospital and National Yu-Li Hospital.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Itokawa M, Arinami T, Futamura N, Hamaguchi H, Toru M. A structural polymorphism of human dopamine D2 receptor, D2(Ser311 → Cys) Biochem Biophys Res Commun. 1993;196:1369–1375. doi: 10.1006/bbrc.1993.2404. [DOI] [PubMed] [Google Scholar]
- 2.Arinami T, Itokawa M, Enguchi H, Tagaya H, Yano S, Shimizu H, et al. Association of dopamine D2 receptor molecular variant with schizophrenia. Lancet. 1994;343:703–704. doi: 10.1016/s0140-6736(94)91581-4. [DOI] [PubMed] [Google Scholar]
- 3.Verga M, Macciardi F, Pedrini S, Cohen S, Smeraldi E. No association of the Ser/Cys311 DRD2 molecular variant with schizophrenia using a classical case control study and the haplotype relative risk. Schizophr Res. 1997;25:117–121. doi: 10.1016/S0920-9964(97)00013-3. [DOI] [PubMed] [Google Scholar]
- 4.Glatt SJ, Faraone SV, Tsuang MT. Meta-analysis identifies an association between the dopamine D2 receptor gene and schizophrenia. Mol Psychiatr. 2003;8:911–915. doi: 10.1038/sj.mp.4001321. [DOI] [PubMed] [Google Scholar]
- 5.Glatt SJ, Jönsson EG. The Cys allele of the DRD2 Ser311Cys polymorphism has a dominant effect on risk for schizophrenia: evidence from fixed- and random-effects meta-analyses. Am J Med Genet B Neuropsychiatr Genet. 2006;141:149–154. doi: 10.1002/ajmg.b.30273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seeman P, Schwarz J, Chen JF, Szechtman H, Perreault M, McKnight GS, et al. Psychosis pathways converge via D2 high dopamine receptors. Synapse. 2006;60:319–346. doi: 10.1002/syn.20303. [DOI] [PubMed] [Google Scholar]
- 7.Cravchik A, Sibley DR, Gejman PV. Functional analysis of the human D2 dopamine receptor missense variants. J Biol Chem. 1996;271:26013–26017. doi: 10.1074/jbc.271.42.26013. [DOI] [PubMed] [Google Scholar]
- 8.Hwu HG, Faraone SV, Liu CM, Chen WJ, Liu SK, Shieh MH, et al. Taiwan schizophrenia linkage study: the field study. Am J Med Genet B Neuropsychiatr Genet. 2005;134:30–36. doi: 10.1002/ajmg.b.30139. [DOI] [PubMed] [Google Scholar]
- 9.Cloninger CR. Schizophrenia: genetic etiological factors. In: Kaplan HI, Sadock BJ, editors. Comprehensive Textbook of Psychiatry. Williams and Wilkins; Baltimore: 1989. [Google Scholar]
- 10.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- 11.Chen WJ, Faraone SV. Sustained attention deficits as markers of genetic susceptibility to schizophrenia. Am J Med Genet. 2000;97:52–57. doi: 10.1002/(sici)1096-8628(200021)97:1<52::aid-ajmg7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 13.NIMH Genetics Initiative. Family Interview for Genetic Studies. National Institute of Mental Health; Rockville: 1992. [Google Scholar]
- 14.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 15.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Oeth P, Beaulieu M, Park C, Kosman D, del Mistro G, van den Boom D, et al. Sequenom Application Note. Sequenom Inc; San Diego, CA: 2005. iPLEX™ assay: increased plexing efficiency and flexibility for MassARRAY system through single base primer extension with mass-modified terminators; pp. 1–12. [Google Scholar]
- 18.Beaulieu M, Hong P. Sequenom Application Note. Sequenom Inc; San Diego, CA: 2004. Multiplexing the homogeneous MassEXTEND™ assay; pp. 1–10. [Google Scholar]
- 19.Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy–Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 22.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype—phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 24.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 25.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 26.Lange C, DeMeo DL, Laird NM. Power and design considerations for a general class of family-based association tests: quantitative traits. Am J Hum Genet. 2002;71:1330–1341. doi: 10.1086/344696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whittaker JC, Lewis CM. The effect of family structure on linkage tests using allelic association. Am J Hum Genet. 1998;63:889–897. doi: 10.1086/302008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, Demeo DL, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet. 2005;37:683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 29.Murphy A, McQueen MB, Su J, Kraft P, Lazarus R, Laird NM, et al. Genomic screening in family-based association testing. BMC Genet. 2005;6(Suppl 1):S115. doi: 10.1186/1471-2156-6-S1-S115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulayeva KB, Glatt SJ, Bulayev OA, Pavlova TA, Tsuang MT. Genome-wide linkage scan of schizophrenia: a cross-isolate study. Genomics. 2007;89:167–177. doi: 10.1016/j.ygeno.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21–22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3–24 and 20q12.1–11.23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.