Abstract
In Escherichia coli and Bacillus subtilis, essentiality, not expressivity, drives the distribution of genes between the two replicating strands. Although essential genes tend to be coded in the leading replicating strand, the underlying selective constraints and the evolutionary extent of these findings have still not been subject to comparative studies. Here, we extend our previous analysis to the genomes of low G + C firmicutes and γ-proteobacteria, and in a second step to all sequenced bacterial genomes. The inference of essentiality by homology allows us to show that essential genes are much more frequent in the leading strand than other genes, even when compared with non- essential highly expressed genes. Smaller biases were found in the genomes of obligatory intracellular bacteria, for which the assignment of essentiality by homology from fast growing free-living bacteria is most problematic. Cross-comparisons used to assess potential errors in the assignment of essentiality by homology revealed that, in most cases, variations in the assignment criteria have little influence on the overall results. Essential genes tend to be more conserved in the leading strand than average genes, which is consistent with selection for this positioning and may impose a strong constraint on chromosomal rearrangements. These results indicate that essentiality plays a fundamental role in the distribution of genes in most bacterial genomes.
INTRODUCTION
The concept of an ‘essential’ gene is ambiguous, since all genes needed for the basic supplies of indispensable cell metabolites are essential under special circumstances. However, when one uses a medium enriched in all presumably necessary components, essentiality pertains to the core of the cell’s functions, in particular to the processes involved in information transfer and compartmentalisation. The availability of complete genome sequences and high-throughput techniques has allowed the definition of the set of essential genes in several bacteria. This has been achieved through different techniques. Saturation transposon mutagenesis, followed by sequencing, allowed the definition of essential genes in Mycoplasma genitalium (1) and Haemophilus influenzae (2). Antisense RNA has been used to specifically silence genes in Staphylococccus aureus (3). These techniques are fast and easy to implement, especially in bacteria that are hard to cultivate. However, they require important resources, and tend to over-represent the number of essential genes because of polarity of transcription in operons and insertion biases. For example, these studies identified as many as 658 essential genes in S.aureus and 478 in H.influenzae, whereas precise systematic gene inactivation in the larger genome of the free-living Bacillus subtilis resulted in the identification of only 277 essential genes (4). In Escherichia coli, an equivalent published work is still unavailable, but the PEC database at the Japan National Institute of Genetics provides extensive information on essentiality for about two-thirds of its genes. This database classifies 206 genes as essential in E.coli, of which 82% are also classified as essential in B.subtilis. The study of essentiality provides important information on cell functioning; it can be used to develop new antibiotic targets, and is of substantial evolutionary interest. For example, it is currently a subject of intense debate whether essentiality plays an important role in constraining the evolutionary rate of proteins (5–8). Also, the patterns of loss of ‘essential’ genes in the process of genome degradation can be important in the understanding of the co-evolution of bacteria and their hosts (9).
The existence of a larger number of genes in the leading strand, relative to the lagging strand, has been known for two decades. It was first observed that rDNA and ribosomal proteins are systematically coded in the leading strand of the E.coli chromosome (10). This was speculated to result from selection to prevent collisions between DNA polymerase (DNAP) and RNA polymerase (RNAP) (11). DNA replication and transcription occur simultaneously on the same DNA molecule. DNAP in E.coli proceeds 10–20 times faster than RNAP (12), and both head-on and co-oriented collisions will often occur in replicating bacteria. However, the outcome of the collision is different depending on whether the polymerases are co-oriented or not, resulting in transcription abortion and more severe replication slow-down when genes are in the lagging strand (13,14). Thus, the preferential positioning of translation-related highly expressed genes (rDNA and ribosomal proteins) in the leading strand has been widely deemed to result from selection for lower transcription abortion and higher replication rates. The observations that genes coding for rDNA and ribosomal proteins are in the leading strand of most bacterial genomes seemed to confirm this hypothesis (15,16). As selection would only be effective for highly expressed genes and acts on the positioning of genes in the leading strand, this would create a gene strand bias proportional to the expression level in replicating bacteria. The bias should also be proportional to the frequency of replication. Fast-growing bacteria should therefore have a higher strand bias.
Several lines of evidence have recently challenged the emphasis of this model on replication slow-down as the basis of gene strand bias. First, gene strand bias strongly depends on the composition of DNAP, with bacteria containing two different α-subunits exhibiting much higher strand biases (17). In these bacteria, where each α-subunit seems to be dedicated to the replication of one DNA strand (18), an average of 78% of the genes are coded in the leading strand, many of which are not highly expressed. Secondly, some of the largest gene strand biases are found in slow-growing bacteria. Among bacteria containing two different DNAP α-subunits, the slow-growing M.genitalium has one of the highest biases (80% of genes in the leading strand), whereas the fast-growing B.subtilis has one of the lowest (74%). For genomes being replicated with only one gene coding for the DNAP α-subunit, the slow-growing Borrelia burgdorferi shows the highest bias (65%). Comparing equally sized chromosomes, one finds that the slow-growing Mycobacterium tuberculosis (59%) is more biased than the fast-growing E.coli (55%). Thirdly, we have recently shown that gene strand bias in B.subtilis and E.coli stems from the ‘essentiality’ of the genes. Controlling for essentiality revealed no significant role for expression levels in gene strand bias (19). This suggests that anti-oriented collisions are selected against due to problems associated with the existence of aborted transcripts, not replication pausing. It also suggests that essentiality is a major determinant of the chromosome structure. Rearrangements resulting in important inversions of leading strands are often extremely deleterious (20,21). The strand switch of essential genes may be partly responsible for these observations.
To substantiate our previous observation concerning essential genes, which was limited to two fast-growing bacteria with similar genome sizes, we tested if essentiality plays a similar role in other bacterial genomes. Putative essentiality was identified by homology from the genomes of B.subtilis (for low G + C firmicutes) or E.coli (for γ-proteobacteria). As a working hypothesis, we made several simplifying assumptions. First, we defined a set of putative orthologues in all genomes, using reciprocal best hits. Secondly, we assumed that the orthologues of essential genes are likely to be also essential in the other genomes. Thirdly, we defined putative highly expressed genes in each genome with codon adaptation index (CAI) values computed from ribosomal proteins (22). At this point, we tested four different hypotheses concerning the higher frequency of genes in the leading strand. First, we tested whether essential genes are more frequent in the leading strands than non-essential genes. Secondly, we tested if among the non-essential genes, highly expressed genes are more frequent in the leading strand than the other genes. Thirdly, we tested if non-highly expressed essential genes are more biased than highly expressed non-essential genes. Fourthly, we tested if the conservation of essential genes in the leading strand is more important than that of the other genes. To check if the difficulties associated with the assignment of essentiality by homology were biasing our results, we made a set of tests and cross-comparisons with the available experimental data on B.subtilis, E.coli, M.genitalium and H.influenzae.
MATERIALS AND METHODS
Data
Sequence data and the corresponding annotations were taken from GenBank Genomes (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). The following genomes of low G + C firmicutes were analysed: B.subtilis 168, Bacillus halodurans, Clostridium acetobutylicum ATCC824, Clostridium perfringens 13, Lactococcus lactis IL1403, Listeria inocua Clip11262, Listeria monocytogenes EGD, Oceanobacillus iheyensis, Staphylococcus aureus N315, Streptococcus agalactiae serotype V, Streptococcus pneumoniae TIGR4, Streptococcus pyogenes M1 and Thermoanaerobacter tengcongensis. The following genomes of γ-proteobacteria were analysed: E.coli MG1655, Buchnera Ap, H.influenzae Rd, Pasteurella multocida PM70, Pseudomonas aeruginosa PA01, Pseudomonas putida KT2440, Salmonella enterica Typhimurium LT2, Shewanella oneidensis MR-1, Xanthomonas axonopodis, Xanthomonas campestris, Xylella fastidiosa 9a5c and Yersinia pestis CO92. Because different strains of the same species are mostly co-linear, only one strain for each species was analysed. Essentiality data were taken from the literature for the following genomes: B.subtilis (4), M.genitalium (1), H.influenzae (2) and E.coli (http://www.shigen.nig.ac.jp/ecoli/pec/). Identification of origins of replication and leading strands was done through the use of linear discriminant analysis and GC skews, as detailed elsewhere (23).
Analysis of orthology
Orthologues were identified through reciprocal best hits. First, a BlastP search identified the 10 best hits of each protein in the other genome. These proteins were then aligned using a global alignment where the end gaps facing the largest sequence are ignored. Two proteins were regarded as orthologues if they are reciprocal best hits with at least 40% similarity in amino acid sequence and <20% of difference in protein length (24). Essentiality was derived by homology from B.subtilis to the genomes of low G + C firmicutes and from E.coli to the genomes of γ-proteobacteria. In a second step, we analysed the set of complete bacterial genomes for which an origin and terminus of replication can be reliably identified. In this case, essentiality was derived by homology from the B.subtilis subset. The classifications of the E.coli and B.subtilis genes that were used as reference can be obtained at http://wwwabi.snv.jussieu.fr/~erocha/essential/ and as Supplementary Material at NAR Online.
Analysis of codon usage
CAI values were computed using the EMBOSS package (http://www.uk.embnet.org/Software/EMBOSS). The reference values of codon usage in highly expressed genes (understanding high expression as that when cells are growing exponentially) were computed using ribosomal proteins (22). It has recently been shown in yeast that the correlation between CAI (defined using ribosomal proteins) and transcriptome data was good, and as good as the different transcriptome data sets among themselves (under the same experimental conditions) (25). Another recent work indicated that CAI values are very robust to small changes in the learning set (26). We have previously checked that transcriptome and proteome data, when available, confirmed our analysis of gene strand bias made with CAI values (19). Here, a gene was regarded as potentially highly expressed if its CAI is among the 10% highest values of the genome. The use of a 10% threshold is somewhat arbitrary, although it reflects the results of classifications made by several authors, using CAI or factorial analysis, indicating that 5–10% of genes are particularly highly expressed (27–29). We have shown previously that varying this threshold in the range 5–15% did not change the results significantly (19).
Tests on CAI
Codon usage bias may not be a good estimator of gene expression in some bacteria (30). Hence, we designed a set of tests to verify if the codon usage of ribosomal proteins is significantly biased, and if the codon usage classification of the genome using this set is more discriminant than using a random set of genes. For this, we made 100 random experiments. In each, we randomly took 50 genes (corresponding to the number of ribosomal proteins used in the previous analysis) to build the reference table of codon usage. Using this table, we computed CAI values for all the genes and then the frequency of the 50 initial genes among the 10% highest CAI. A CAI indicating significant codon usage bias should have high consistency (i.e. most of the genes initially hypothesised to be highly expressed should be classed as highly expressed) and a genome average value smaller than expected by chance (to discriminate the minority of highly expressed genes from the rest of the genome). Significant codon usage bias in ribosomal proteins met three conditions. First, >40% of ribosomal proteins are classed as highly expressed in the top 10% CAI values. Secondly, the frequency of ribosomal proteins classed in the top 10% CAI values must be significantly higher than the one obtained in the random experiments for the genes used to build the codon index (significant difference was taken to be >3 SDs away from the mean of the random experiments). Thirdly, the average CAI of all genes, when the CAI is computed using ribosomal proteins, must be significantly lower than that obtained in the random experiments (>3 SDs away).
Statistical tests
We have performed several types of statistical tests adapted to different situations. To test if two groups have different frequency of leading strand genes in a genome, we carried out two-tailed χ2 tests on 2 × 2 contingency tables. Strand conservation of groups of genes between genomes was computed as the frequency of orthologues that are in the same replicating strand in the genomes. We then tested if this value is different from that expected given the average strand conservation of the orthologues. This test consisted of two-tailed χ2 tests on 2 × 2 contingency tables where the comparisons were made between the class of orthologous essential genes versus all remaining orthologues. Because for each type of the two latter tests there are multiple tests being done (one for each genome), we used a Bonferroni correction within each taxonomic group. Thirdly, to test if among all sequenced genomes one group of genes is more biased than another, we performed one single paired t-test. In this case, only one test is carried out for each hypothesis and thus no Bonferroni correction was used.
Results
Distribution of essential genes between replicating strands
Low G + C firmicutes. We explored the link between essentiality and gene strand bias in genomes of the closely related low G + C firmicutes (excluding the more distant Mycoplasmas). For this, we identified orthologues between B.subtilis and the other genomes, assigning essentiality according to the B.subtilis classification. Then we performed three tests. First, we tested if essential genes are more frequently found in the leading strand than non-essential genes. This was found to be the case for all genomes (Table 1). Therefore, essentiality plays an important role in gene strand bias in low G + C firmicutes. Secondly, we tested if among the non-essential genes, highly expressed genes are more frequent in the leading strand than non-highly expressed genes. This test only identified one case of significantly higher frequency of highly expressed genes in the leading strand (C.acetobutylicum) (Table 1). Therefore, expression levels do not seem to contribute significantly to gene strand bias, when essentiality is taken into account. Thirdly, we tested whether essential non-highly expressed genes are more biased than non-essential but highly expressed genes. With the above-mentioned exception of C.acetobutylicum, for which the difference is not statistically significant, this test showed that this was generally true.
Table 1. Distribution of genes in the leading strands of the genomes.
| Genome | Genes | % Genes lead | Statistical tests | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EH | EnH | nEH | nEnH | EH | EnH | nEH | nEnH | Ess | Exp | EvH | SC | CB | |
| B.subtilis | 78 | 199 | 119 | 3710 | 94 | 94 | 72 | 74 | + | 0 | + | + | |
| B.halodurans | 240 | 15 | 1503 | 402 | 93 | 93 | 81 | 79 | + | 0 | + | + | + |
| C.acetobutylicum | 218 | 10 | 918 | 251 | 93 | 91 | 91 | 80 | + | + | 0 | + | + |
| C.perfringens | 208 | 9 | 770 | 196 | 95 | 93 | 88 | 85 | + | 0 | + | + | + |
| L.lactis | 225 | 11 | 698 | 186 | 88 | 89 | 74 | 81 | + | 0 | + | + | + |
| L.inocua | 242 | 16 | 1069 | 255 | 95 | 94 | 75 | 80 | + | 0 | + | + | + |
| L.monocytogenes | 243 | 18 | 1035 | 243 | 96 | 93 | 73 | 79 | + | 0 | + | + | + |
| O.iheyensis | 248 | 14 | 1366 | 395 | 96 | 93 | 79 | 76 | + | 0 | + | + | + |
| S.agalactiae | 219 | 11 | 665 | 186 | 93 | 92 | 76 | 79 | + | 0 | + | + | + |
| S.aureus | 237 | 13 | 956 | 261 | 95 | 89 | 75 | 75 | + | 0 | + | + | + |
| S.pneumoniae | 214 | 10 | 654 | 153 | 93 | 90 | 72 | 80 | + | 0 | + | + | + |
| S.pyogenes | 208 | 9 | 575 | 131 | 92 | 93 | 78 | 80 | + | 0 | + | + | + |
|
T.tengcongensis |
212 |
11 |
771 |
166 |
100 |
94 |
90 |
87 |
+ |
0 |
+ |
0 |
0 |
| E.coli | 112 | 94 | 249 | 1955 | 80 | 71 | 57 | 56 | + | 0 | + | + | |
| BuchneraAp | 127 | 33 | 169 | 116 | 70 | 47 | 73 | 61 | 0 | 0 | – | 0 | 0 |
| H.influenzae | 149 | 44 | 389 | 289 | 79 | 67 | 52 | 51 | + | 0 | + | + | + |
| P.multocida | 152 | 44 | 450 | 318 | 67 | 72 | 65 | 54 | + | 0 | + | + | + |
| P.aeruginosa | 150 | 40 | 628 | 437 | 75 | 82 | 51 | 59 | + | – | + | + | + |
| P.putida | 145 | 39 | 615 | 422 | 75 | 78 | 54 | 61 | + | – | + | + | + |
| S.enterica | 154 | 49 | 967 | 718 | 80 | 70 | 57 | 57 | + | – | + | + | + |
| S.oneidensis | 148 | 40 | 569 | 393 | 81 | 72 | 62 | 57 | + | + | + | + | + |
| X.axonopodis | 145 | 35 | 474 | 323 | 79 | 77 | 56 | 60 | + | 0 | + | + | + |
| X.campestris | 141 | 34 | 482 | 321 | 79 | 79 | 53 | 59 | + | 0 | + | + | + |
| X.fastidiosa | 139 | 35 | 342 | 216 | 99 | 63 | 98 | 51 | + | + | – | + | + |
| Y.pestis | 156 | 46 | 730 | 567 | 80 | 74 | 59 | 53 | + | + | + | + | + |
The classification of low G + C firmicutes was made by homology from B.subtilis and that of γ-proteobacteria from E.coli. The genes were classed according to expression level and essential phenotype into highly expressed (H) versus non-highly expressed (nH) and essential (E) versus non-essential (nE). The acronyms were concatenated to name the classes (e.g. EnH indicates essential non-highly expressed genes). The columns under ‘Statistical tests’ (two-tail χ2 tests on contingency tables) indicate the significance of the test at P < 0.05, after applying a Bonferroni correction. The + and – signs indicate rejection of the hypothesis (right- and left-side, respectively), and 0 indicates no rejection. The tests correspond to: E ≠ nE (Ess), nEH ≠ nEnH (Exp), EnH ≠ nEH (EvH), larger strand conservation of essential genes (SC) and significant codon usage bias in ribosomal proteins (CB).
γ-Proteobacteria. The same analysis was done for the γ-proteobacteria, starting from the PEC database of E.coli essential genes. This database contains about 1700 genes for which the essentiality character is unknown, and they were removed for simplicity. The resulting sample contains 2410 E.coli genes, which were used to identify orthologous genes in the other γ-proteobacteria. Because of the incomplete classification and the lack of an extensive experimental gene disruption programme in E.coli, this analysis may be more error prone. Still, the results are qualitatively similar to those observed among the low G + C firmicutes (Table 1). With the exception of Buchnera, all other genomes show high relative frequencies of leading strand essential genes. When analysing non-essential genes for gene strand bias as a function of expression levels, three genomes show higher biases among highly expressed genes, three genomes show lower biases among highly expressed genes and four genomes show non-significant differences. Thus, among non-essential genes, expression does not seem to play a significant role in gene strand bias. Finally, nine out of eleven genomes showed a significantly higher frequency of leading strand essential but non-highly expressed genes than non-essential but highly expressed genes. The exceptions to this trend are the two non-free-living Bacteria Buchnera and X.fastidiosa, the former not showing sufficient codon usage bias to reliably determine expression levels.
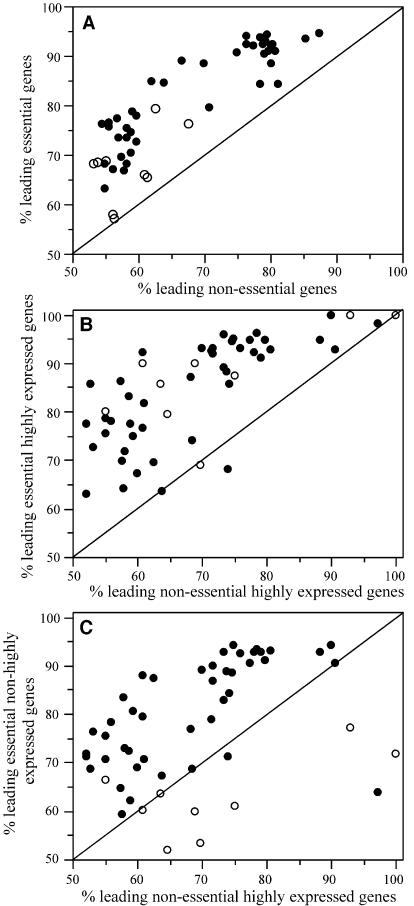
Extension of the procedure. The assignment of essentiality by homology into very distant phylogenetic domains is prone to more important errors, because essentiality is relative to a set of functions and lifestyle and because the definition of orthology becomes increasingly error prone as evolutionary distances increase. In some of these cases, the definition of high expression based on the CAI is also problematic. Still, we made a preliminary analysis of the entire set of sequenced bacteria, including 30 other fully sequenced genomes of different species. For each genome, we determined all orthologues to B.subtilis. These orthologues were then classed as essential or non-essential according to their classification in B.subtilis. The results were qualitatively similar to those presented above. First, essential genes are more biased than non-essential genes in nearly all genomes (Fig. 1). Secondly, among highly expressed genes, essential genes are always more biased than non-essential ones. Thirdly, highly expressed non-essential genes are less biased than essential but not highly expressed genes. In the latter analysis, we observed some exceptions, mostly concerning intracellular bacteria (Buchnera, Chlamydia, Spirochetes and Rickettsia), but also one obligatory pathogen (X.fastidiosa).
Figure 1.
Frequency of leading strand genes in 53 genomes of bacteria. For the bacteria with more than one chromosome, we pooled together the chromosomes. Essentiality is assigned by homology from B.subtilis, and only genes presenting an orthologous gene in B.subtilis are considered in the analysis. (A) Essential versus non-essential genes. (B) Essential versus non-essential genes among putative highly expressed genes. (C) Highly expressed non-essential genes versus essential non-highly expressed genes. The diagonal line indicates a similar frequency in the two sets. Intracellular obligatory pathogens or symbionts are indicated as open circles and the other genomes as full circles. In the three panels, essential genes are more biased than the other set (P < 0.001, paired t-tests).
The positioning of essential genes in the leading strand is highly conserved
These observations suggest that essentiality is important in both low G + C firmicutes and γ-proteobacteria, and possibly in most bacterial domains. However, closely related genomes may resemble E.coli and B.subtilis because of phylogenetic inertia, not because of a common selective pressure for a positioning of essential genes in the leading strand. Some close genomes, such as those of E.coli and Y.pestis (31) or S.agalactiae and S.pneumoniae (32), have been severely shuffled since speciation. Interestingly, genome rearrangements tend to be symmetrical around the origin of replication and thus do not change the replicating strand where the gene is coded, even though they disrupt gene order (33,34). Therefore, we have performed a further test to determine if strand conservation is stronger among essential genes than among other genes (SC column in Table 1). The results indicate that in 21 out of 23 genomes, the conservation of the leading strand character of essential genes is significantly more important that that of the other genes (P < 0.05, after applying the Bonferroni correction for multiple tests) (Table 1). The two exceptions concern Buchnera and T.tengcongensis. In the latter, the conservation is important but, because this genome shows the highest overall strand bias (35), the difference is not significant after application of the Bonferroni correction.
Tests
The previous analyses strongly suggest that in the majority of bacteria, essentiality drives gene strand bias. Yet, assigning essentiality by homology is prone to some errors. For example, all genomes lack some orthologues of the essential B.subtilis genes, indicating their non-essentiality in these bacteria. Methodological problems may also lead to erroneous inference of orthology because of gene duplication or non-orthologous gene displacement (36). Hence, it is important to test the robustness of our assumptions concerning essentiality. The low G + C firmicutes sharing fewer orthologues with B.subtilis essential genes are S.pyogenes and C.acetobutylicum (Table 1). Yet, in both cases, we could identify orthologues to 78% of B.subtilis essential genes. Among γ-proteobacteria, the genome containing fewer orthologues to E.coli essential genes is Buchnera. Buchnera is relatively close to E.coli, but its evolutionary history of extensive genome reduction led to a genome with only approximately 500 genes (37). Although only 13% of the E.coli non-essential genes have an orthologue in Buchnera, 77% of the essential genes have such an orthologue. These results indicate that the majority of essential genes are conserved at these evolutionary distances, even when very few genes are conserved. We then turned to the comparison of the very distantly related E.coli and B.subtilis genomes.
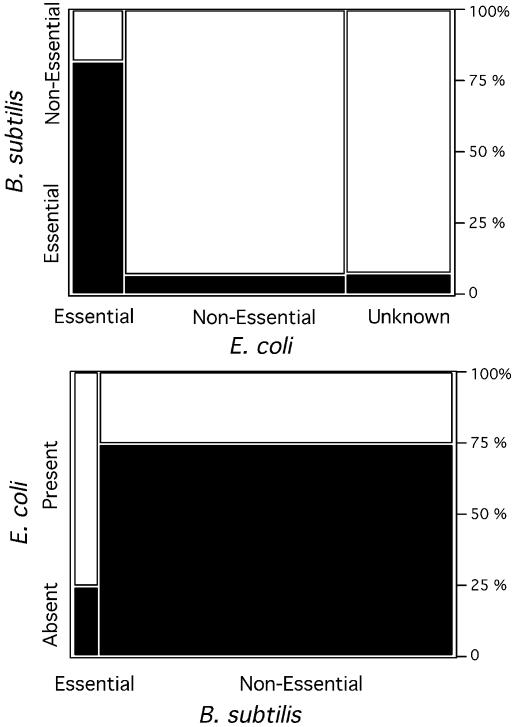
Comparison of essential genes in B.subtilis and E.coli. Among the 1129 orthologues between E.coli and B.subtilis, we found 75% of the B.subtilis essential genes and only 26% of the non-essential genes (Fig. 2). Thus, as expected, the presence of essential genes is much more conserved than that of the other genes at such large evolutionary distances. Further, 82% of the E.coli essential genes are also classed as essential in B.subtilis (and only 7% of non-essential genes are regarded as essential in B.subtilis) (Fig. 2). Given the very large evolutionary distance between these bacteria, the finding that such a large fraction of essential genes is conserved in the two genomes suggests that assigning essentiality by homology is reasonable, at least between free-living bacteria.
Figure 2.
Comparison of the essential genes data set from E.coli and B.subtilis. Upper panel: assignment of essentiality in orthologues (black squares represent essential genes in B.subtilis). Lower panel: presence or absence of orthologues depending on the classification of essentiality in the two genomes (black squares represent genes present in B.subtilis but absent in E.coli). The area of the blocks is proportional to the relative frequency of each category.
Comparisons between experimental data sets. We used the experimental data sets of essential genes in B.subtilis, E.coli, M.genitalium and H.influenzae to make a set of cross-comparisons. In each comparison, we made three different analyses (Table 2). The first analysis consists simply of computing the frequency of leading strand essential and non-essential genes in the genome, where essentiality is taken from the experimental data set [e.g. from the M.genitalium data set in the analysis ‘M.genitalium (Bs)’]. The second analysis consists of computing the frequency of leading strand essential genes in the genome where essentiality is assigned by homology from the other genome [e.g. assigned from homology to B.subtilis in the analysis ‘M.genitalium (Bs)’]. The third analysis consists of computing the frequency of leading strand essential genes where essentiality is assigned only if the gene is essential in the experimental data set and if its orthologue is essential in the other genome [e.g. if the gene is experimentally found to be essential both in M.genitalium and in B.subtilis in the analysis ‘M.genitalium (Bs)’]. The three analyses give consistently the same results, with essential genes being more frequent in the leading strand independently of the information used to define essentiality. Only the analysis of H.influenzae’s own experimental data set indicates no significant difference between essential and non-essential genes. The experimental analysis of H.influenzae identified 474 essential genes among 1272 analysed (i.e. ∼70% of the genome) (2), which is more than twice the essential genes found in B.subtilis (4), and predicted in H.influenzae from computational genome analysis (38). In H.influenzae, essentiality was determined by transposon mutagenesis, which is likely to overestimate the number of essential genes. Unsurprisingly, the use of the E.coli data set or the intersection between the two data sets indicates a significant bias (73% of essential leading strand genes for the latter). This is a general trend: the highest biases are found with the intersection of the data sets (Table 2), probably because in this case the likehood of falsely assigning essentiality is minimised.
Table 2. Cross-comparisons of the frequency of genes in the leading strand when using experimental data sets of essential genes in four genomes.
| Experimental data | Homology | Both | ||||
|---|---|---|---|---|---|---|
| Essential | Non-essential | Essential | Non-essential | Essential | Non-essential | |
| M.genitalium (Bs) | 82% | 75% | 85% | 78% | 86% | 77% |
| H.influenzae (Ec) | 55% | 55% | 70% | 51% | 73% | 53% |
| B.subtilis (Ec) | 94% | 72% | 97% | 83% | 97% | 80% |
| E.coli (Bs) | 76% | 56% | 75% | 56% | 84% | 56% |
Three types of information on essentiality were used to compute the frequency of leading strand essential genes: (i) using the experimental data set for the genome; (ii) by homology to the data set of another genome (Bs, B.subtilis; Ec, E.coli); and (iii) making the intersection of (i) and (ii). For example, M.genitalium shows 82, 85 and 86% of essential genes in the leading strand when essentiality is defined respectively from its experimental data, from homology to essential genes in B.subtilis, and using the intersection of the two criteria (essential in both B.subtilis and M.genitalium).
Testing the degree of gene strand conservation. We also used the four experimental data sets to test the conservation of the leading strand character of essential genes. For this, we compared B.subtilis with the genomes of M.genitalium and E.coli, and the latter with that of H.influenzae. We found that genes essential in both genomes tend to co-occur in the leading strand at a much higher frequency than the other genes (Table 3). For example, the comparison of B.subtilis and E.coli, among which there is almost no gene order conservation (39), shows 73% of essential genes in the leading strand in both genomes. The similar analysis applied to the remaining genes indicates 44% of genes conserved in the leading strand, whereas the expected frequency, taking into account the average gene strand composition of each genome, is 41%. Between B.subtilis and M.genitalium, the difference is smaller (85 versus 70%), but the expected value is also much larger (60%). This conservation of the positioning of essential genes in the leading strand is consistent with the existence of selective forces against the displacement of essential genes to the lagging strand.
Table 3. Percentage of genes in the leading strand of both genomes, according to essentiality.
| Comparison | Leading essential | Leading others | Expected |
|---|---|---|---|
| B.subtilis versus M.genitalium | 85% | 70% | 60% |
| B.subtilis versus E.coli | 73% | 44% | 41% |
| E.coli versus H.influenzae | 60% | 35% | 35% |
A gene is regarded as essential if it is found experimentally to be essential in both genomes.
DISCUSSION
Essentiality drives gene strand bias
As discussed above, several lines of evidence indicate that the positioning of certain genes in the leading strand is subject to selection. Brewer had proposed that such biases were caused by selection for avoidance of frequent head-on collisions between DNAP and RNAP (13). Here, we confirmed our previous analyses, showing that in bacteria, essentiality, not expressiveness, plays the major role in gene strand bias (19). This is compatible with the previous model if its emphasis is shifted from the rate of collisions between polymerases to the transcript resulting from such collisions. Most importantly, it is the gene product, not its expression rate, which is the important selective factor. Thus, understanding the fate of the transcript may cast light on the causes of gene strand bias. A possibility is that, because transcription abortion occurs at lower rates in co-oriented collisions than in head-on collisions (14), truncated transcripts may lead to truncated, and thus non-functional, peptides. When such peptides are parts of large complexes (e.g. the ribosome or DNAP), they are typically the basis for dominant-negative phenotypes. This is naturally most counter-selected among essential functions.
We extended our previous observations concerning gene strand bias in three directions. First, we have shown that in spite of the substantial rearrangements between E.coli and B.subtilis, the leading strand positioning of essential genes is conserved. It is also more conserved than that of the average orthologue. Secondly, our results show that the putatively identified essential genes are preferably located in the leading strand of almost all the analysed genomes of low G + C firmicutes and γ-proteobacteria. Hence, the collision problems associated with gene strand bias are likely to be of general importance in the bacterial world. Thirdly, we show that when essentiality is taken into account, expression levels do not significantly contribute to explain gene strand bias in these groups. Although not all essential genes are synthesised at high levels, the frequency of highly expressed genes among essential genes is higher than average (e.g. it is nearly 30% among B.subtilis essential genes). It is thus quite natural that previous analyses using rDNA and ribosomal proteins have revealed expressiveness to be the basis of gene strand bias: these genes are simultaneously essential and highly expressed. Here, we show that when essentiality is taken into account, expression plays a small, if any, role in determining the distribution of genes between replication strands. All these results are derived from analyses of orthology and assignment of essentiality by such orthologous relationships. As we have shown, this is prone to some errors. Yet, the degree of consistency of the results throughout the two phylogenetic groups and the cross-validations we have performed suggest that such errors are not strongly biasing our conclusions.
Understanding differences in gene strand bias intensity
There are significant differences in terms of overall gene strand bias among bacteria. As discussed elsewhere (17), the composition of DNAP may explain some of these differences. This effect seems to be independent of genome size and species ecology and may relate to the differential stability of the replication fork. Bacillus subtilis has both a higher general gene strand bias (75%) and a higher essential gene strand bias (94%) than E.coli (55 and 76%, respectively). Hence, the effect of essentiality in gene strand bias adds to that caused by differences in DNAP composition, and the two effects are likely to be related. For example, one might speculate that having two DNAP α-subunits leads to an increased asymmetry in the outcome of collisions between DNAP and RNAP. This would lead to a higher frequency of leading strand genes in general, and of essential genes in particular.
Except for Buchnera, we found that essential genes are more frequent in the leading strand than are the remaining genes (Fig. 1A). When one plots, among putative highly expressed genes, the distribution of essential versus non-essential genes, the differences are also typically high. There are, however, several exceptions in the analysis of highly expressed genes versus essential but not highly expressed genes, and these all concern obligatory parasites/symbionts, most of them obligate intracellular (Fig. 1C). One could propose that expression levels are more important in these genomes. This is unlikely for several reasons. First, these bacteria grow very slowly and therefore expression levels should not be as important as in fast growers. Secondly, we have used CAI to determine high levels of expression. Yet, the genomes of spirochetes (30) and Chlamydia (40) exhibit weak (although significant) codon usage biases among highly expressed genes, and Buchnera genomes are nearly devoid of such biases (41). Our tests on the CAI show that only two genomes among γ-proteobacteria and low G + C firmicutes lack significant codon usage bias among ribosomal proteins: T.tengcongensis and Buchnera. If CAI is a bad predictor of expression levels in these genomes, then one cannot safely attribute their atypical bias to expression levels. Thirdly, gene strand bias between putative highly expressed non-essential genes is relatively small. Therefore, expression is unlikely to play a much more important role in these bacteria than in B.subtilis or E.coli. Genome rearrangements may change the leading strand character of a gene and, if too frequent, they lead to lower gene strand bias. Although the genomes of obligatory intracellular bacteria are very stable, Buchnera suffered extensive rearrangements associated with the process of genome reduction, which may have lowered its gene strand bias. It is also possible that in these bacteria, the leading strand positioning of essential genes is under weaker selection, e.g. because of the drift associated with their small population sizes. Many of these bacteria suffer relaxed selection of housekeeping functions (9). Also, they depend on the host for many functions otherwise deemed as essential. As such, the assignment of essentiality by homology may be more error prone for many functions, especially among metabolic ones. On the other hand, intracellular bacteria may have some essential genes (e.g. transporters for some metabolites) that may be essential for their survival, which are not essential in B.subtilis. A clearer insight on gene strand bias in these genomes will require the availability of experimental data sets for essential genes in these bacteria (note that M.genitalium does not belong to this category because it is not an intracellular bacterium).
In addition to suggesting lower biases in obligatory intracellular bacteria, our data indicate different levels of gene strand bias in different bacteria. The causes of this may have to do with different stabilities of the replication forks, as discussed above, different RNAP processivity, leading to RNAP being more robust to collisions, or different efficiency of the mechanisms degrading truncated peptides. When an aborted transcript results in stalled ribosomes, tmRNA tags the truncated peptide and directs it to the protease degradation machinery (42,43). tmRNA does not direct the elimination of the corresponding mRNA and it is likely to become saturated in replicating bacteria, due to the large number of collisions. Nevertheless, if tmRNA is intrinsically more efficient, one might expect lower levels of gene strand bias in these bacteria. Experimental work will be necessary to tackle all these questions.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Joël Pothier for comments on this manuscript, and the scientists who disrupted and studied all the genes of the B.subtilis genome, in particular those who made the enterprise happen S. D. Ehrlich, F. Kunst, N. Ogasawara and H. Yoshikawa.
REFERENCES
- 1.Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B.J., Rubin,E.J., Novick,V.L., Amaya,K., Judson,N. and Mekalanos,J.J. (2002) A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl Acad. Sci. USA, 99, 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsyth R.A., Haselbeck,R.J., Ohlsen,K.L., Yamamoto,R.T., Xu,H., Trawick,J.D., Wall,D., Wang,L., Brown-Driver,V., Froelich,J.M. et al. (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol., 43, 1387–1400. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi K., Ehrlich,S.D., Albertini,A., Amati,G., Andersen,K.K., Arnaud,M., Asai,K., Ashikaga,S., Aymerich,S., Bessieres,P. et al. (2003) Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA, 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan I.K., Rogozin,I.B., Wolf,Y.I. and Koonin,E.V. (2002) Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res., 12, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal C., Papp,B. and Hurst,L.D. (2003) Rate of evolution and gene dispensability. Nature, 421, 496–497. [DOI] [PubMed] [Google Scholar]
- 7.Hirsh A.E. (2003) Rate of evolution and gene dispensability—reply. Nature, 421, 497–498. [DOI] [PubMed] [Google Scholar]
- 8.Rocha E.P.C. and Danchin,A. (2003) An analysis of determinants of protein substitution rates in bacteria. Mol. Biol. Evol., in press. [DOI] [PubMed] [Google Scholar]
- 9.Ochman H. and Moran,N.A. (2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science, 292, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 10.Ellwood M. and Nomura,M. (1982) Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J. Bacteriol., 149, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomura M. and Morgan,E.A. (1977) Genetics of bacterial ribosomes. Annu. Rev. Genet., 11, 297–347. [DOI] [PubMed] [Google Scholar]
- 12.Hirose S., Hiraga,S. and Okazaki,T. (1983) Initiation site of deoxyribonucleotide polymerization at the replication origin of the Escherichia coli chromosome. Mol. Gen. Genet., 189, 422–431. [DOI] [PubMed] [Google Scholar]
- 13.Brewer B. (1988) When polymerases collide: replication and the transcriptional organization of the E.coli chromosome. Cell, 53, 679–686. [DOI] [PubMed] [Google Scholar]
- 14.French S. (1992) Consequences of replication fork movement through transciption units in vivo. Science, 258, 1362–1365. [DOI] [PubMed] [Google Scholar]
- 15.Zeigler D.R. and Dean,D.H. (1990) Orientation of genes in the Bacillus subtilis chromosome. Genetics, 125, 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLean M.J., Wolfe,K.H. and Devine,K.M. (1998) Base composition skews, replication orientation and gene orientation in 12 prokaryote genomes. J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 17.Rocha E.P.C. (2002) Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol., 10, 393–396. [DOI] [PubMed] [Google Scholar]
- 18.Dervyn E., Suski,C., Daniel,R., Bruand,C., Chapuis,J., Errington,J., Janniere,L. and Ehrlich,S.D. (2001) Two essential DNA polymerases at the bacterial replication fork. Science, 294, 1716–1719. [DOI] [PubMed] [Google Scholar]
- 19.Rocha E.P.C. and Danchin,A. (2003) Essentiality, not expressiveness, drives gene strand bias in bacteria. Nature Genet., 34, 377–378. [DOI] [PubMed] [Google Scholar]
- 20.Louarn J.M., Bouche,J.P., Legendre,F., Louarn,J. and Patte,J. (1985) Characterization and properties of very large inversions of the E.coli chromosome along the origin-to-terminus axis. Mol. Gen. Genet., 201, 467–476. [DOI] [PubMed] [Google Scholar]
- 21.Segall A., Mahan,M.J. and Roth,J.R. (1988) Rearrangement of the bacterial chromosome: forbidden inversions. Science, 241, 1314–1318. [DOI] [PubMed] [Google Scholar]
- 22.Sharp P.M. and Li,W.H. (1987) The codon adaptation index—a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocha E.P.C. and Danchin,A. (2001) Ongoing evolution of strand composition in bacterial genomes. Mol. Biol. Evol., 18, 1789–1799. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov R.L. and Koonin,E.V. (1997) A genomic perspective of protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 25.Coghlan A. and Wolfe,K.H. (2000) Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast, 16, 1131–1145. [DOI] [PubMed] [Google Scholar]
- 26.Jansen R., Bussemaker,H.J. and Gerstein,M. (2003) Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res., 31, 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharp P.M. and Li,W.-H. (1986) An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol., 24, 28–38. [DOI] [PubMed] [Google Scholar]
- 28.Andersson S.G.E. and Kurland,C.G. (1990) Codon preferences in free-living microorganisms. Microbiol. Rev., 54, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Médigue C., Rouxel,T., Vigier,P., Henaut,A. and Danchin,A. (1991) Evidence for horizontal gene transfer in E.coli speciation. J. Mol. Biol., 222, 851–856. [DOI] [PubMed] [Google Scholar]
- 30.Perriere G. and Thioulouse,J. (2002) Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res., 30, 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkhill J., Wren,B.W., Thomson,N.R., Titball,R.W., Holden,M.T., Prentice,M.B., Sebaihia,M., James,K.D., Churcher,C., Mungall,K.L. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 32.Glaser P., Rusniok,C., Buchrieser,C., Chevalier,F., Frangeul,L., Msadek,T., Zouine,M., Couve,E., Lalioui,L., Poyart,C. et al. (2002) Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol., 45, 1499–1513. [DOI] [PubMed] [Google Scholar]
- 33.Tillier E.R. and Collins,R.A. (2000) Genome rearrangement by replication-directed translocation. Nature Genet., 26, 195–197. [DOI] [PubMed] [Google Scholar]
- 34.Eisen J.A., Heidelberg,J.F., White,O. and Salzberg,S.L. (2000) Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol., 1, RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao Q., Tian,Y., Li,W., Xu,Z., Xuan,Z., Hu,S., Dong,W., Yang,J., Chen,Y., Xue,Y. et al. (2002) A complete sequence of the T.tengcongensis genome. Genome Res., 12, 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin E.V., Mushegian,A.R. and Bork,P. (1996) Non-orthologous gene displacement. Trends Genet., 12, 334–336. [PubMed] [Google Scholar]
- 37.Shigenobu S., Watanabe,H., Hattori,M., Sakaki,Y. and Ishikawa,H. (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [DOI] [PubMed] [Google Scholar]
- 38.Mushegian A.R. and Koonin,E.V. (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA, 93, 10268–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh T., Takemoto,K., Mori,H. and Gojobori,T. (1999) Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol., 16, 332–346. [DOI] [PubMed] [Google Scholar]
- 40.Romero H., Zavala,A. and Musto,H. (2000) Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res., 28, 2084–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wernegreen J.J. and Moran,N.A. (1999) Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol. Biol. Evol., 16, 83–97. [DOI] [PubMed] [Google Scholar]
- 42.Muto A., Ushida,C. and Himeno,H. (1998) A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci., 23, 25–29. [DOI] [PubMed] [Google Scholar]
- 43.Gillet R. and Felden,B. (2001) Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol. Microbiol., 42, 879–885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.