Abstract
The mechanism by which Hsp40 and other molecular chaperones recognize and interact with non-native polypeptides is a fundamental question, as is how Hsp40 co-operates with Hsp70 to facilitate protein folding. Years of structural studies of Hsp40 from yeast and other species, conducted using X-ray protein crystallography, NMR and small-angle X-ray scattering, have shed light on the mechanisms how Hsp40 functions as a molecular chaperone and how Hsp40-Hsp70 pair promotes protein folding, protein transport and degradation. This review provides a discussion of recent structural studies of Hsp40s and their functional implications.
Keywords: molecular chaperone, Hsp40, Hsp70, J-domain, peptide-binding fragment, protein structure, Sis1, Ydj1
Introduction
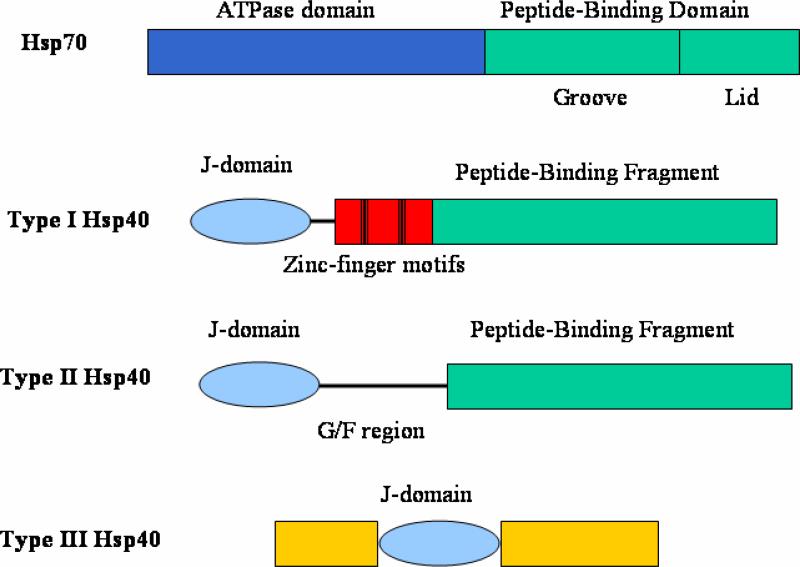
Molecular chaperones encompass a large group of proteins that bind and stabilize non-native polypeptides. Molecular chaperones can suppress protein aggregation and facilitate protein folding into the native conformations that are specified by their primary sequences [1-3]. Most of the molecular chaperones identified to date are heat shock proteins. Among them, Heat shock protein 70 (Hsp70), probably the most well-studied molecular chaperone, plays essential roles in cell physiology [1-3]. Hsp70 contains an N-terminal ATPase domain and a C-terminal peptide-binding domain (Fig. 1). The Hsp70 peptide-binding domain can be sub-divided into a peptide-binding groove and a lid domain. Members of the Hsp70 and Hsp40 (DnaJ-like) families function in specific pairs that form transient complexes with non-native regions of polypeptides to promote protein folding, assembly and transport of proteins within the cell [1-9].
Fig. 1.
Domain structures for Hsp70 and Hsp40 family members. In type I Hsp40 protiens, the Zinc-finger motifs are indicated by the black bars. The G/F rich linker regions in type I and type II Hsp40 are shown in black lines. In type III Hsp40 proteins, the J-domain can be located anywhere in the protein sequence.
Hsp40 family members are categorized into type I, II and III (Fig. 1). All types of Hsp40 proteins contain a J-domain which has about 70 amino acid residues. In Types I and II Hsp40 proteins, the J-domain is at the N-terminus. In Type III Hsp40 family members, it can be located at any position within the protein sequence. The J-domain of Hsp40 can make contacts with Hsp70 ATPase domain and then stimulate the ATPase activities of Hsp70.
Both type I and type II Hsp40s have a peptide-binding fragment located at the C-terminus of the proteins. The N-terminal J-domains are connected to the peptide-binding fragments via a G/F rich linker in both type I and type II Hsp40s. Type I Hsp40 such as E. coli DnaJ, yeast Ydj1 and human Hdj2 contain two Zinc-finger motifs between the J-domain and the C-terminal peptide-binding fragment. In contrast, type II Hsp40 proteins such as yeast Sis1 and human Hdj1 do not [10-12]. It was proposed that Hsp40 binds non-native polypeptide first and then delivers the non-native polypeptide to Hsp70 for folding [1, 13, 14]. The ability to bind non-native polypeptides for the cytosolic Hsp40 is an essential function in vivo [15].
As mentioned above, Type III Hsp40 family members contain the J-domain, but it can be located at any position within the protein. Type III Hsp40 members lack the other conserved domains found in type I and II Hsp40 members. By searching the ORFs in the Saccharomyces cerevisiae genome, 22 Hsp40 proteins have been identified. Five (Ydj1, Xdj1, Apj1, Mdj1 and Scj1) have been classified as type I Hsp40 and four (Sis1, Djp1, Caj1 and Hlj1) have been classified as type II Hsp40. The remaining 13 (Zuo1, Swa2, Jjj1-3, Cwc23, Mdj2, Pam18, Jac1, Jid1, Jem1, Sec63, Erj5) have been classified as type III Hsp40 [16, 17]. Type III Hsp40 members can recruit Hsp70 through the J-domains to achieve many diversified functions. In this review, we will focus on the recent structural and mechanistic studies of the more conserved type I and II Hsp40s.
The working model for Hsp40-Hsp70 system
Hsp40 protein functions in specific pair with Hsp70 protein to promote protein folding, protein transport and degradation [1, 18]. The J-domain of Hsp40 can make direct contacts with Hsp70 N-terminal ATPase domain. The binding can regulate and stimulate the Hsp70 intrinsic ATPase activity[19]. Within Hsp70, the ATP hydrolysis in the N-terminal domain mediates conformational changes in the C-terminal peptide-binding domain. When ATP is bound to the N-terminal domain, the peptide-binding domain is in an “open” conformation with a low affinity for non-native polypeptides. Upon hydrolysis of ATP to ADP in the N-terminal domain, the peptide-binding domain switches to a “closed” conformation with a high affinity for non-native polypeptide [20]. A recent crystal structure of Hsp70 indicated that a small helix between the Hsp70 N-terminal and C-terminal domains may function as a hinge by which N-terminal ATP hydrolysis transmits a conformational change in the C-terminal peptide-binding domain [21]. The crystal structure of the Hsp110, which is a homologue of Hsp70, suggested a working model how Hsp70 achieves the “open” conformation at ATP-bound state [22].
Hsp40 C-terminal peptide-binding fragment interacts with non-native polypeptides and suppresses the protein aggregations. It has been proposed that Hsp40 may bind the non-native polypeptides and then transfer them to Hsp70 [3, 5, 6, 23, 24]. The Hsp40s function as dimers and may interact with the non-native polypeptide through hydrophobic interactions. The binding of Hsp40 may prime the non-native polypeptide into an extended conformation in preparation for the subsequent Hsp70 recognition and binding [6, 23-25].
It has been showed that the last four amino acid residues EEVD within human Hsp70 play regulatory roles in Hsp40-Hsp70 functions [4]. The recent in vivo data also supported the bi-partite interactions between Hsp40 and Hsp70 [26]. The crystal structure of Hsp40 and Hsp70 C-terminus complex showed that the charge-charge interactions were primarily responsible for maintaining this interaction [7]. It is possible that the Hsp70 C-terminal EEVD motif recruits Hsp40 and ensures efficient non-native polypeptide transfer from Hsp40 to Hsp70 [7].
The J-domain structures of Hsp40s
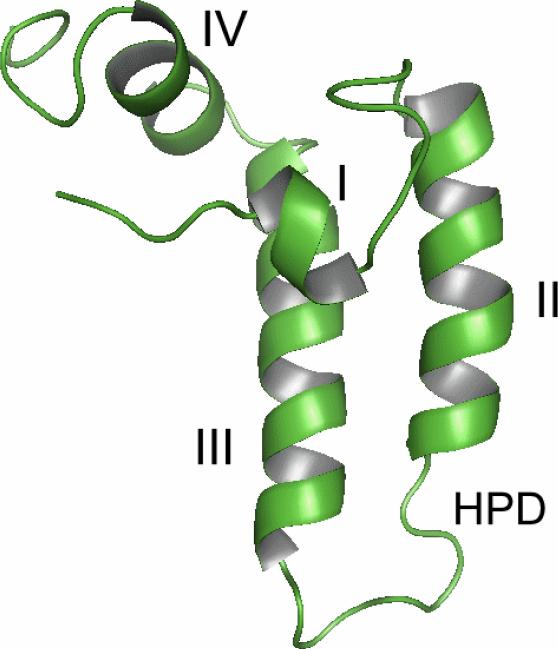
The first solved structures of the J-domains were NMR solution structures from E.coli Hsp40 DnaJ and human Hsp40 Hdj1 [27-29]. These solution structures were followed by J-domain crystal structures from other Hsp40 proteins, such as E. coli Hsc20, bovine auxilin, and yeast Pam16 [18, 30-32]. All of these structures revealed that Hsp40 J-domain contains four helices (I-IV) and a conserved HIS-PRO-ASP motif located between helix II and III (Fig. 2). Binding inhibition assays using synthetic peptides have suggested that the N-terminal portion of the J-domain (helix I, II and the HIS-PRO-ASP motif) interacts with the Hsp70 N-terminal ATPase domain [33]. The J-domains could be flexible in solution, and the binding of Hsp40 J-domain to Hsp70 may stabilize the J-domain structures [34].
Fig. 2.
Ribbon drawings of representative J-domain structures [41]. The helices I-IV are labeled and the conserved HIS-PRO-ASP motif are also shown. a) The NMR structure of the J-domain from E.coli Hsp40 DnaJ [29]. b) The crystal structure of the J-domain from E.coli Hsc20 [30].
The peptide-binding fragments of the type I and II Hsp40 members
The Type I and II Hsp40 proteins contain the conserved C-terminal peptide-binding fragments, and they function as ATP-independent molecular chaperones for the non-native polypeptides [3, 5]. The major difference between type I and type II Hsp40 C-terminal fragments is the presence of two well conserved Zinc-finger motifs within type I Hsp40 members [23, 24]. Type I Hsp40s such as yeast Hdj1 can suppress protein aggregation by themselves while type II Hsp40s require Hsp70 to achieve this goal [35].
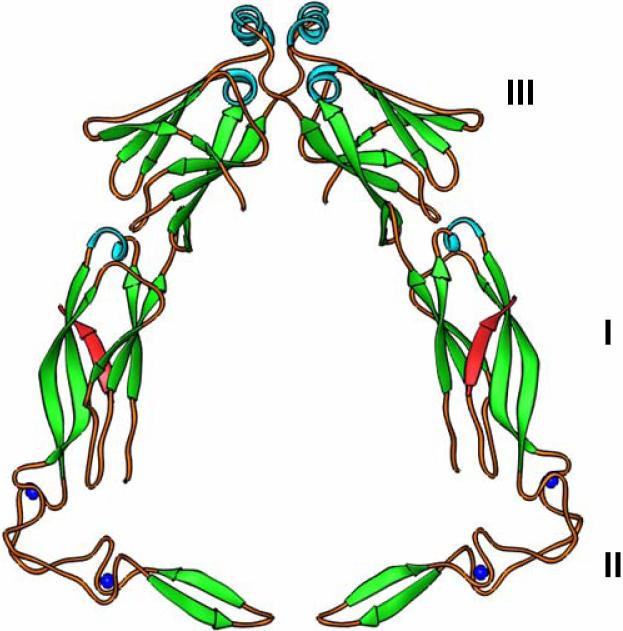
To investigate the mechanisms behind how Hsp40 peptide-binding fragments interact with their non-native polypeptide substrates, we carried out extensive structural studies for both Type I and Type II Hsp40 C-terminal peptide-binding fragments. We have determined the crystal structure of the peptide-binding fragment of Sis1, an essential type II Hsp40 family member from Saccharomyces cerevisiae [23]. The 2.7Å structure reveals that Sis1 forms a homo-dimer in the crystal by a crystallographic two-fold axis (Fig. 3 and Supplemental materials/movies). Sis1 monomers are elongated and consist of two domains (I and II) with similar folds. Both domain I and II contain two layers of ß-strands. Sis1 dimerizes through a short C-terminal stretch. Deletion of the Sis1 C-terminal dimerization motif may generate severe defects in chaperone activity. Thus, dimer formation is critical for Sis1 chaperone function [23]. The Sis1 dimer has a U-shaped architecture with a large cleft between the two elongated monomers. Hydrophobic surface drawing revealed a hydrophobic depression located in Domain I of each monomer, and we suspect that these depressions might be involved in binding the exposed hydrophobic amino acid side chains that are characteristic of non-native polypeptide structures. Simultaneous polypeptide binding by each Sis1 monomer might maintain the substrate in the extended conformation preferred for Hsp70 recognition.
Fig. 3.
Hsp40 peptide-binding fragment structures. a) The modeled dimer structure of yeast type I Hsp40 Ydj1 peptide-binding fragment complexed with its peptide substrate. Domain I, II and III of Ydj1 are labeled. The bound peptide substrates are in red. b) Crystal structure of yeast type II Hsp40 Sis1 peptide-binding fragment dimer. Domain I and II are labeled. c) The surface potential presentations for the crystal structure of Ydj1 and the peptide substrate complex. The right panel shows the surface potential drawing of the Ydj1 monomer with no C-terminal dimerization motif. Blue and red denote positively and negatively charged regions, respectively. The bound peptide GWLYEIS is shown in rod model. The area within the red box of the right panel is magnified in the left panel. The residues of the peptide substrate GWLYEIS are labeled in black.
Very recently, we solved the crystal structure of the peptide-binding fragment from human Type II Hsp40 Hdj1 [36]. The Hdj1 peptide-binding fragment structure resembled the one from Sis1, except that Domain I of Hdj1 was rotated by 7.1° from the main body of the molecule, which made the cleft between the two Hdj1 monomers smaller than the cleft of Sis1. This structural variability suggested that Domain I of the Hsp40 monomers may possess significant flexibility, which might help Hsp40 proteins regulate the size of the cleft in the dimer.
We also determined the crystal structure of the type I Hsp40 Ydj1 peptide-binding fragment from S. cerevisiae [24, 37]. In this structure, however, the Ydj1 peptide-binding fragment is complexed with the peptide substrate GWLYEIS (Fig. 3, Supplemental materials/movies). The Ydj1 peptide substrate GWLYEIS was identified through the combination of phage display library screening and isothermal titration calorimetry (ITC) technique [24]. The 2.7Å resolution complex structure reveals that Ydj1 peptide-binding fragment forms an L-shaped molecule constituted by three domains. The domain I exhibits a similar protein fold as domain III, while the domain II contains two Zinc-finger motifs. The peptide substrate interacted with the peptide-binding fragment by forming an extra β-strand with Domain I.
Domain II, which forms the shorter arm of the L-shaped structure, contains two zinc-finger motifs and a β-hairpin. The zinc-finger motifs display few contacts with the other domains within Ydj1, and they constitute the 90° turn between the long and short arms of the L-shaped Ydj1 peptide-binding fragment. The NMR solution structure of the Zinc-finger motifs of E. coli type I Hsp40 DnaJ revealed that these motifs formed a V-shaped molecule [38], an observation that is consistent with the Ydj1 crystal structure. It is interesting to note that the Zinc finger motifs of Ydj1 are not directly involved in binding the peptide substrate.
Both type I and type II Hsp40 proteins function as dimers [5]. The crystal structure of type I Ydj1 peptide-binding fragment complexed with the peptide substrate was determined in the monomeric form without the C-terminal dimerization motif. In the crystal structure, the Ydj1 peptide-binding fragment contains domain I, II and III, but lacks the C-terminal dimerization motif [24]. Later, we determined the crystal structure of Ydj1 C-terminal fragment that comprises of the dimain III and the C-terminal dimerization motif [39]. Based on the above-mentioned two structures, the Ydj1 peptide-binding fragment dimer structure can be accurately modeled [39]. The modeled Ydj1 peptide-binding fragment dimer revealed that a cleft was formed between the two Ydj1 monomers (Fig. 3, supplemental materials/movies). The two monomers within the dimer lay almost within the same plane. At the bottom of the modeled dimer, the short arm of the L-shaped molecule containing the Zinc-finger motifs point directly against each other. The tips of the domain IIs from the two monomers are about 10Å away. Therefore the Ydj1 peptide-binding fragment dimer forms a bell-shaped molecule.
When the structure of the type I Hsp40 Ydj1 dimer is compared with that of the type II Hsp40 Sis1 dimer, both Hsp40 proteins contain a large cleft within the homo-dimer, which is presumably the docking site for Hsp70 (Fig. 3, Supplemental materials/movies). In Sis1, domain I and domain II utilized similar folds. The folding topology is also utilized in Ydj1 domain I and III. However, the Ydj1 structure has a short arm containing two Zinc-finger motifs (domain II) at the bottom of the molecule, a structural element not present in the Sis1 structure. Moreover, the two monomers within the Ydj1 dimer lay almost within one plane. But in Sis1 dimer structure, the domain I is twisted about 130° from domain II, meaning that the two monomers do not fit in the same plane (Fig. 3, supplemental materials/movies). In addition, the C-terminal dimerization motif in Ydj1 is much longer than that of Sis1, generating more extensive interactions between Ydj1 monomers than Sis1 [39]. The structural differences between the type I and type II Hsp40s revealed in these studies may help to explain their distinct cellular functions.
The Hsp40 and peptide substrate interactions
The bound peptide substrate GWLYEIS forms an anti-parallel β-strand with the β-strand of the domain I in Ydj1 peptide binding fragment (Fig. 3). Within the 7-mer peptide substrate, the main chain atoms of the six residues (GWLYEI) form the typical β-sheet hydrogen bond networks with Ydj1. The terminal Ser in the peptide substrate does not make significant interactions with the protein Ydj1.
Among the six amino acid residues that did form contacts with Ydj1, the leucine residue in the middle of the GWLYEIS substrate made the most significant contacts. The side chain of this leucine was fully buried in a hydrophobic pocket formed by a number of hydrophobic residues (I116, L135 L137, L216, V247 and F249) on the surface of Domain I of Ydj1 (Fig. 3c). The hydrophobicity of these residues is nicely conserved among the type I Hsp40 family members, indicating that this hydrophobic pocket may be a feature common to this molecular chaperone family. Mutations of these hydrophobic residues may reduce non-native polypeptide binding and chaperone activity for Ydj1 [25].
This hydrophobic pocket of Ydj1 may also define the peptide substrate specificity for the molecular chaperone family of Hsp40. Binding studies using synthetic peptides indicated that the middle leucine of GWLYEIS may be replaced by other hydrophobic residues, such as tryptophan or alanine, without affecting the binding affinity of Ydj1. However, replacing the leucine residue with a polar glutamine residue abolished the Ydj1 binding affinity, even though the glutamine and leucine side chains were similar in size. Synthetic peptide studies also showed that type I Hsp40 Ydj1 may interact with the peptide substrates with the minimum length of six amino acid residues [24].
The Hsp40-Hsp70 molecular chaperone system
Hsp40 and Hsp70 stay in close contact to achieve these diverse functions such as protein folding and protein transport activities. In addition to the contacts between Hsp40 J-domain and Hsp70 ATPase domain, the conserved C-terminal EEVD motif in Hsp70 mediates and regulates the interaction between Hsp40 and Hsp70 by a mechanism that may also play important roles in Hsp40-Hsp70 functions [4, 6].
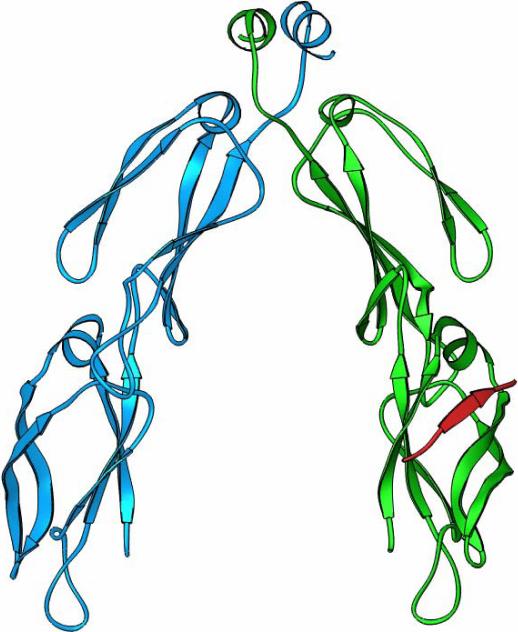
To investigate the mechanism how Hsp40 interacts with Hsp70 C-terminus, we determined the crystal structure of yeast Hsp40 Sis1 peptide-binding fragment complexed with the Hsp70 Ssa1 C-terminus [7]. The eight residues at the Ssa1 extreme C-terminus (GPTVEEVD) form a ß-strand with the domain I of Sis1 peptide-binding fragment (Fig. 4). Surprisingly, the Ssa1 C-terminus binds Sis1 at the same site where Sis1 interacts with the non-native polypeptides. The negatively charged residues within EEVD motif in Ssa1 C-terminus form extensive charge-charge interactions with the positively charged residues in Sis1 (Fig. 4b). The nature of this interaction contrasts sharply with the hydrophobic interaction between Hsp40 Ydj1 and its peptide substrate [7, 24].
Fig. 4.
The crystal structure of the complex between Hsp40 Sis1 peptide-binding fragment and the Hsp70 Ssa1 C-terminal EEVD motif. a) the ribbons drawing of the Sis1-Ssa1 complex. Sis1 peptide-binding fragment monomers are in blue and green and the bound Hsp70 Ssa1 C-terminus is in red. b) The charge-charge interactions between the EEVD motif of Ssa1 and Sis1. The Ssa1 C-terminus is shown in red and Sis1 is shown in silver. The K199, K202, K214 and K256 from Sis1 are labeled in red and E638, E639 and D641 from Ssa1 EEVD motif are labeled in blue.
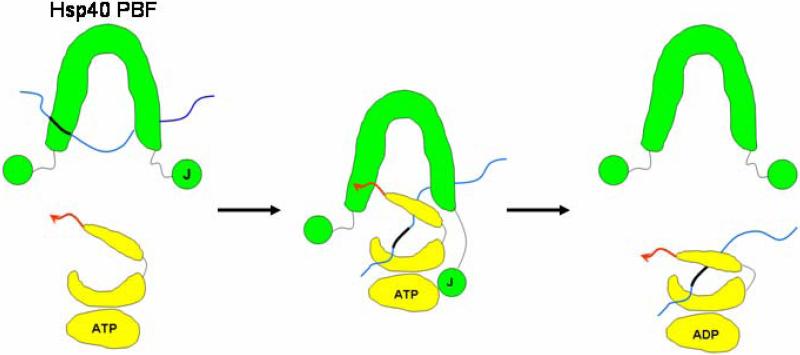
Based on the available structural and functional data, we propose an “anchoring and docking” model how Hsp40 interacts with Hsp70 (Fig. 5). This model illustrates our proposed mechanism by which Hsp40 achieves both stimulation of the Hsp70 ATPase and transfer of non-native polypeptides to Hsp70 [6, 7, 23, 36]. In this model, the cleft between Hsp40 dimer serves as the docking site for the Hsp70 peptide-binding domain and the C-terminal EEVD motif of Hsp70 may act as the anchor to initiate the docking (Fig. 5).
Fig. 5.
Schematic representation of the proposed “anchoring and docking” mechanism by which Hsp40 (green) delivers a non-native peptide (thin blue line) to Hsp70 (yellow). The cartoon drawing depicts an Hsp70 molecule divided into its ATPase domain, peptide-binding groove, and lid domain. The J-domain (J) and peptide-binding fragment (PBF) of Hsp40 are shown schematically. The C-terminal “anchor” region (EEVD motif) of Hsp70 is shown as a red arrow. The thick black region within the thin blue non-native polypeptide indicates the hydrophobic region that is recognized by the Hsp40 peptide-binding fragment and the Hsp70 peptide-binding domain.
Other Hsp40 and Hsp70 structural data are consistent with this “anchoring and docking” model. The low resolution quaternary structures of full-length type I and type II Hsp40s have been determined by small angle X-ray scattering [40]. These studies demonstrated that the J-domains within the Hsp40 dimers are loosely connected to the peptide-binding fragment of Hsp40. X-ray crystallography studies have also clarified the relative positioning of Hsp70 N-terminal domain and C-terminal domain [21]. According to these current structural data, it is possible that the J-domain of Hsp40 interacts with the ATPase domain of Hsp70, while the C-terminal EEVD motif of Hsp70 anchors the C-terminal peptide-binding fragment of Hsp40.
Type I and II Hsp40 proteins pair with Hsp70 to achieve distinct cellular functions. For example, yeast type I Hsp40 Sis1 is required for protein translation initiation by interacting with nascent polypeptides while yeast type II Hsp40 Ydj1 is responsible for protein translocations across membranes. Both Sis1 and Ydj1 work with the cytosolic Hsp70 Ssa1 for their different functions [11, 12]. Therefore, the structural differences within type I and type II Hsp40s may account for their distinct cellular functions. Both type I and II Hsp40 need to interact with the non-native polypeptides and prime them for subsequence binding and folding by Hsp70 [14]. However, type I and II Hsp40s appear to have different substrate specificity because type I Hsp40 Sis1 doe not recognize and bind the peptide substrate of type II Hsp40 Ydj1 [24]. The different structural property at the peptide-binding site of each type of Hsp40 may define the peptide substrate specificity.
Summary
Hsp40 can function as a molecular chaperone to bind non-native polypeptide. Hsp40 pairs with Hsp70 to achieve its function of protein folding, transport and degradation functions. The J-domain of Hsp40 may stimulate the ATPase activity of Hsp70 and the C-terminal peptide-binding fragment of Hsp40 can load the bound non-native polypeptide to Hsp70.
The structural studies revealed, in both type I and II Hsp40 proteins, that a cleft is formed between the two elongated monomers. However, the Type I Hsp40 dimer contains a bell-shaped cleft with zinc-finger motifs comprising the bottom of the bell, while the Type II Hsp40 dimer contains a U-shaped cleft with no zinc-finger motifs present. The surface of Domain I in the peptide-binding site of Hsp40 interacts with non-native polypeptides through hydrophobic interactions, as long as the non-native polypeptide is at least six residues in length. The peptide-binding site of Hsp40 can also serve as the binding site for the C-terminal EEVD motif of Hsp70 through charge-charge interactions. Based on the most current structural and biochemical data of Hsp40-Hsp70 system, an “anchoring and docking” model has been proposed to illustrate the mechanism how Hsp40 to interact with Hsp70 to ensure the efficient protein folding.
Supplementary Material
Supplemental materials/movies
S1. A movie of the yeast type I Hsp40 Ydj1 and the peptide substrate complex dimer structure. Ydj1 dimer is in blue and the bound peptide substrate is in red.
S2. A movie of the yeast type II Hsp40 Sis1 crystal structure. The Sis1 dimer is in silver.
References
- 1.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92(3):351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 2.Craig EA, Weissman JS, Horwich AL. Heat shock proteins and molecular chaperones: mediators of protein conformation and turnover in the cell. Cell. 1994;78(3):365–372. doi: 10.1016/0092-8674(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 3.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 4.Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. Embo J. 1995;14(10):2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356(6371):683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 6.Qian X, Hou W, Zhengang L, Sha B. Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. Biochem J. 2002;361(Pt 1):27–34. doi: 10.1042/0264-6021:3610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Wu Y, Qian X, Sha B. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem J. 2006;398(3):353–360. doi: 10.1042/BJ20060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright CM, Fewell SW, Sullivan ML, Pipas JM, Watkins SC, Brodsky JL. The Hsp40 molecular chaperone Ydj1p, along with the protein kinase C pathway, affects cell-wall integrity in the yeast Saccharomyces cerevisiae. Genetics. 2007;175(4):1649–1664. doi: 10.1534/genetics.106.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. Rna. 2007;13(9):1570–1581. doi: 10.1261/rna.585007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19(4):176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 11.Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71(7):1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73(6):1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 13.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 14.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381(6583):571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JL, Craig EA. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J Cell Biol. 2001;152(4):851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5(6):567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14(7):1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42(17):4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20(4):513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131(1):106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure Fold Des. 2000;8(8):799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure (Camb) 2003;11(12):1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Sha B. Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J. 2005;386(Pt 3):453–460. doi: 10.1042/BJ20041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169(4):1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill RB, Flanagan JM, Prestegard JH. 1H and 15N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coli DnaJ(1-78). Biochemistry. 1995;34(16):5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- 28.Qian YQ, Patel D, Hartl FU, McColl DJ. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol. 1996;260(2):224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 29.Pellecchia M, Szyperski T, Wall D, Georgopoulos C, Wuthrich K. NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J Mol Biol. 1996;260(2):236–250. doi: 10.1006/jmbi.1996.0395. [DOI] [PubMed] [Google Scholar]
- 30.Cupp-Vickery JR, Vickery LE. Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J Mol Biol. 2000;304(5):835–845. doi: 10.1006/jmbi.2000.4252. [DOI] [PubMed] [Google Scholar]
- 31.Jiang J, Taylor AB, Prasad K, Ishikawa-Brush Y, Hart PJ, Lafer EM, Sousa R. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry. 2003;42(19):5748–5753. doi: 10.1021/bi034270g. [DOI] [PubMed] [Google Scholar]
- 32.Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. Embo J. 2006;25(19):4675–4685. doi: 10.1038/sj.emboj.7601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271(16):9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 34.Landry SJ. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry. 2003;42(17):4926–4936. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- 35.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97(14):7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, Wu Y, Li J, Qian X, Fu Z, Sha B. The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1. BMC Struct Biol. 2008;8(1):3. doi: 10.1186/1472-6807-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Sha B. Peptide substrate identification for yeast Hsp40 Ydj1 by screening the phage display library. Biol Proced Online. 2004;6:204–208. doi: 10.1251/bpo90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Yamout M, Legge GB, Zhang O, Wright PE, Dyson HJ. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J Mol Biol. 2000;300(4):805–818. doi: 10.1006/jmbi.2000.3923. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Li J, Jin Z, Fu Z, Sha B. The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J Mol Biol. 2005;346(4):1005–1011. doi: 10.1016/j.jmb.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 40.Borges JC, Fischer H, Craievich AF, Ramos CH. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J Biol Chem. 2005;280(14):13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- 41.Carson M. Ribbons 2.0. “Ribbon models for macromolecule.”. J Mol Graphics. 1987;5:103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials/movies
S1. A movie of the yeast type I Hsp40 Ydj1 and the peptide substrate complex dimer structure. Ydj1 dimer is in blue and the bound peptide substrate is in red.
S2. A movie of the yeast type II Hsp40 Sis1 crystal structure. The Sis1 dimer is in silver.