Abstract
Among the large family of C/D methylation guide RNAs, the intron of euryarchaeal pre-tRNATrp represents an outstanding specimen able to guide in cis, instead of in trans, two 2′-O-methylations in the pre-tRNA exons. Remarkably, both sites of methylation involve nucleotides within the bulge–helix–bulge (BHB) splicing motif, while the RNA-guided methylation and pre-tRNA splicing events depend on mutually exclusive RNA folding patterns. Using the three recombinant core proteins of archaeal C/D RNPs, we have analyzed in vitro RNP assembly of the pre-tRNA and tested its site-specific methylation activity. Recognition by L7Ae of hallmark K-turns at the C/D and C′/D′ motifs appears as a crucial assembly step required for subsequent binding of a Nop5p–aFib heterodimer at each site. Unexpectedly, however, even without L7Ae but at a higher concentration of Nop5p–aFib, a substantially active RNP complex can still form, possibly reflecting the higher propensity of the cis-acting system to form guide RNA duplex(es) relative to classical trans- acting C/D RNA guides. Moreover, footprinting data of RNPs, consistent with Nop5p interacting with the non-canonical stem of the K-turn, suggest that binding of Nop5p–aFib to the pre-tRNA–L7Ae complex might direct transition from a splicing-competent structure to an RNA conformer displaying the guide RNA duplexes required for site-specific methylation.
INTRODUCTION
In both Archaea and Eukarya, a large family of box C/D RNAs directs the site-specific 2′-O-methylation of a few ribonucleotide sugars within definite cellular RNA species. In addition to rRNAs, their target RNAs include tRNAs and spliceosomal snRNAs in Archaea and vertebrates, respectively (1–6). Archaeal and eukaryal box C/D RNAs both contain two short consensus motifs designated box C (RUGAUGA) and D (CUGA) located near the 5′ and 3′ ends of the molecule, respectively. They also contain a second copy of each box motif, C′ and D′, in the center of the molecule. However, while in eukaryal C/D RNAs the C′ and D′ boxes often display some deviations (7), in archaeal C/D RNAs they fit each motif consensus much more strictly (4,8,9). A hallmark feature of box C/D methylation guides is the 10–21 nt sequence complementary to the site of 2′-O-methylation in target RNA. Located immediately upstream from the D or D′ box, this sequence, termed the antisense element, directs the 2′-O-methylation onto the nucleotide paired to the fifth nucleotide upstream from the D or D′ box within the RNA duplex formed by the guide RNA and its RNA target (10–12). Box C/D RNAs function as small RNP particles, each one consisting of a site-specific guide RNA associated with a small set of common proteins (3–6).
Eukaryal methylation guide C/D RNPs include four different core proteins (13): fibrillarin, which catalyzes the methyltransfer reaction; Nop56p and Nop58p, which are structurally related to each other; and Snu13p/15.5 kDa which belongs to a family of RNA-binding proteins including human ribosomal proteins L7a and S12 as well as yeast ribosomal protein L30 (13–23). The Snu13p/15.5 kDa protein was first characterized as a component of the U4/U6·U5 tri-snRNP that directly binds to the 5′ stem–loop of U4 snRNA (24,25). In the tri-snRNP particle as well as box C/D RNPs, the protein recognizes a characteristic RNA secondary structure motif, the kink-turn (or K-turn) (26). In C/D RNAs, the K-turn is formed by a specific interaction between the C and D boxes (20,25,26). Characterized by two helical stems, a canonical stem I and a non-canonical stem II, separated by an internal three-base loop, the K-turn imposes a kink of ∼120° in the phosphate backbone. The internal loop is always asymmetric with a flipped-out U, while stem II exhibits two tandem sheared GA pairs usually followed by a UU pair and a Watson–Crick base pair. Stem II of the box C/D K-turn is essential for nucleolar localization of C/D snoRNAs and, along with Snup13p/15.5 kDa, for the hierarchical assembly of the C/D snoRNP (27). Site-specific in vivo cross-linking analyses of Xenopus U25 snoRNA and nucleotide analog interference mapping argue that eukaryal box C/D snoRNPs are asymmetric, with the C′ box contacting Nop56 and fibrillarin, the C box interacting with Nop58p, and the D and D′ boxes contacting fibrillarin (28,29).
Archaeal C/D RNPs contain only three different core proteins, aFib, L7Ae and Nop5p (6,30). aFib and ribosomal protein L7Ae represent homologs of eukaryal fibrillarin and Snu13p/15.5 kDa, respectively. Remarkably, the third protein component of archaeal C/D RNPs, Nop5p, is structurally related to both eukaryal C/D RNP proteins Nop56p and Nop58p (30). Recent crystal structure analysis has revealed that archaeal Nop5p and fibrillarin form a complex of two heterodimers within bipartite box C/D RNPs (31). The 2-fold symmetry observed between two fibrillarin–Nop5p heterodimers is consistent with the symmetric structure exhibited by archaeal box C/D RNAs (4,8,9,30), suggesting the possibility that each fibrillarin–Nop5p heterodimer interacts with one box C/D (or C′/D′) motif via L7Ae protein interaction (31). Recently, in vitro reconstitution of archaeal sRNP complexes containing a single box C/D or C′/D′ motif has demonstrated that the terminal box C/D RNP is the minimal methylation-competent particle and that all three archaeal core proteins bind both motifs symmetrically (32). However, efficient methylation requires that both boxes C/D and C′/D′ RNPs function within the full-length RNA molecule. Moreover, while archaeal L7Ae represents a functional homolog of the human 15.5 kDa protein as confirmed by in vitro RNA binding experiments (33), the human 15.5 kDa binds only to the terminal C/D, not the C′/D′, box motif of eukaryal snoRNAs, suggesting that the box C/D methylation guide complex has evolved into structurally distinct RNP particles in Eukarya and Archaea (29,32,34).
Outstanding among archaeal box C/D sRNAs is the precursor to tRNATrp in most Euryarchaea (35). Its unusually long intron unambiguously harbors the C/D hallmarks as well as two antisense elements predicted to direct in cis- two 2′-O-methylations in the 5′ and 3′ exons of pre-tRNATrp, at positions 34 and 39 of the mature tRNA sequence, respectively, through formation of two intramolecular RNA duplexes phylogenetically conserved in Euryarchaea (35). The presumptive methylation guide activity of the C/D pre-tRNATrp intron has been experimentally confirmed in vitro, through incubation of a pre-tRNATrp transcript in the presence of an S100 extract prepared from halophilic euryarchaeon Haloferax volcanii and supplemented with S-adenosyl methionine (SAM). These experimental conditions allow for faithful splicing of pre-tRNATrp in vitro (36). Remarkably, both sites of 2′-O-methylation directed by the pre-tRNA intron involve nucleotides within the bulge–helix–bulge (BHB) splicing motif recognized by the archaeal tRNA endonuclease (37). Moreover, processes of RNA-guided methylation and pre-tRNA splicing appear to depend on mutually exclusive patterns of pre-tRNATrp folding (35). The cis-acting C/D tRNATrp intron might represent an ancestral form of the trans-acting C/D methylation guides now widespread in Archaea and Eukarya (35). Functional dissection of the archaeal pre-tRNATrp RNP complex might therefore provide novel insights into the structure and function of C/D methylation guides as well as reveal diversification of crucial features of their molecular organization in the evolution of the two fundamental groups of organism. In the present study, we have analyzed the in vitro reconstitution of an active, cis-acting methylation guide C/D box RNP using H.volcanii and Pyrococcus abyssi pre-tRNATrp, together with the three recombinant archaeal C/D sRNP core proteins, L7Ae, aNop5p and aFib, obtained from P.abyssi. In a first stage, we have investigated the association of the three proteins with pre-tRNAs by gel retardation assays. We have then analyzed the structure of the RNP complexes containing the pre-tRNA by RNA footprint experiments which have provided detailed information about the L7Ae- and Nop5p-binding sites and confirmed the occurrence of an overall RNA conformational change upon protein assembly. We also observed substantial methylation activity of a pre-tRNP complex assembled in the absence of L7Ae, revealing unexpected features of this cis-acting guide system as compared with classical trans-acting C/D RNAs.
MATERIALS AND METHODS
Unless otherwise noted, all techniques for cloning and manipulating nucleic acids were performed according to standard protocols (38).
PCR primer pairs
L7-5′His, CGCCATATGGAGGGATGGATGATGGC; and L7-3′His, CGCGGATCCTCACTTCATGAGCTCCC; Nop-5′His, CGCCATATGAAGGCGTTCATAGCTG; and Nop-3′His, CGCGGATCCTCACCTCCTCTTTCCC; Fib-5′His, CGCCATATGGTCGAGGTTAAGAAGC; and Fib-3′His, CGCGGATCCTTAAGTTTTCCTAACGACG; 1-CCGGAATTCTAATACGACTCACTATAGGGGGCGTGGTGTAGCCTGGTC; and 2-CGCGGATCCTGGTGGGGGCGCGGG GATTTG; 3-CCAGAGGCCATTCGGACAGGCGGG and 4-CCCGCCTGTCCGAATGGCCTCTGG; 5-CCTTTGGAGCCCCTCGGGCCGGAG and 6-CTCCGGCCCGAGGGGCTCCAAAGG; 7-GACCTCGGGCGTTCTGAACCTTTGG and 8-CCAAAGGTTCAGAACGCCCCGAGGTC; and 9-GCGGGTTTGCTTCCCTCGGGGCG and 10-CGCCCCGAGGGAAGCAAACCCGC.
Cloning, expression and purification of recombinant proteins
Genes encoding L7Ae, Nop5p and aFib (GenBank accession nos C75109, A75192 and H75191, respectively) were amplified from P.abyssi genomic DNA using primer pairs L7-5′His–L7-3′His, Nop-5′His–Nop-3′His and Fib-5′His–Fib-3′His, respectively, which included NdeI and BamHI restriction sites (underlined) at the 5′ and 3′ gene boundaries. After subsequent cloning in a pET15b vector resulting in N-terminally His-tagged proteins, all constructions were verified by DNA sequencing. Proteins were expressed in Escherichia coli BL21 (DE3) pLys codons+ (Novagen) at 37°C for 4 h. The His-tagged proteins were purified by nickel affinity chromatography using Ni-NTA–agarose (Qiagen) according to the manufacturer’s protocol. After concentration and dialysis against buffer A containing 20 mM HEPES-KOH pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol in Centricon YM-30 (for Nop5p) or YM-3 (for L7Ae and aFib) (Millipore), affinity tags were removed with thrombin cleavage for 2 h at room temperature. After a thermoprecipitation at 70°C for 10 min, the supernatant containing the thermoresistant cleaved proteins was recovered after centrifugation for 15 min at 16 110 g.
DNA template constructions and RNA synthesis
All H.volcanii pre-tRNATrp constructs and the full-length P.abyssi pre-tRNATrp DNA template were previously described (35). Mutant P.abyssi pre-tRNA templates were generated as reported for H.volcanii using pairs of appropriate mutagenic oligonucleotides: (1–2), (3–4), (5–6), (7–8) and (9–10) for Pa mutC, Pa mutD, Pa mut C′ and Pa mut D′ DNA templates, respectively.
RNAs were synthesized by in vitro transcription of the cognate DNA template. Large-scale synthesis of RNA transcripts was carried out in a 100 µl volume reaction containing 10 µg of BamHI-digested DNA template in 0.04 M dithiothreitol (DTT), 40 mM Tris pH 8.0, 24 mM MgCl2, 2 mM spermidine with 40 U of RNasin (Promega), 5 mM of each rNTP (Amersham Pharmacia) and 160 U of T7 RNA polymerase (Promega). The reaction was performed overnight at 37°C. After ethanol precipitation in the presence of 12 µl of 0.5 M EDTA, RNA was gel purified. 5′ End labeling with T4 polynucleotide kinase was performed on dephosphorylated RNA in the presence of [γ-32P]ATP and 3′ end labeling was carried out with [5′-32P]pCp and T4 RNA ligase. Uniformly 32P-labeled RNAs were synthesized as previously described (35). All labeled RNAs were repurified onto a 6% denaturating polyacrylamide gel. Prior to all RNP assembly experiments, the purified RNA was submitted to denaturation (1 min at 90°C in sterile water and cooled quickly on ice) and renaturation (room temperature for 10 min in buffer A or acetate buffer).
RNA–protein interaction by EMSA
About 0.25 fmol of radiolabeled RNA, mixed with 5 µg of E.coli tRNA, was incubated with increasing amounts of L7Ae, Nop5p or aFib in a 10 µl reaction for binding analysis. RNP complexes assembled by incubation at room temperature for 10 min were transferred to 70°C for 5 min, resolved by electrophoretic mobility shift assay (EMSA) on an 8% (19:1) polyacrylamide gel containing 0.5× TBE and 5% glycerol at 250 V in 0.5× TBE–5% glycerol buffer and visualized by autoradiography. After quantification of the signals by a Fuji-Bas-1000 imager, the dissociation constant and numbers of binding sites were determined by Hill plot analysis.
Footprint of proteins on RNA
RNP complexes were formed in the same conditions as for RNA–protein interaction in a 10 µl reaction at 70°C for 10 min in an acetate buffer (50 mM HEPES-KOH pH 7.2, 5 mM Mg acetate, 50 mM K acetate) with 0.1 pmol of 5′- or 3′-end-labeled pre-tRNA in the presence of 1 µg of tRNA and 10 pmol of each recombinant protein (1 µM). Prior to chemical probing, the RNP complex was cooled rapidly on ice. Lead-induced cleavages with 4 or 12 mM lead acetate freshly prepared in sterile water were performed at room temperature for 10 min. Reactions were stopped by adding 5 µl of 0.1 M EDTA before ethanol precipitation, as previously described (39). Chemical probing of naked RNA was performed in parallel in the same conditions. Mapping of accessible sites was achieved by utilization of an RNase T1 ladder [100 000 c.p.m. of transcript, 1 µg of E.coli tRNA (Sigma) in 20 mM Na citrate pH 5, 1 mM EDTA, 7 M urea and 0.1 U of RNase T1 for 10 min at 55°C] and a hydroxyl ladder (100 000 c.p.m. of transcript in 50 mM Na2CO3/NaHCO3, 1 mM EDTA, for 10 min at 70°C). Samples were resolved on a 12% denaturating polyacrylamide gel.
In vitro methyl incorporation
RNP complexes were assembled as described above in buffer A without glycerol in a reaction containing 200 pmol of RNA, 60 pmol of L7Ae, 30 pmol of Nop5p and 30 pmol of aFib. The mixtures were then added to 800 pmol of SAM (S-adenosyl-l-methionine dihydrogen sulfate) and 3.3 µCi of [3H]SAM (80 Ci/mmol; Amersham) in a final volume of 60 µl. After incubation at 70°C, 10 µl aliquots were removed at 0, 5, 15, 30 and 60 min, precipitated in 6 ml of 5% trichloroacetic acid (TCA; Sigma) at 4°C for 15 min and filtered on glass microfiber filters (GF/C, Whatman), followed by three 5% TCA washes, and then dried. Incorporation of [3H]methyl was determined by scintillation counting. Initial rates of methyl incorporation and extents of reaction were determined from at least three independent experiments.
Analysis of modified nucleotides
RNP complexes were assembled with 150 fmol of transcript labeled with [α-32P]ATP, [α-32P]UTP or [α-32P]CTP, 1 µg of E.coli tRNA, 10 pmol of each recombinant protein and 200 pmol of SAM. After gel purification of the RNP complex, analysis of modified nucleotides by two-dimensional thin-layer chromatography (2D-TLC) using 10 cm × 10 cm cellulose plates (Merck) was performed on an RNase T2 digest as previously described (35).
RESULTS
Binding of the Pyrococcus abyssi L7Ae protein to Haloferax volcanii and Pyrococcus abyssi pre-tRNATrp
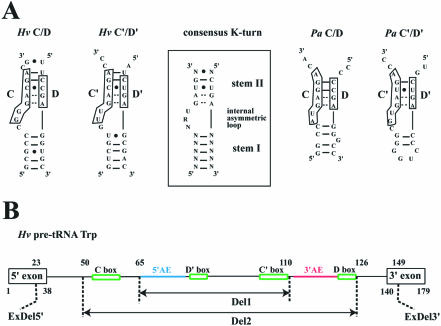
The L7Ae proteins of Sulfolobus solfataricus and Archeoglobus fulgidus have been shown to interact directly with the archaeal sR1 guide sRNA and the pre-tRNATrp, respectively (30,31). However, it was not clear if the two structural C/D and C′/D′ motifs of these archaeal sRNA guides were binding sites for the L7Ae protein. Predicted K-turns for C/D and C′/D′ pre-tRNA introns of H.volcanii and P.abyssi are shown in Figure 1A. The C/D and C′/D′ motifs of H.volcanii pre-tRNA as well as the C′/D′ motif of P.abyssi pre-tRNA can be structured according to the K-turn consensus motif even if stem II is shorter and if some deviations are observed in the internal loop. In contrast, the C/D motif of P.abyssi substantially deviates from the consensus, exhibiting a bulge of only two nucleotides and a Watson–Crick base pair GC instead of a non-canonical base pair UU in stem II.
Figure 1.
(A) Predicted K-turns motifs formed at the C/D and C′/D′ boxes of the pre-tRNATrp intron in H.volcanii and P.abyssi (left and right, respectively). The consensus K-turn structural motif derived for C/D RNAs in Eukarya and Archaea (boxed, middle part) harbors a canonical stem I, a non-canonical stem II extending from tandem sheared GA pairs, and an internal asymmetric loop of three nucleotides (20,25). (B) Structure of the two large intronic deletion mutants, Hv Del1 and Hv Del2, and of the exonic deletion Hv ExDel. The intact H.volcanii pre-tRNATrp structure is schematized (top line) with indication of sequence coordinates and of major sequence features (5′AE and 3′AE: 5′ and 3′ antisense elements, in blue and red, respectively).
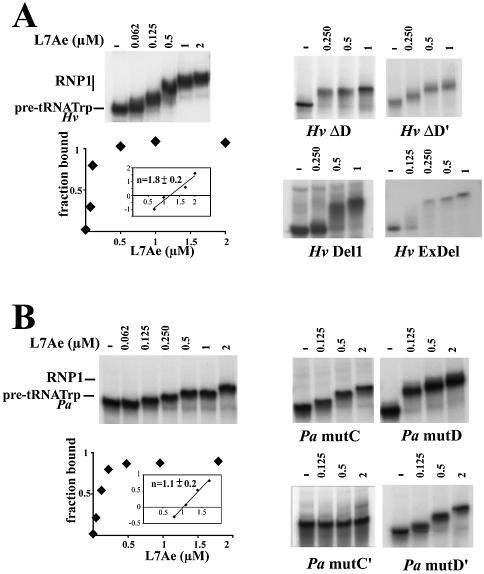
In a first stage, we tested the potential of the P.abyssi protein L7Ae to bind the H.volcanii (Hv) and P.abyssi (Pa) pre-tRNATrp introns. The in vitro 32P-labeled transcripts were incubated with increasing concentrations of protein and then analyzed by gel electrophoresis retardation assay (Fig. 2A and B). Hv pre-tRNATrp was clearly displaced in the presence of L7Ae into an RNP complex, RNP1 (Fig. 2A, left). In contrast, the L7Ae protein failed to bind in vitro transcripts corresponding to either the mature H.volcanii tRNATrp or a pre-tRNATrp transcript deleted of its central, 76 nt portion spanning its intronic box C/D hallmarks (mutant Hv Del2, Fig. 1B) [data not shown (35)]. Consistent with the role determined for boxes D/D′ in the recognition of trans-acting methylation guides by L7Ae (32), Hv pre-tRNATrp transcripts deleted from only one of the two D/D′ motifs, Hv ΔD or Hv ΔD′ RNAs, still formed an RNP complex but the mobility of the complex was substantially altered. Moreover, deletion of a larger intronic region spanning the 5′ antisense element (5′AE) and downstream boxes D′ and C′ (mutant Hv Del1, Fig. 1B) still allowed RNP complex formation [Fig. 2A, right (35)]. This indicates that while D and D′ boxes are both involved in L7Ae binding, recognition of the terminal C/D K-turn can take place in the absence of the internal C′/D′ K-turn. Finally, we tested a mutant pre-tRNA transcript, Hv ExDel (Fig. 1B), containing an intact intron and still able to form the BHB splicing motif but exhibiting dramatic 5′ and 3′ truncations of the tRNA 5′ and 3′ exons, respectively (35). In this case, again RNP complex formation was not suppressed, suggesting that canonical D- and T-stem–loops of the tRNA do not cooperate in L7Ae recognition (Fig. 2A, right).
Figure 2.
Binding of L7Ae to archaeal pre-tRNATrp in vitro assayed by gel mobility shift. The 32P-labeled RNA was incubated in the presence of increasing concentrations of the P.abyssi protein. Dissociation constants (shown below) for the complex, termed RNP1, were determined by measuring the fraction bound with respect to the total amount of protein. The number of binding sites, n, was determined from the slope of the Hill plot analysis in which the log[fraction bound/fraction non-bound] was plotted versus the log[protein]. Kd and n values were derived from a minimum of three independent experiments. (A) Haloferax volcanii pre-tRNATrp. Binding curve fitted to a sigmoidal binding curve for the wild-type RNA (left). Binding was studied for deletion mutants lacking box D, box D′, boxes C′ and D′, or the 5′ and 3′ regions of the 5′ and 3′ tRNATrp exons, respectively (right). (B) Pyrococcus abyssi pre-tRNATrp. Binding curve fitted to a binding isotherm for the wild-type RNA (left). Bindings were also studied for four different box mutants (right).
Likewise, the L7Ae protein could bind the in vitro transcribed P.abyssi pre-tRNATrp (Fig. 2B). To test the significance of the K-turns predicted for the P.abyssi specimen (Fig. 1), we systematically tested the effects of box point mutations abolishing formation of the tandem sheared G·A pairs. Mutation in the C′ box (GUGAUGA to GUCUUGA) abolished binding (mutant Pa mut C′). However, mutation of the GA dinucleotide of the D′ box (CUGA to CUUC) did not affect binding (mutant Pa mutD′). This unexpected difference between the two mutants might reflect the fact that the box D′ mutations, unlike the box C′ mutations, may somehow adopt an alternative geometry, favorable to L7Ae binding. Introduction of the same mutations within the C or D boxes (mutants Pa mutC and Pa mutD, respectively) did not affect the capacity of Pa pre-tRNATrp to bind to the L7Ae protein. This result is consistent with the notion that boxes C and D of Pa pre-tRNATrp do not form a canonical K-turn recognized by L7Ae and that the sole L7Ae-binding site in the Pa pre-tRNATrp corresponds to the C′/D′ K-turn. Overall dissociation constants (Kd) of binary complexes were evaluated by quantitative analysis of the mobility shift assays. Roughly similar values were derived for H.volcanii and P.abyssi pre-tRNATrp (120 and 160 nM, respectively) as well as the various box mutants allowing complex formation, except for Pa mutD and HvDel1 which exhibited increased (Kd = 60 nM) and decreased (Kd = 300 nM) affinities, respectively. These values are in the range of binding affinities previously observed for L7Ae and diverse RNAs harboring a K-turn motif (27,40–42), except for the recently reported Methanococcus jannaschii box C/D sR8 for which a substantially stronger binding was observed (32). The number of binding sites for L7Ae derived by Hill plot analysis of the quantification values of gel shift assays was one and two for P.abyssi and H.volcanii, respectively. In the case of H.volcanii, the absence of an additional shifted band of intermediate mobility between the naked pre-tRNATrp and the RNP1 complex suggests that the two L7Ae-binding sites are occupied almost simultaneously, probably reflecting a cooperative process (Fig. 2A, left) as reported for trans-acting M.jannaschii box C/D sR8 (32). However, we could not exclude that an instability of the RNA–protein complexes during electrophoresis at lower protein concentration could also account for a gradual increase in size for the shifted band. Altogether, analysis of L7Ae binding to Pa and Hv pre-tRNAs points to the presence of two K-turns in Hv pre-tRNATrp and a single K-turn in Pa tRNATrp (Fig. 1A).
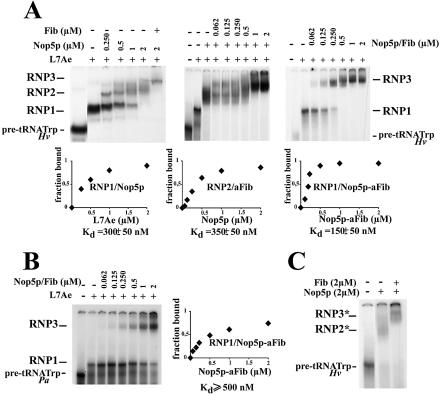
The two protein components of P.abyssi small C/D RNPs, Nop5p and aFib, bind specifically the pre-formed L7AE–pre-tRNATrp complex and the pre-tRNATrp alone at higher concentration
The aFib–Nop5p heterodimer of A.fulgidus has been shown to shift the pre-formed L7Ae–pre-tRNATrp complex in a gel retardation assay (31). Likewise, in our conditions, the complex formed between P.abyssi L7Ae and the pre-tRNATrp from either H.volcanii or P.abyssi underwent a marked mobility shift in the presence of increasing concentrations of the two P.abyssi proteins Nop5p and aFib to give rise to higher order complexes: Nop5p–L7Ae–pre-tRNATrp complex (RNP2) and aFib–Nop5p–L7Ae–pre-tRNATrp complex (RNP3), respectively (Fig. 3A and B). However, in contrast to the L7Ae protein, neither aFib nor Nop5p alone, nor the two proteins together in the absence of L7Ae were able to shift the mobility of the pre-tRNATrp at concentrations <1 µM (data not shown). These results are consistent with the assembly order onto the RNA substrate reported in the case of the sR1 Sulfolobus C/D RNA higher order complex: L7Ae, Nop5p and aFib (30). Interestingly, however, at concentrations >1 µM, Nop5p had by itself or in the presence of aFib the potential to displace the H.volcanii pre-tRNATrp (but not the mature tRNATrp) to give rise to higher order complexes, RNP2* and RNP3*, respectively (Fig. 3C). Binding of Nop5p to the pre-formed L7Ae–Hv pre-tRNATrp complex (Fig. 3A) corresponded to a dissociation constant 2-fold higher than previously reported for binding of the pre-mixed Nop5p–aFib to the pre-formed L7Ae–Hv pre-tRNATrp complex (31). If this increase is significant, it might reflect a substantial conformational change (with an entropic contribution) of Nop5p in the presence of aFib prior to its binding to the L7Ae–Hv pre-tRNATrp complex, in agreement with the formation of a Nop5p–aFib heterodimer in the crystal structure (31). Moreover, the shifted bands corresponding to complex RNP3 appeared generally less diffused than the bands corresponding to complex RNP2, probably reflecting a stabilization of the RNP complex in the presence of aFib (Fig. 3A). For the association of the Nop5p–aFib complex with L7Ae–Hv pre-tRNATrp, the number of binding sites determined by Hill plot analysis was undoubtedly two. However, because of the instability of the Nop5p–L7Ae–Hv pre-tRNA complex on the native gel, the number of binding sites for Nop5p and for aFib separately was difficult to ascertain. Interestingly, in the case of P.abyssi, the interaction of aFib–Nop5p with the pre-formed L7Ae–pre-tRNATrp complex was much weaker than for the H.volcanii pre-tRNA, possibly reflecting the absence of a functional K-turn at the box C/D motif (Fig. 3B).
Figure 3.
(A) Formation of higher order RNP complexes with H.volcanii pre-tRNATrp in the presence of the P.abyssi L7Ae concentration sufficient to shift >95% of the RNA into the RNP1 complex, together with increasing concentrations of Nop5p and/or aFib. The Nop5p–L7Ae–pre-tRNATrp and aFib–Nop5p–L7Ae–pre-tRNATrp complexes are termed RNP2 and RNP3, respectively. The numbers of binding sites were determined as in Figure 2. (B) Same as in (A), but with P.abyssi pre-tRNATrp. (C) Higher order complexes of H.volcanii pre-tRNATrp formed with Nop5p and aFib in the absence of L7Ae. The Nop5p–pre-tRNATrp and aFib–Nop5p–pre-tRNATrp complexes are termed RNP2* and RNP3*, respectively.
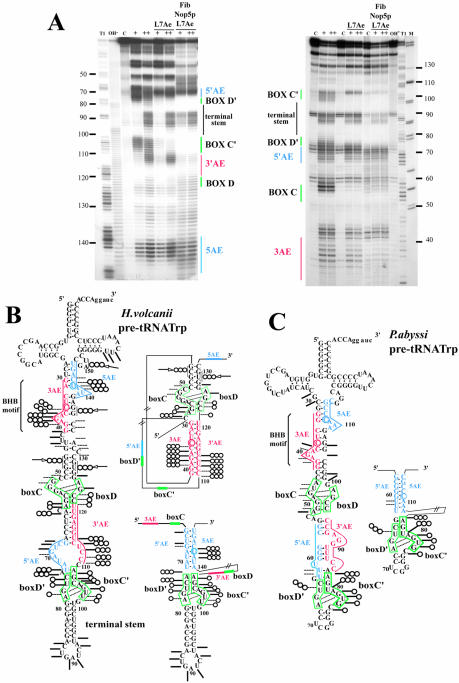
Footprinting of L7Ae, Nop5p and aFib on the pre-tRNATrp
In an attempt to map RNA–protein interactions and RNA conformational changes involved in the formation of higher order RNP complexes, we chemically probed the pre-tRNA molecule with lead acetate. This probe is able to induce cleavages in dynamic regions of the RNA phosphate backbone, such as interhelical or loop regions and bulged nucleotides (39). Cleavage patterns were determined for the naked RNA and for an RNA complexed with either L7Ae alone or the three proteins, aFib, Nop5p and L7Ae, using 5′- or 3′-end-labeled pre-tRNATrp transcripts (Fig. 4A). Cleavage patterns observed for naked pre-tRNAs were found to be in quite good agreement with the pre-tRNA secondary structure predicted from the presence of the hallmark BHB motif required for splicing (43,44) as well as K-turn motifs (25) involving the C/D and C′/D′ boxes (Fig. 4B and C). Thus, the phosphate backbones of nucleotides in the two 3-nt bulges of the BHB motif and in the bulges of the C/D and C′/D′ motifs were exposed to the probe, reflecting their flexibility. For the L7Ae–pre-tRNA binary complex, footprints of L7Ae revealed that L7Ae-binding regions were centered on both C/D and C′/D′ box motifs in the H.volcanii pre-tRNATrp (Fig. 4A and B), while they were restricted to the C′/D′ motif in the P.abyssi homolog ( Fig. 4C). This discrepancy between H.volcanii and P.abyssi is in agreement with the respective numbers of L7Ae-binding sites reported above for the two Archaea, i.e. two and one, respectively. The reactivity observed with the naked pre-tRNA in presumptive stems of the C/D and C′/D′ motifs for H.volcanii and the C′/D′ motif for P.abyssi suggests that the corresponding K-turns are not stably pre-formed and that L7Ae favors their formation, in line with recent conclusions about the snoRNA U3–Snu13p interaction (42). In the next stage, we analyzed the pattern of lead acetate accessibilities of the pre-tRNATrp complexed with the three proteins, L7Ae, Nop5p and aFib. Some new protections were observed as well as a few additional sensitive sites, as schematized on the two mutually exclusive pre-tRNATrp intron folding patterns competent for splicing and 2′-O-methylation guide activity, respectively (Fig. 4B and C). Interestingly, the P.abyssi pre-tRNATrp C/D motif differs from the three other predicted K-turns in the P.abyssi and H.volcanii homologs by a lack of protection of its stem II element. In contrast, stem II nucleotides U73 and C74 (in box D′) and C60 (downstream of box C) of the H.volcanii pre-tRNATrp as well as U66 (in box D′) and U81 (in box C′) of the P.abyssi pre-tRNATrp become fully protected from lead acetate-induced cleavage. Meanwhile, targets of both antisense elements as well as the 3′ antisense element of the H.volcanii intron were clearly protected in the presence of the three proteins (Fig. 4A). Conversely, additional cleavage sites were detected in the H.volcanii pre-tRNA, especially within the interval between the target of the 3′AE and the C box (positions 46, 47 and 48), downstream from the D box (positions 127, 128 and 129), upstream from the 5′AE (positions 136–137) and within the tRNA variable loop (position 151 and 152). Results of RNase T1 footprinting experiments (data not shown) were in full agreement with those observations, revealing that G57 (in box C), G77 (in box D′) and G105 (in box C′) were protected in the L7AE–pre-tRNA complex of H.volcanii, whereas G89 in the terminal stem remained exposed (data not shown). Conversely, the accessibility of G45 to RNase T1 increased in the complex formed with the three proteins (not shown). Moreover, the accessibility pattern observed for the Nop5p–L7Ae–pre-tRNATrp complex (data not shown) was indistinguishable from the aFib–Nop5p–L7Ae–pre-tRNATrp complex (Fig. 4B and C), suggesting that aFib was not directly involved in the footprint variations upon assembly of the higher order complex. In conclusion, for both types of structural probes, variations in the accessibility patterns among the different RNP complexes are consistent with the occurrence of an overall conformational change of the H.volcanii pre-tRNA driven by Nop5p binding. This change could correspond to the transition from the splicing-competent structure (Fig. 4B and C, left) to an RNA conformer in which RNA duplexes guiding the 2′-O-methylation of Cm34 and Um39 are formed (Fig. 4B and C, right). Alternatively, the modified accessibilities might reflect direct binding of Nop5p to the pre-tRNA at both target sites as well as stem II in both K-turns.
Figure 4.
Footprinting analysis of the L7Ae–pre-tRNATrp and aFib–Nop5p–L7Ae–pre-tRNATrp complexes using lead acetate as structural probe. (A) Mapping of the lead acetate cleavage sites along H.volcanii pre-tRNATrp using a 5′- or 3′-end-labeled transcript (left and right, respectively). After pre-incubation of the transcript at 70°C and formation of the complex as in Figure 3 (see Materials and Methods), lead acetate was used at two different concentrations: 4 mM (+) and 12 mM (++). An RNase T1 ladder and an alkaline hydrolysis pattern (T1 and OH-, respectively) as well as a control experiment (C) without lead acetate were analyzed in parallel. M, molecular weight markers (pBR322 digested with HaeIII and TaqI). The location of major sequence features in the pre-tRNATrp is indicated along the gel (5′AE and 3′AE: 5′ and 3′ antisense elements, in blue and red, respectively; 5AE and 3AE: sequences matched by the 5′AE and 3′AE, in blue and red respectively). (B and C) Schematic representation of the lead acetate probing data obtained for H.volcanii and P.abyssi pre-tRNATrp, respectively. For each species, the pre-tRNA splicing-competent structure is represented on the left, while an alternative secondary structure involving formation of cis-acting RNA duplex(es) guiding a 2′-O-methylation is schematized on the right. For H.volcanii, the two guide RNA duplexes which might form concomitantly are shown on two separate structures (on top of each other) for clarity, while a single guide duplex involving the C′/D′ motif is shown for P.abyssi pre-tRNA. Strong and weak cleavages are denoted by bars and dots, respectively (data were confirmed by a minimum of three independent experiments). Phosphate backbone positions accessible in the naked pre-tRNATrp and protected from cleavage in the RNP1 or RNP3 complexes are indicated by one (O) and three circles (OOO), respectively. Three circles accompanied by an arrow (→OOO) indicate new cleavages appearing in the RNP3 complex only.
Methylation activity of in vitro reconstituted C/D box pre-tRNP
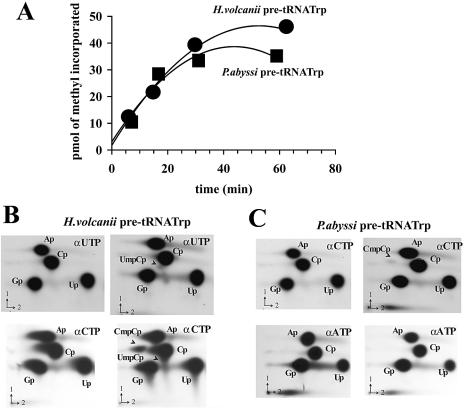
While the 2′-O-methylations at positions C34 and U39 in the H.volcanii tRNATrp have been experimentally verified (45), the presence of Cm34 and Cm39 in P.abyssi tRNATrp has been merely inferred from the conservation of the cognate antisense elements in its C/D intron (35). In an in vitro splicing/RNA modification assay using H.volcanii S100 cellular extract, ribose methylations of the H.volcanii pre-tRNATrp at both positions were faithfully reproduced and the predicted guide function of the box C/D intron was confirmed (35). We therefore tested the potential of the in vitro assembled aFib–Nop5p–L7Ae–pre-tRNATrp complex (RNP3) to sustain a site-specific methyl transferase activity in the presence of added SAM. In a first stage, using SAM radiolabeled at the methyl position, we assessed the incorporation of radioactivity into the P.abyssi and H.volcanii pre-tRNATrp (Fig. 5A). The rapid incorporation of labeled methyl groups reaching a plateau after 30–40 min observed in both cases was strictly dependent on the presence of aFib in the complex (data not shown). The amount of methyl groups incorporated after 60 min, i.e. ∼50 pmol for Hv pre-tRNA, was nearly twice the molar amount of aFib present (30 pmol) in a reaction carried out in the presence of a vast excess of pre-tRNA and SAM (200 and 825 pmol, respectively). This could indicate one turnover of the enzyme, assuming that the three recombinant proteins remained fully active in our conditions. In agreement with previous observations on a Sulfolobus reconstitution system (30), the plateau reached after ∼40 min appears to be largely a consequence of the degradation of the RNA and SAM cofactor at 70°C (data not shown), possibly combined with a slow product release.
Figure 5.
In vitro 2′-O-methylation activity of the reconstituted RNP complex (RNP3). (A) Kinetics of labeled methyl incorporation into the H.volcanii (circles) or P.abyssi (squares) pre-tRNATrp transcripts. (B) Characterization of the 2′-O-methylation sites by 2D-TLC of an RNase T2 digest of in vitro synthesized H.volcanii pre-tRNATrp. The transcript was labeled by incorporation of [α-32P]UTP and [α-32P]CTP (top and bottom, respectively) in the absence and presence (left and right panels, respectively) of the three recombinant proteins supplemented with SAM. Chromatographic separation was performed with system B. (C) As in (B) using in vitro synthesized P.abyssi pre-tRNATrp. The transcript was labeled by incorporation of [α-32P]CTP and [α-32P]ATP (top and bottom, respectively) in the absence and presence (left and right panels, respectively) of the three recombinant proteins supplemented with SAM.
Assignment of the sites of ribose methylation within the pre-tRNA assembled in the higher order RNP complex was performed by using H.volcanii pre-tRNATrp labeled by incorporation of [α-32P]NTP. After a pre-incubation at 70°C with an excess of the three P.abyssi recombinant proteins, the labeled RNA was incubated in the presence of SAM, and sites of 2′-O-methylation assessed by 2D-TLC of an RNase T2 digest of gel-purified pre-tRNA, as has been described (35). Using an [α-32P]UTP-labeled H.volcanii pre-tRNA, a single labeled dinucleotide spot was detected, UmpCp, whereas two labeled dinucleotide spots, UmpCp and CmpCp, were detected by using either an [α-32P]CTP-labeled or an [α-32P]ATP-labeled pre-tRNA from H.volcanii (Fig. 5B and data not shown). These data are in full agreement with the in vitro formation of Cm34 and Um39, as previously observed following incubation of the pre-tRNATrp with an H.volcanii cellular extract (35). However, the CpmCp spot was at least 10 times more intense than UpmCp, a ratio in line with the discrepancy in the initial rate of the reaction determined for both mutants Hv ΔD′ RNA and Hv ΔD RNA (see Table 1). When the same assay was performed with an [α-32P]CTP-labeled P.abyssi pre-tRNATrp, a single labeled dinucleotide spot was detected, CpmCp, indicating that the modification occurred at position 39 only (Fig. 5C). Moreover, as expected, no methyl incorporation was detected after deletion of the entire C– D′–C′–D-containing region in the H.volcanii intron (mutant Hv Del2). In contrast, methyl incorporation was not adversely affected by the extensive 5′ and 3′ truncations of the Hv pre-tRNATrp 5′ and 3′ exons, respectively, in mutant Hv ExDel (Table 1). The deletion of box D only (mutant Hv ΔD RNA) decreased the initial rate of methyl incorporation only slightly. Conversely, deletion of box D′ (mutant Hv ΔD′ RNA) or deletion of the internal C′/D′ containing part of the intron (mutant Hv Del1 RNA) resulted in an increase (>2-fold) of both the initial rate and extent of the reaction (Table 1). In these two mutant transcripts containing only the C/D motifs, the presence of an RNA conformer with a free energy closer to the methylation-competent structure might facilitate product release leading to a turnover of the enzyme. As for the P.abyssi pre-tRNA, while the integrity of C′ and D′ boxes was essential for methyl incorporation (Table 1), mutations in the C and D boxes preventing formation of the tandem sheared G·A pairs in the K-turn motif (i.e. a GA to CU change) had no or reduced effect, respectively, on the initial rate of methyl incorporation, again consistent with the absence of a functional K-turn at the P.abyssi pre-tRNA C/D motif.
Table 1. Initial rates and extents of methyl incorporation at the plateau value for wild-type and mutant pre-tRNATrp from H.volcanii and P.abyssi.
| RNA | Initial rate (pmol methyl/min) | Extent (pmol methyl) |
|---|---|---|
| Hv pre-tRNATrp | 1.5 ± 0.4 | 50 ± 5 |
| Hv ΔD RNA | 0.5 ± 0.2 | 7 ± 3 |
| Hv ΔD′ RNA | 4 ± 0.5 | 113 ± 15 |
| Hv Del1 RNA | 3 ± 0.5 | 120 ± 20 |
| Hv Del2 RNA | 0 | 0 |
| Hv ExDel | 1.5 ± 0.4 | 60 ± 10 |
| Hv tRNATrp | 0 | 0 |
| Pa pre-tRNATrp | 1.8 ± 0.5 | 30 ± 10 |
| Pa mutC | 1.2 ± 0.5 | 29 ± 10 |
| Pa mutD | 0.5 ± 0.1 | 11 ± 5 |
| Pa mutC′ | 0 | 0 |
| Pa mutD′ | <0.1 | <4 |
| Pa mutC′/D′ | 0 | 0 |
Initial rates correspond to the slope of the curve for the 0–30 min interval.
Substantial methylation activity of in vitro reconstituted aFib–Nop5p–H.volcanii pre-tRNATrp complex (RNP3*)
Unexpectedly, we observed a very significant methyl incorporation in the aFib–Nop5p–H.volcanii pre-tRNATrp complex (termed RNP3* in Fig. 3C) reconstituted in the absence of L7Ae and incubated in the presence of radiolabeled SAM. The initial rate of incorporation reached 0.3 ± 0.1 pmol of methyl/min, i.e. ∼20% of the rate observed for the RNP3 complex assembled in the presence of L7Ae (Table 1), while no significant methyl incorporation could be observed when using mature H.volcanii tRNATrp as a control. Mapping of the site(s) of 2′-O-methylation within the pre-tRNATrp assembled in the RNP3* complex was performed by 2D-TLC as before (Fig. 6). A CmCp spot indicative of Cm34 formation was clearly detected in the RNP3* complex, but the UmCp spot previously observed with the RNP3 complex assembled in the presence of L7Ae was missing, reflecting the lack of Um39 formation within the detection limits of our assay.
Figure 6.
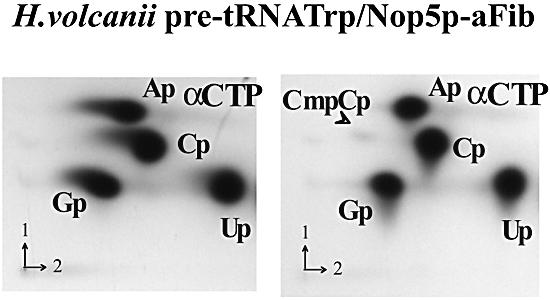
In vitro 2′-O-methylation activity of the reconstituted aFib–Nop5p–H.volcanii pre-tRNATrp complex (or RNP3* complex). The pre-tRNATrp transcript was labeled by incorporation of [α-32P]CTP in the absence and presence (left and right panels, respectively) of the two recombinant proteins, aFib and aNop5p, supplemented with SAM, and the sites of 2′-O-methylation were identified by 2D-TLC of an RNase T2 digest of the RNA, as in Figure 5B.
DISCUSSION
The archaeal C/D pre-tRNATrp was used as a model to further examine both the assembly of C/D RNPs and mechanisms of the guided ribose methylation reaction. Using the purified cloned protein components of C/D RNP particles in an in vitro assembly system, we have analyzed the interactions of these proteins with the intronic C/D RNA and the requirements for formation of methylation-competent RNA conformers. Clearly the two K-turns involving the C/D and C′/D′ motifs of the H.volcanii pre-tRNA intron are equivalent sites for L7Ae binding (i.e. they exhibit the same affinity) but not for ribose methylation, since significantly different initial rates of methyl incorporation were measured for Cm34 and Um39. Interestingly, the C/D motif is even more efficient for methylation in the absence of the C′/D′ motif, in line with the notion of a terminal box C/D constituting the minimal methylation-competent particle in trans-acting guides (32). A less rigid RNP complex with only one L7Ae bound to the C/D motif could account for the increased activity. Concerning the C/D motif, however, our present data seem somewhat at odds with results obtained with the in vitro splicing/RNA modification assay using an S100 extract which showed a complete block of box D-directed methylation activity in various mutants exhibiting box D′ deletions (35). Still unidentified factors in the cellular extract could account for this discrepancy and for the difference in the yield of Cm34 and Um39 modifications. Remarkably, in contrast to H.volcanii, the only functional K-turn in the P.abysii pre-tRNATrp intron is the C′/D′ motif, and the affinity of aFib–Nop5p for a pre-formed L7Ae–pre-tRNA complex is significantly lower for P.abyssi than for H.volcanii (Fig. 3A and B).
L7Ae binds primarily to the internal loop of the K-turn formed by the C/D (C′/D′) motif, whereas Nop5p binds to the non-canonical stem II of the K-turn, thereby probably favoring the formation of the intramolecular RNA duplex required for ribose methylation in cis. The non-canonical UU base pair in stem II of the P.abyssi C′/D′ K-turn, which is protected in our experiments by Nop5p, has been shown in eukaryotes to cross-link with Nop58p and fibrillarin during in vivo snoRNP assembly in Xenopus oocytes (28). Moreover, this base pair and the conserved GC base pair of stem II required for methylation activity of archaeal pre-tRNATrp are essential for the association of eukaryal C/D proteins Nop56/Nop58, fibrillarin, TIP48 and TIP49 and critical for the nucleolar localization of U14 box C/D snoRNA injected into HeLa cell nuclei (27). Altogether, binding sites for the stem II-dependent protein, Nop5p in Archaea or Nop56p and Nop58p in Eukarya, seem to be generated upon L7Ae binding to the C/D motif, which probably stabilizes the kink introduced into the RNA backbone.
Although the internal C′/D′ motif in trans-acting archaeal guide sRNAs has been recently considered as a variant of the classical K-turn fold exhibited by the C/D motif (32), the P.abyssi pre-tRNATrp intron is clearly able to form an active K-turn at its C′/D′ motif. While its stem I is particularly short (only two GC base pairs extended by a 4 nt loop), the internal loop matches perfectly the C/D K-turn consensus, which might make up for a shorter stem I, as suggested in recent U3 C/D snoRNA studies (42). It is noteworthy that the C′/D′ motif of most Pyrococcus sRNAs guiding rRNA methylations does not harbor a stem I, within only 3 nt separating the C′ and D′ boxes (8). A consensus GNG can be defined for these 3 nt (B.Clouet-d’Orval, unpublished data) which might compensate somehow for the absence of stem I.
Our analysis of various mutants of the pre-tRNATrp intron has confirmed the key importance of ribose methylation of the D and D′ boxes downstream from each antisense element. In the two phylogenetically supported intramolecular RNA duplexes (35), ribose methylated nucleotides 34 and 39 both map at the fifth position upstream from box D and D′, pointing to a nucleotide targeting mechanism closely related to the snoRNA-guided methylation of eukaryotic rRNA and snRNA (1–3,5). However, the eukaryotic asymmetric C/D snoRNP assembly model departs from the archaeal symmetric model. The latter is based on the observed 2-fold symmetry between two Archeoglobus fibrillarin–Nop5p heterodimers, suggesting that each fibrillarin–Nop5p heterodimer interacts with one box C/D (or C′/D′) motif in the presence of L7Ae (31), and on in vitro assembly studies of the terminal box C/D core and internal C′/D′ RNA half-molecules (32). In eukaryotes, the notion that the fibrillarin–Nop56–Nop58–15.5 kDa complex interacts with box C/D RNA asymmetrically is supported by chemical cross-linking studies and nucleotide analog interference mapping (28,29). This is also in agreement with the presence of a single guide sequence upstream from either box D or D′ in most eukaryal box C/D RNAs (7,12,46,47), in contrast to the majority of archaeal C/D sRNAs which exhibit two antisense elements (8,9). In this hypothesis, the eukaryal particle would comprise two different heterodimers of fibrillarin with Nop58p and Nop56p, respectively, and one 15.5 kDa, while the archaeal particle appears to contain two identical fibrillarin–Nop5p heterodimers and two L7Ae (31,32). However, among Eukarya, plant C/D snoRNAs are remarkable by the widespread occurrence of two antisense elements (48–50), suggesting that the asymmetric protein distribution of C/D RNPs might not apply to all eukaryal lineages. In agreement with this notion, a striking example of functional diversification of a C/D snoRNA in the evolution of Eukarya has been recently reported, regarding Drosophila U14 (51). Further analysis of C/D RNP assembly focused on a plant model might illuminate this point. In line with the symmetrical model of archaeal sRNP assembly, our present results strongly suggest that the pre-tRNATrp of H.volcanii harbors a selective binding site for L7Ae at each of its two K-turn motifs, thus providing the basis for the binding of one Nop5p–aFib heterodimer at each site. In contrast, the P.abyssi pre-tRNATrp must exhibit a markedly different RNP organization resulting from the non-canonical structure of its C/D motif, suggesting that it can guide only one of the two predicted tRNATrp 2′-O-methylations, Cm39 but not Cm34. However, the presumably methylation-incompetent antisense element associated with the C/D motif in Pyrococcus exhibits a perfect complementarity to its cognate RNA target, preserved through several compensatory base changes among distant Euryarchaea (35). This might reflect a distinct function of the RNA duplex structure, possibly in chaperoning pre-tRNA folding, in addition to (or instead of) guiding a tRNA 2′-O-methylation. As stressed previously, box C/D and H/ACA snoRNAs have the potential to act as intrinsic RNA chaperones controlling the folding of their RNA targets, in addition to guiding RNA modification (52). While the two functions seem inextricably linked in most cases, the Pyrococcus pre-tRNATrp C/D intron might represent a unique example of dissociated RNA chaperone and modification guide functions in Archaea.
Molecular mechanisms of the intramolecularly guided methylations of archaeal pre-tRNATrp and their potential coupling with archaeal tRNA splicing remain elusive. Both types of reaction depend on mutually exclusive folding patterns of the pre-tRNA intron and its stepwise structural rearrangements during processing. A single, methylation-competent conformer displaying canonical guide RNA duplexes at both target sites can be drawn, which would include at least one pseudoknot (Fig. 4B and C). However, formation of different conformers harboring a single guide RNA duplex cannot be ruled out at this stage. The pre-tRNA bound to the C/D core proteins must assume a conformation dramatically different from the one including the BHB structural motif. A difference in free energy between the methylation-competent and splicing-competent conformers could be compensated by protein contacts involving the three archaeal C/D RNP proteins as well as the splicing endonuclease. In the classical trans-acting modification guide system, an important structural reorganization of both guide and target RNAs must take place upon formation/dissociation of the binary RNA duplex at the modification site. Recently, an outstanding example of RNA conformational reorganization of tRNA upon binding to a tRNA modification enzyme, the archeosine tRNA-guanine transglycosylase (ArcTGT), was observed in a co-crystal (53). The buried target site of the substrate must undergo a profound conformational change resulting in an alternative tRNA structure, the so-called λ-form, drastically different from the L form, in which the D stem and canonical tertiary interactions are disrupted.
Our present results suggest that Nop5p might play a crucial role in stabilizing the methylation-competent form of pre-tRNATrp folding, thereby favoring directly or indirectly the formation of the cognate intramolecular RNA duplexes. Indeed, a pre-tRNATrp assembled with Nop5p and the methylase aFib but devoid of L7Ae still exhibits a substantial site-specific methylation activity (Fig. 6). Consistent with the possibility that Nop5p might recognize a double-helical RNA, plant homologs of Nop56 and Nop58 proteins bind matrix attachment regions of the chromosome through the recognition of the minor groove of A/T-rich DNA sequences (54), and in the crystal structure of the Nop5p–aFib heterodimer a patch of positively charged surface in the C-terminal domain delineates a site for binding negatively charged RNA backbone (31).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to A. Mougin for helpful discussions and Y. De Préval for oligodeoxynucleotide synthesis. We thank B. Charpentier and C. Branlant for providing us with a batch of recombinant P.abyssi GST-tagged L7Ae protein used in initial stages of this study. This work was supported by laboratory funds from the Centre National de la Recherche Scientifique and Université Paul-Sabatier, Toulouse, and by a grant from the Ministère de l’Education Nationale, de la Recherche et de la Technologie to J.P.B. (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, 2001–2002).
REFERENCES
- 1.Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 255–272. [Google Scholar]
- 2.Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachellerie J.P., Cavaille,J. and Huttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. [DOI] [PubMed] [Google Scholar]
- 4.Dennis P.P., Omer,A. and Lowe,T. (2001) A guided tour: small RNA function in Archaea. Mol. Microbiol., 40, 509–519. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 6.Omer A.D., Ziesche,S., Decatur,W.A., Fournier,M.J. and Dennis,P.P. (2003) RNA-modifying machines in archaea. Mol. Microbiol., 48, 617–629. [DOI] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z., Henry,Y. and Kiss,T. (1998) Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaspin C., Cavaille,J., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 9.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 10.Cavaille J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- 11.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 12.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 13.Galardi S., Fatica,A., Bachi,A., Scaloni,A., Presutti,C. and Bozzoni,I. (2002) Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol., 22, 6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.H. (2000) Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafontaine D.L. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyman S.K., Gerace,L. and Baserga,S.J. (1999) Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA, 5, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watkins N.J., Segault,V., Charpentier,B., Nottrott,S., Fabrizio,P., Bachi,A., Wilm,M., Rosbash,M., Branlant,C. and Luhrmann,R. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- 21.Wu P., Brockenbrough,J.S., Metcalfe,A.C., Chen,S. and Aris,J.P. (1998) Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao H. and Williamson,J.R. (1999) Local folding coupled to RNA binding in the yeast ribosomal protein L30. J. Mol. Biol., 292, 345–359. [DOI] [PubMed] [Google Scholar]
- 23.Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Luhrmann,R. (1999) Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidovic I., Nottrott,S., Hartmuth,K., Luhrmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- 26.Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins N.J., Dickmanns,A. and Luhrmann,R. (2002) Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol., 22, 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cahill N.M., Friend,K., Speckmann,W., Li,Z.H., Terns,R.M., Terns,M.P. and Steitz,J.A. (2002) Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J., 21, 3816–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szewczak L.B., DeGregorio,S.J., Strobel,S.A. and Steitz,J.A. (2002) Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol., 9, 1095–1107. [DOI] [PubMed] [Google Scholar]
- 30.Omer A.D., Ziesche,S., Ebhardt,H. and Dennis,P.P. (2002) In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 99, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aittaleb M., Rashid,R., Chen,Q., Palmer,J.R., Daniels,C.J. and Li,H. (2003) Structure and function of archaeal box C/D sRNP core proteins. Nature Struct. Biol., 10, 256–263. [DOI] [PubMed] [Google Scholar]
- 32.Tran E.J., Zhang,X. and Maxwell,E.S. (2003) Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J., 22, 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn J.F., Tran,E.J. and Maxwell,E.S. (2002) Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kDa/Snu13p snoRNP core protein. Nucleic Acids Res., 30, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fatica A. and Tollervey,D. (2003) Insights into the structure and function of a guide RNP. Nature Struct. Biol., 10, 237–239. [DOI] [PubMed] [Google Scholar]
- 35.Clouet-d’Orval B., Bortolin,M.L., Gaspin,C. and Bachellerie,J.P. (2001) Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res., 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armbruster D.W. and Daniels,C.J. (1997) Splicing of intron-containing tRNATrp by the archaeon Haloferax volcanii occurs independent of mature tRNA structure. J. Biol. Chem., 272, 19758–19762. [DOI] [PubMed] [Google Scholar]
- 37.Nieuwlandt D.T., Carr,M.B. and Daniels,C.J. (1993) In vivo processing of an intron-containing archael tRNA. Mol. Microbiol., 8, 93–99. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 39.Brunel C. and Romby,P. (2000) Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol., 318, 3–21. [DOI] [PubMed] [Google Scholar]
- 40.Rozhdestvensky T.S., Tang,T.H., Tchirkova,I.V., Brosius,J., Bachellerie,J.P. and Huttenhofer,A. (2003) Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res., 31, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang T.H., Rozhdestvensky,T.S., Clouet-d’Orval,B., Bortolin,M.L., Huber,H., Charpentier,B., Branlant,C., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res., 30, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marmier-Gourrier N., Clery,A., Senty-Segault,V., Charpentier,B., Schlotter,F., Leclerc,F., Fournier,R. and Branlant,C. (2003) A structural, phylogenetic and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA, 9, 821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lykke-Andersen J. and Garrett,R.A. (1997) RNA–protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J., 16, 6290–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lykke-Andersen J., Aagaard,C., Semionenkov,M. and Garrett,R.A. (1997) Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci., 22, 326–331. [DOI] [PubMed] [Google Scholar]
- 45.Gupta R. (1984) Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids and the sequences of 33 class I tRNAs. J. Biol. Chem., 259, 9461–9471. [PubMed] [Google Scholar]
- 46.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 47.Bachellerie J.P. and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- 48.Barneche F., Gaspin,C., Guyot,R. and Echeverria,M. (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- 49.Brown J.W., Clark,G.P., Leader,D.J., Simpson,C.G. and Lowe,T. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 50.Marker C., Zemann,A., Terhorst,T., Kiefmann,M., Kastenmayer,J.P., Green,P., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol., 12, 2002–2013. [DOI] [PubMed] [Google Scholar]
- 51.Yuan G., Klambt,C., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2003) RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res., 31, 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachellerie J.P., Cavaille,J. and Qu,L.H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNA guides revisited. In Grosjean,H. and Benne,R. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203. [Google Scholar]
- 53.Ishitani R., Nureki,O., Nameki,N., Okada,N., Nishimura,S. and Yokoyama,S. (2003) Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell, 113, 383–394. [DOI] [PubMed] [Google Scholar]
- 54.Hatton D. and Gray,J.C. (1999) Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J., 18, 417–429. [DOI] [PubMed] [Google Scholar]