Abstract
Several single nucleotide polymorphisms (SNPs) have been linked to antimalarial drug resistance in Plasmodium falciparum . However, standard polymerase chain reaction (PCR) methods to detect these polymorphisms are unable to detect SNPs in variants representing < 20% of the parasites in a mixed infection, nor can they detect polymorphisms at nearby loci. Here we use heteroduplex tracking assays (HTAs) to analyze dhps540 mutations in 96 samples from Peru and pfcrt76 mutations in 70 samples from China. All samples had been previously analyzed by standard PCR. We detected drug-resistant minority variants and two novel non-synonymous pfcrt mutations in China. In Peru, we found no drug-resistant minority variants and a synonymous mutation in dhps. Thus, even in regions of low malaria transmission, HTA assays are more informative than PCR with agarose gel electrophoresis.
Drug-resistant Plasmodium falciparum malaria is a growing worldwide problem. Molecular markers promise to be important surveillance tools.1 Single nucleotide polymorphisms (SNPs) in genes such as dhfr, dhps, cytb, and pfcrt have been associated with antimalarial resistance in vitro and in vivo. However, there are two problems with the current methods. First, standard polymerase chain reaction (PCR) methods, such as restriction length polymorphism (RFLP), allele-restricted PCR (AR-PCR), and real-time PCR, can misclassify patients with multiclonal infections because PCR cannot reliably detect variants representing < 20% of the parasite population in a single host (minority variants).2–4 Second, these methods are designed to target only specific mutations and consequently do not provide information about unknown or unsuspected mutations in variant alleles. Heteroduplex tracking assays (HTAs) for drug resistance SNPs, on the other hand, are sensitive to minority variants and may detect polymorphisms at positions near known resistance mutations.2,3,5,6 In this report, we describe a new HTA for dhps540 that is sensitive for minority variants. We use this assay and a previously described HTA for pfcrt76 to evaluate clinical isolates for novel and previously described SNPs. Our findings include the presence of two novel non-synonymous polymorphisms in pfcrt and one novel synonymous polymorphism in dhps.
Clinical samples obtained from two cohorts were evaluated by HTA. For the pfcrt analysis, genomic DNA was used from 70 samples that were collected in 2001 from consenting symptomatic uncomplicated malaria patients in Hainan, People’s Republic of China.7 For the dhps analysis, genomic DNA from 96 consenting patients with symptomatic uncomplicated P. falciparum malaria was provided by the US Naval Medical Research Center Detachment, Lima, Peru. A single replicate of the dhps540 HTA was carried out on these samples.
The pfcrt76 HTA was carried out in duplicate on these samples as previously described in the literature.2 For the dhps540 HTA, PCR amplification of the dhps540 gene was carried out using a Peltier thermal cycler (MJ Research, Waltham, MA) in a volume of 50 µL. The reaction consisted of 5 µL DNA, 1.25 units HotStar Taq DNA polymerase (Qiagen, Valencia, CA), 5 µL 10× PCR buffer, 1 µL 10 mmol/L dNTP mix (Promega, Madison, WI), and 300 nmol/L of forward and reverse primers, which were previously described.8 This reaction was amplified by preheating to 95°C for 15 minutes, followed by 35 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 1 minute. The reaction was completed with a 10-minute hold at 72°C. Probe development, screening, evaluation, and harvesting were carried out as previously described.2,3,6 The final probe contained the following mutations relative to the dhps540 mutation of interest (A to G in the first position of codon 540): −3 G to A, −1 T to G, +1 A to T, and +3 C to T.
The probe was radiolabeled, and HTAs were performed under the conditions noted by Ngrenngarmlert and others 6 with some modifications. An annealing reaction consisting of 8 µL PCR product (either a control or sample DNA) was mixed with 1 µL 10× annealing buffer (1 mol/L NaCl, 100 mmol/L Tris-HCL, pH 7.5, 20 mmol/L EDTA), 2 µL 6× loading dye, 0.5 µL 0.1 mmol/L F primer, and 0.5 µL radiolabeled probe in a total volume of 12 µL. The annealing reaction and electrophoresis were carried out under the conditions previously described.2,3 All HTA gels included the following controls: a non-template control (NTC) PCR reaction and PCR reactions from the appropriate wild-type and mutant DNA stocks. Gels were analyzed as previously described.2,3
Malaria DNA samples and controls used to develop assays were acquired from MR4 (www.mr4.org). For pfcrt76 assays, P. falciparum strain 3D7 (MRA-102G, from strain MRA-102 deposited by DJ Carucci) was used for wild-type DNA and strain K1 (MRA-159G, from strain MRA-159 deposited by DE Kyle) for mutant DNA. Plasmids (∼5,100 bp in size) containing wild-type (MRA-189) and mutant (MRA-190) dhps DNA were used for the dhps540 assays.
The sensitivity of the dhps540 HTA to detect minority variants was tested using artificial mixtures of control DNA (wildtype and mutant plasmids) in duplicate as described previously in the literature.2,3 The plasmid mixtures were maintained at a total concentration of 1 × 10−5 ng/µL DNA (roughly equivalent to 0.1 ng genomic DNA based on a 22.8−MB genome). Bands were only counted as positive if they were visible to the eye. The HTA probe formed heteroduplexes with different mobilities when annealed to P. falciparum DNA amplicons from wild-type parasites and from parasites containing the resistance mutations of interest. It was possible to accurately and reproducibly detect drug-resistant minority variants comprising as little as 1% of the total population (Table 1).
TABLE 1.
Sensitivity of the dhps540 HTA for drug-resistant minority variants
| Percent mutant DNA* | Percent mutant detected (replicate 1)† |
Percent mutant detected (replicate 2)† |
|---|---|---|
| 50 | 46.1 | 45.5 |
| 20 | 21.1 | 16.2 |
| 10 | 8.7 | 7.3 |
| 5 | 3.9 | 3.8 |
| 1 | 1.2 | 1.3 |
| 0.1 | 0 | 0 |
These values represent the known percentage of mutant DNA in the mixture.
These values represent the percent of mutant DNA detected by the HTA as quantified by the intensity of the mutant heteroduplex band by PhosphorImager compared with the intensity of both the wild-type and mutant heteroduplex bands.
The pfcrt76 HTA successfully detected DNA in 55/70 (79%) of the P. falciparum isolates from Hainan, China. Because of depleted DNA stocks, repeat evaluation with more sample DNA was not possible. Twenty-two (40%), 28 (51%), and 5 (9%) samples contained pure wild-type, pure mutant, and mixed parasites, respectively, giving a prevalence similar to previous reports.7 However, among the mixed samples, three samples contained minority-variant drug-resistant parasites that would have been missed by conventional genotyping.2 These represented 11.3%, 3.4%, and 14.4% of the parasite DNA. Thus, 60% of samples would be classified as resistant (pure or mixed mutant) by HTA, whereas only 55% would be expected by ARPCR.2
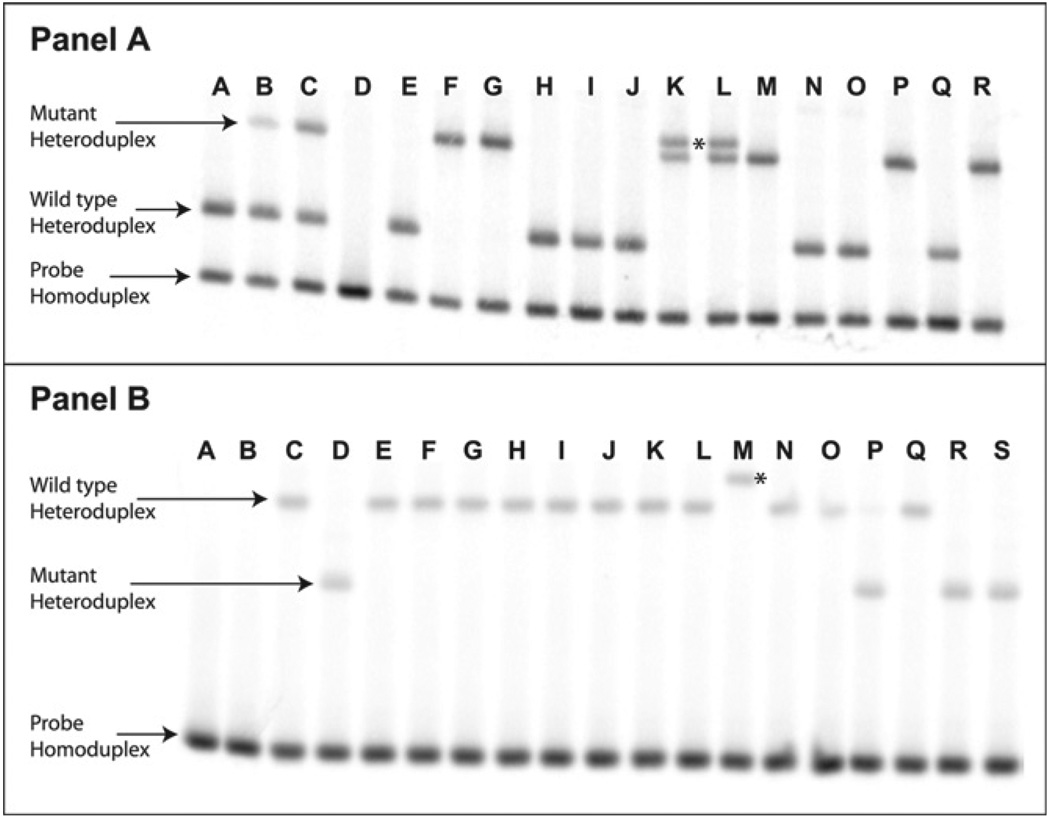
Among these samples, two contained a novel heteroduplex band that migrated more slowly than the typical mutant heteroduplex. Both were mixed infections, also containing a heteroduplex that migrated to the typical mutant location (Figure 1A, Lanes K and L). The novel variant (the higher band in the figure) represented on average 14.4% and 41.7% of the parasite DNA in the samples, respectively. To confirm the sequence of the variants in the clinical s amples, the PCR product was cloned using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA). The pfcrt–plasmid construct was amplified by colony PCR under the conditions noted for the assay, and the products were screened by HTA. The plasmid from colonies containing each variant was purified using the Promega Wizard Miniprep kit (Promega), and the pfcrt insert from the plasmid was sequenced at the UNC Automated Sequencing Facility. Both PCR reactions from each of these two samples and both PCRs from two wild-type samples were evaluated to give four independent sequences per variant. The wild-type sequence matched the 3D7 consensus sequence (CVMNK haplotype from amino acids 72–76; Figure 2. Both mutant variants contained the CVIET haplotype, thus showing the SNP for the K76T mutation. In addition, a non-synonymous SNP (A to G) was found to cause a serine to glycine change at PfCRT amino acid 90 (S90G; GenBank accession no. FJ424266). BlastN analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of the sequence and review of SNPs in PlasmoDB (http://plasmodb.org, GeneID: MAL7P1.27) showed no sequence match for this SNP. The novel variant sequence contained a non-synonymous SNP (A to G) coding an asparagine to serine switch at amino acid 58 (N58S). This variant did not contain the S90G mutation (Genbank accession no. FJ424267). Sequence review confirmed this as a novel SNP.
FIGURE 1.
Detection of heteroduplexes containing novel polymorphisms in patient samples. A, pfcrt76 HTA. Lane D represents a NTC reaction. Lanes E and F contain wild-type and mutant PCR controls, respectively. Lanes A, H, I, J, N, O, and Q represent infections with pure wild-type DNA from patient samples. Lanes B and C represent mixed mutant and wild-type DNA. Lanes G, M, P, and R contain pure mutant parasite DNA. Lanes K and L contain a second mutant heteroduplex (above the usual mutant heteroduplex and marked with an * between them) that contains a novel polymorphism. B, dhps540 HTA. Lanes A and B represent a probe alone lane and a NTC PCR reaction, respectively. Lane C contains wild-type PCR control and Lane D contains a mutant PCR control. Lanes E–L, N, O, and Q contain pure wild-type DNA from patient samples. Lane P contains a mixed infection, with 92% mutant DNA and 8% wild-type DNA. Lanes R and S contain pure mutant DNA. Lane M shows a wild-type heteroduplex containing the novel polymorphism (marked with an * to the right), which migrated slower than the standard wild-type heteroduplex.
FIGURE 2.
Partial amino acid sequence of pfcrt-containing novel polymorphisms The pfcrt amino acid sequence alignment for the 3d7 consensus (Genbank accession no. XM_001348968.1), the Chinese wild-type variant, the majority mutant variant (containing the S90G polymorphism; (Genbank accession no. FJ424266), and the minority mutant variant (containing the N58S polymorphism; (Genbank accession no. FJ424267).
The dhps540 HTA successfully detected 96/96 (100%) of the P. falciparum isolates from Peru. None of these samples contained drug-resistant minority variants. One sample did contain a wild-type minority variant (Figure 1B, Lane P) representing 8% of the parasite DNA. However, the HTA identified four that contained a heteroduplex that migrated more slowly than the typical wild-type variant (Figure 1B, Lane M). Three of the four samples that contained this novel variant were pure. The fourth contained a mixture of novel, representing 7% of the DNA, and typical wild-type variants. Two samples containing pure wild-type, pure mutant, and pure novel variants were each cloned and sequenced. The wild-type sequence matched the 3D7 consensus sequence. The mutant sequence contained the A to G SNP in the first position of the codon associated with the dhpsK540E mutation. The novel variant contained a SNP (A to G) in the third position of the codon and was synonymous to the wild-type (Genbank accession no. FJ424265). Sequence review (PlasmoDB GeneID: PF08_0095) showed this polymorphism to be novel.
This report suggests that surveillance for known drug resistance mutations by HTA may have two advantages: 1) the assays are sensitive to minority variants that may change estimates of the prevalence of a mutation in the population and allow for earlier detection of the emergence of drug resistance,2,3 and 2) it may provide insights into genetic variability in the region closely associated with these loci. This is because HTAs are typically sensitive to SNPs in the region to which the probe binds (typically a 150- to 200-bp region).2,3 We show here that these advantages are preserved over geographically distinct regions and at different genetic loci. The non-synonymous mutations found in pfcrt are particularly interesting because they may represent continued selection caused by drug pressure, because chloroquine is still used to treat P. vivax in this region. However, HTA also has disadvantages. Not all mutations change the motility of the heteroduplex in unique ways. A complete picture of in-host diversity may only be possible using high-throughput sequencing technologies.
Acknowledgments
The authors thank Thomas E.Wellems for providing advice and assistance on the manuscript. One of the authors is a military service member (DJB). This work was prepared as part of his official duties.
Financial support: This project was funded in part by NIH 1R21AI07-6785 and the 2007 IDSA ERF/NFID Merle A. Sande/Pfizer Fellowship in International Infectious Diseases, KL2RR025746 from the National Center for Research Resources, and funds provided by the Depart ment of Defense–Global Emerging Infectious System (DoD-GEIS) under Unit 847705.82000.25GB.B0016. These agencies had no involvement in the design, collection, analysis, or interpretation of data in this study or in writing this paper or submitting it for publication.
Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of the Navy, Department of Defense, nor the US government. Title 17 U.S.C. § 105 provides that “Copyright protection under this title is not available for any work of the US Government.” Title 17 U.S.C. § 101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person’s official duties. The study was conducted following a protocol approved by the Walter Reed Army Institute of Research Human Use and Review Committee (Protocol 719).
REFERENCES
- 1.Plowe CV, Roper C, Barnwell JW, Happi CT, Joshi HH, Mbacham W, Meshnick SR, Mugittu K, Naidoo I, Price RN, Shafer RW, Sibley CH, Sutherland CJ, Zimmerman PA, Rosenthal PJ. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar J. 2007;6:121. doi: 10.1186/1475-2875-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 3.Juliano JJ, Trottman P, Mwapasa V, Meshnick SR. Detection of the dihydrofolate reductase−164L mutation in Plasmodium falciparum infections from Malawi by heteroduplex tracking assay. Am J Trop Med Hyg. 2008;78:892–894. [PMC free article] [PubMed] [Google Scholar]
- 4.Liu S, Mu J, Jiang H, Su XZ. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am J Trop Med Hyg. 2008;79:178–184. [PMC free article] [PubMed] [Google Scholar]
- 5.Kwiek JJ, Alker AP, Wenink EC, Chaponda M, Kalilani LV, Meshnick SR. Estimating true antimalarial efficacy by het eroduplex tracking assay in patients with complex Plasmodium falciparum infections. Antimicrob Agents Chemother. 2007;51:521–527. doi: 10.1128/AAC.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngrenngarmlert W, Kwiek JJ, Kamwendo DD, Ritola K, Swanstrom R, Wongsrichanalai C, Miller RS, Ittarat W, Meshnick SR. Measuring allelic heterogeneity in Plasmodium falciparum by a heteroduplex tracking assay. Am J Trop Med Hyg. 2005;72:694–701. [PubMed] [Google Scholar]
- 7.Wang X, Mu J, Li G, Chen P, Guo X, Fu L, Chen L, Su X, Wellems TE. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- 8.Alker AP, Mwapasa V, Meshnick SR. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2004;48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]