Abstract
In response to bacterial infection, the neutrophil NADPH oxidase assembles on phagolysosomes to catalyze the transfer of electrons from NADPH to oxygen, forming superoxide and downstream reactive oxygen species (ROS). The active oxidase is composed of a membrane-bound cytochrome together with three cytosolic phox proteins, p40phox, p47phox and p67phox, and the small GTPase Rac2, and is regulated through a process involving Protein Kinase Cs, MAP kinases, and PI 3-kinases. The role of p40phox remains less well defined than those of p47phox and p67phox. We investigated the biological role of p40phox in differentiated PLB-985 neutrophils, and show that depletion of endogenous p40phox using lentiviral shRNA reduces ROS production and impairs bacterial killing under conditions where p67phox levels remain constant. Biochemical studies using a cytosol-reconstituted permeabilized human neutrophil cores system that recapitulates intracellular oxidase activation revealed that depletion of p40phox reduces both the maximal rate and total amount of ROS produced without altering the KM of the oxidase for NADPH. Using a series of mutants, p47PX-p40phox chimeras, and deletion constructs, we found that the p40phox PX domain has PtdIns(3)P-dependent and independent functions. Translocation of p67phox requires the PX domain but not 3-phosphoinositide binding. Activation of the oxidase by p40phox, however, requires both PtdIns(3)P binding and an SH3 domain competent to bind to poly-Pro ligands. Mutations that disrupt the closed auto-inhibited form of full-length p40phox can increase oxidase activity ∼2.5-fold above that of wild-type p40phox, but maintain the requirement for PX and SH3 domain function. We present a model where p40phox translocates p67phox to the region of the cytochrome and subsequently switches the oxidase to an activated state dependent upon PtdIns(3)P and SH3 domain engagement.
Neutrophils are phagocytic polymorphonuclear white blood cells (PMNs) of the innate immune system and represent one of the first lines of defense against invading microorganisms (1,2). The neutrophil NADPH oxidase enzyme catalyzes the transfer of electrons from NADPH to oxygen to form superoxide within pathogen-containing phagosomes and at the plasma membrane. The crucial role of NADPH oxidase activity in host defense is evidenced by patients with chronic granulomatous disease (CGD) whose neutrophils lack a functional NADPH oxidase resulting in frequent and persistent infections due to an inability to kill microbes efficiently (3-5).
The NADPH oxidase is a multisubunit enzyme made up of a membrane-spanning heterodimer of gp91phox and p22phox which forms the catalytic core, (cytochrome b558) as well as four cytosolic components, p47phox, p67phox, p40phox and the small G-protein Rac2 (2,6). Upon stimulation, the cytosolic components translocate to and activate the cytochrome (7). p47phox and p67phox have relatively well-established roles in activation: p47phox is essential for localization of the cytosolic phox components to the cytochrome (8); p67phox has been implicated as the cytosolic NADPH binding protein (9,10) and facilitates electron transport by the cytochrome (11). Mutations in either of these proteins results in CGD. In contrast, a definitive role for endogenous p40phox in NADPH oxidase activation and bacterial killing within neutrophils has been difficult to establish, and no CGD mutations within p40phox have yet been reported. Hawkins and colleagues recently generated p40phox−/− mice and reported that murine bone marrow-derived neutrophils from these animals were substantially defective in NADPH oxidase activity in response to some, but not all stimuli examined, as well as in their ability to kill serum-opsonized S. aureus (12). However, the genetic knock-out of p40phox resulted in the concomitant ∼60 % decrease in the levels of p67phox, complicating interpretation of their data. Additional insights into the function of p40phox from in cellulo studies has primarily involved non-neutrophil cell types. For example, Suh et al (13) examined p40phox in a monkey kidney COS7 cell system stably transfected with cytochrome b558, p47phox and p67phox transgenes along with the FcγR receptor (COSphox-FcγR cells), and found that p40phox was necessary for activation of the NADPH oxidase on FcγIIA receptor-induced phagosomes, suggesting that similar functions might occur in neutrophils or neutrophil-like cells. Similarly, Kuribayashi et al. (14) transfected p40phox into K562 erythroleukemia cells that stably express cytochrome b558, p47phox and p67phox, and observed increased superoxide production in the p40phox transfected cells compared to vector transfected controls upon stimulation with PMA or a muscarinic receptor agonist peptide, while Sathyamoorthy et al (15) reported that similar transfection studies of p40phox into K562 cells resulted in an ∼40% decrease in NADPH oxidase activity following PMA stimulation. Biochemical analysis of p40phox function has been equally difficult since classical cell-free systems for analyzing the NADPH oxidase either do not require p40phox for activity (16,17) or show only minor effects of p40phox under typical assay conditions (18). Thus, many details of how p40phox functions are still unclear.
p40phox contains an N-terminal PtdIns(3)P-binding PX domain (19,20), a central SH3 domain capable of interacting with a proline-rich region in p47phox in vitro (21,22) and a C-terminal PB1 domain which is required for constitutive interaction with p67phox (23). Recently, the structure of full length p40phox was solved (24). The structure, together with a companion cell biology-based study, showed that p40phox exists in an “closed” state where the PX and PB1 domains interact with each other, preventing the PX domain from interacting with PtdIns(3)P-containing membranes (25). Mutating residues in the interface between the PX and PB1 domains resulted in an “open” conformation that allowed the protein to bind to PtdIns(3)P. However, the consequences of this conformational change in the activation of the oxidase and the generation of superoxide remain unknown.
In this study we have investigated the role of p40phox in the activation of the NADPH oxidase utilizing two separate systems. First we use RNA interference in human PLB-985-derived neutrophils to show that p40phox is important for maximum superoxide production and that p40phox has a physiologically important role in the killing of pathogenic bacteria under conditions where the levels of p67phox are unchanged. We then used a permeabilized neutrophil system developed previously in our laboratory to biochemically investigate the mechanism by which p40phox acts on the NADPH oxidase (26). This system depletes purified human neutrophils of cytosol by permeabilization with the bacterial toxin streptolysin-O, generating cytosol-free neutrophil “cores” which can then be repleted with previously-manipulated neutrophil cytosol. This system offers several advantages for studying the NADPH oxidase compared with traditional recombinant protein-based in vitro assays and with transfected non-neutrophil cell-line-based systems, since the assays are performed using primary human neutrophils. Importantly, the permeabilized neutrophils maintain many of the intracellular structures on which the NADPH oxidase is thought to assemble, and allows for the detection of intracellularly produced superoxide (26). Additionally, since the system requires repletion with prepared cytosol, traditional biochemical techniques can be applied to perform structure-function studies in the setting of the primary neutrophil.
Using this reconstituted cores assay, we report that p40phox positively regulates the NADPH oxidase through all three of its modular signaling domains in a manner that extends beyond its role in facilitating p67phox localization. Finally, we show that the “open” form of p40phox causes the oxidase to generate significantly more superoxide than the “closed” form, and propose a multi-step mechanism of action for how p40phox activates the oxidase in both a PtdIns(3)P-independent and PtdIns(3)P-dependent manner.
Experimental Procedures
Materials
Streptolysin-O, ATP, luminol, DFP, creatine kinase, creatine phosphate, and dimethyl formamide (DMF) were purchased from Sigma. All other protease inhibitors were purchased from American Bioanalytical. Endotoxin-free Dulbecco's Modified PBS without calcium and magnesium (DMPBS) as well as Dulbecco's Modified PBS containing calcium and magnesium (DMPBS+) was purchased from Invitrogen; Protein-A Sepharose beads and Ficoll-Paque from GE Healthcare; GTP, GTPγS, NADPH and Nutridoma-SP from Roche, and wortmannin from EMD Biosciences. Polyclonal antibodies against p40phox and p67phox were generated by peptide-immunization of rabbits, and have been described previously (27). A polyclonal antibody against p22phox was a gift from Dr. Katrin. Rittinger. A mouse monoclonal anti-penta-His antibody was purchased from Qiagen.
Differentiation of PLB-985 cells and RNA interference
PLB-985 cells were obtained from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and were maintained in an undifferentiated state in RPMI media containing 10% FBS and 100U/ml penicillin and 100mg/ml streptomycin at 37°C, 5% CO2. RNA interference was used to make stable p40phox knockdown (p40KD) or luciferase control knockdown cells (LucKD); undifferentiated PLB-985 cells were infected with pLL3.7-derived lentiviruses (28) carrying the p40phox hairpin 5'-TGGAGATTGGGACCAGGAAATTCAAGAG ATTTCCTGGTCCCAATCTCCTTTTTTC-3' or the luciferase hairpin 5'-TCGTACGCGGAATACTTCGATTCAAGAG ATCGAAGTATTCCGCGTACGTTTTTTC-3' (targeting sequences underlined), and selected by growth in media containing 5μg/ml puromycin. To induce neutrophil differentiation, cells were diluted into RPMI media containing 0.5% FBS, 1% Nutridoma-SP, and 0.5% DMF at 2 × 105 cells/ml and cultured for 8 days with one change of media at day 4.
Bacterial Killing Assays
Bacterial killing assays were performed following Silverstein and colleagues (29). In brief, S. aureus Rosenbach (ATCC 25923) was cultured in tryptic soy broth to early log phase, collected by centrifugation, and resuspended at 2×108cfu/ml in DMPBS+ containing 1mg/ml BSA. Bacteria were incubated at a ratio of 5:1 with 1×106 differentiated PLB-985 cells in 75% human serum in a total volume of 1 ml for 90 min at 37°C with intermittent mixing. Following incubation, duplicate aliquots (350 μl) were lysed by mixing with 650 μl of sterile water, pH 11, followed by incubation at 37°C with shaking for 5 min. Lysates were serially diluted, plated on tryptic soy agar plates, grown overnight at 37°C and the number of surviving bacterial colonies counted.
PLB-985 Chemiluminescence Assays
For chemiluminescence assays following S. aureus infection, 2×105 p40KD or LucKD differentiated PLB-985 cells were resuspended in DMPBS containing 100 μg/ml BSA, and 0.15 mM luminol (PBS/B/L) and incubated in 75% human serum. S. aureus was added to the PLB-985 suspension at a ratio of 5:1 and ROS production was monitored as luminol-dependent chemiluminescence (CL), in duplicate, using an AutoLumat LB953 luminometer (Berthold Technologies, Oak Ridge, TN) at 37°C. Unless otherwise indicated, CL is reported as total events recorded during the indicated time after subtracting the machine background, which typically averaged less than 1%−5% of the total signal. When PMA was used as the agonist, experiments were performed by resuspending 1.5×105 differentiated PLB-985 cells in PBS/B/L and the reaction was initiated by the addition of PMA at the indicated final concentrations.
Preparation of Permeabilized Neutrophils and PMN Cytosol
Anticoagulated whole blood was obtained from healthy human volunteers following an IRB approved protocol. PMNs were isolated by centrifugation through Ficoll-Paque followed by dextran sedimentation and water lysis of residual RBCs as described earlier (26). PMNs were resuspended to a final concentration of 1×107/ml in DMPBS containing 4 μg/ml AEBSF, incubated on ice for 10 min, then pelleted and resuspended in RB-EBL buffer (RB buffer [10 mM PIPES pH 7.3 with KOH, 100 mM K+, 3 mM Na+, 3.5 mM Mg2+] containing 0.3 mM EGTA, 100 μg/ml BSA, and 0.15 mM luminol).
Permeabilized PMN cores were prepared by incubating 1×106 freshly isolated cells in 300 μl of RB-EBL buffer containing 1000 U reduced Streptolysin-O for 5 min at 0°C. Cores were recovered by centrifugation at 280 × g for 10 min, resuspended in RB-EBL and used within 60 min.
PMN cytosol was prepared by nitrogen cavitation of 2×108 cells/ml in RB-E buffer containing 5.6 mM DFP by pressurization to 400 psi for 20 min at 0°C prior to release. The cavitate was centrifuged at 2500 × g for 10 min at 4°C followed by centrifugation of the resulting supernatant at 200,000 × g for 1 hr at 4°C. The high-speed supernatant was flash frozen as aliquots in liquid N2 and stored at −80°C. 2×108 PMNs typically yielded approximately 3mg of protein; cytosol was thawed on ice immediately prior to use. To immuno-depelete cytosol of p40phox, anti-p40phox antibody beads were prepared by incubating 50 μl of Protein A Sepharose beads with 100 μg (50 μl) of p40phox antiserum overnight at 4°C. Beads were washed twice with RB buffer, four times with RB containing 1 M NaCl, twice with RB buffer, and added to 60 μl of freshly thawed cytosol. Following a 2.5 hr incubation at 4°C with gentle rocking, the p40phox-depleted cytosol was recovered by pelleting the beads and recovering the supernatant. Mock depleted cytosols were prepared in an identical manner using Protein A Sepharose beads coated with non-specific rabbit IgG.
Expression and purification of recombinant p40phox
For bacterial expression of human p40phox, the full-length cDNA was cloned into the EcoRI and XhoI sites of pET28a to provide an N-terminal His6-tag. Constructs expressing R58Q, W207R, D293K, E259A, as well as R58Q/W207R, E259A/R58Q, E259A/W207R, E259A/D293K p40phox mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene). The ΔPX domain construct lacking amino acids 1−135 and the p47PXp40phox fusion containing amino acids 1−121 of p47phox fused to amino acids 136−339 of p40phox were both constructed using PCR. The p47PX-R43A site mutant was constructed using the QuikChange site-directed mutagenesis kit using the p47PXp40phox construct as a template. All constructs were verified by dideoxy sequencing, and transformed into BL21(DE3) E. coli cells. Recombinant proteins were produced by inducing 1 L of late log-phase cultures (OD 1.0−1.2) with 1 mM IPTG overnight at room temperature. Bacteria were harvested by centrifugation, resuspended in 25 ml of lysis buffer (50mM HEPES pH 8.0, containing 1mM MgCl2, 300mM NaCl, 7mM β-mercaptoethanol, and 4 μg/ml each of leupeptin, pepstatin, AEBSF, and aprotinin), and disrupted by sonication. Lysates were clarified at 150,000 × g for 30−60 min at 4°C and incubated in batch with 1 ml of Ni-NTA beads and mixed end-over-end for 1 hr at 4°C. The beads were then loaded into an FPLC column, washed with 100 ml of lysis buffer containing 10 mM imidazole at a flow rate of 1 ml/min, and eluted with 50 ml of lysis buffer containing 300 mM imidazole. Fractions (8 ml) were analyzed for p40phox content by SDS-PAGE, pooled, and snap frozen at −80°C. Prior to use in core reconstitution reactions, the proteins were thawed and dialyzed against RB buffer.
Reconstitution Reactions and Chemiluminescence Assay
Reconstitution reactions were performed in 100 μl RB-EBL buffer containing 8×104 freshly-prepared neutrophil cores, 30 μg cytosol, 2 mM ATP, 200 μM GTP, 200 μM GTPγS, 10 mM creatine phosphate, and 25 μg/ml creatine kinase. Unless otherwise indicated, reactions were pre-incubated at 37°C for 10 min to induce permeabilization, followed by addition of 400 μM NADPH and 100 ng/ml PMA, and assayed for ROS production by measuring luminol-dependent chemiluminescence (CL) in duplicate using an AutoLumat LB953 luminometer at 37°C. Where appropriate, 400 ng of recombinant p40phox protein was added prior to the pre-incubation step (unless otherwise noted); 400 ng gave the maximal recovery of ROS production in this assay with p40phox-depleted cytosol (data not shown).
Cores Association Assay
To examine movement of p67phox to the cores membranes, 8×105 cores in 1 ml of RB buffer were incubated with 200 μg of p40phox immuno-depleted cytosol, in the presence or absence of 3 μg of purified wt or mutant p40phox, 60 ng/ml PMA, and 120 μM NADPH for 10 minutes at 37°C. Cores were recovered by centrifugation (2,400 × g for 2 min) and rinsed once with RB buffer. To minimize non-specific protein adherence, cores were then incubated in RB buffer containing 5μg/ml of cytochalasin B at 37°C for 10 minutes, then rinsed twice with RB containing 0.1% Brij-35. Proteins were resolved by SDS-PAGE and immunoblotted for p67phox.
Structural Analysis
Structures of the p40phox and p47phox PX domains and full-length p40phox protein were visualized using SwissPB viewer (30) and GRASP (31). Figures were constructed using PyMol (http://www.pymol.org).
Results
p40phox is required for optimal killing of S. aureus and ROS production in differentiated PLB-985 cells
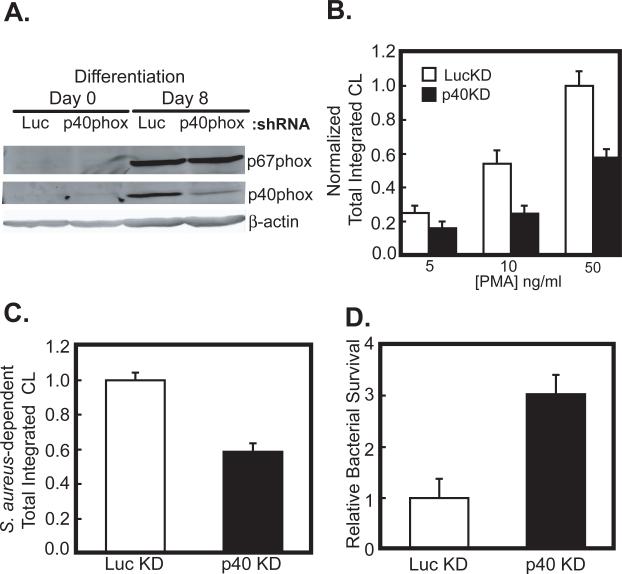
In order to investigate the role of p40phox in the activation of the NADPH oxidase, we used RNA interference to knockdown p40phox in PLB-985 cells, a human myeloid cell-line which can undergo differentiation into neutrophils (32), and examined their ability to generate reactive oxygen species (ROS). Undifferentiated PLB-985 cells were infected with lentiviruses expressing either an shRNA against the 3’ untranslated region of p40phox (p40KD), or a control shRNA against luciferase (lucKD), together with a puromycin resistance gene. Following selection in puromycin-containing media, the cells were induced to undergo granulocyte differentiation. When immunoblotted 8 days following the induction of differentiation, the lucKD control cells showed robust expression of p40phox (Figure 1A), while in the p40KD cells, the levels of p40phox were reduced by 63 ± 2.4%. Importantly, the levels of p67phox in both the differentiated lucKD cells and the p40KD cells were identical (97.8 ± 4.0% in p40KD compared to control).
Figure 1. p40phox is necessary for efficient ROS production and bacterial killing in differentiated PLB-985 cells.
(A) PLB-985 cells were stably infected with lentiviruses expressing a control shRNA against luciferase (LucKD) or an shRNA against the 3’UTR of p40phox (p40KD). Cells were then induced to differentiate into neutrophils by addition of 0.5% DMF to the media. After 8 days, cells were lysed and p40phox and p67phox levels examined by SDS-PAGE and immunoblotting.
(B) LucKD (white bars) or p40KD cells (black bars) were harvested after 8 days of DMF-induced differentiation in culture, and stimulated with the indicated amounts of PMA. Mean levels and S.E.M. for total ROS production over 30 min are shown for n=7 experiments, normalized to the level observed in LucKD PLB-985 cells stimulated with 50ng/ml PMA. Statistically significant differences between the LucKD and p40KD cells were seen at PMA doses of 10 and 50 ng/ml (p<0.01, Student's t-test, 2-tailed).
(C, D) LucKD control and p40KD cells as in panel B were challenged with serum-opsonized S. aureus at an MOI of 5. Total integrated chemiluminescence produced over 30 min following bacterial exposure was measured (C). In parallel, p40KD or LucKD differentiated PLB-985 cells were incubated at 37°C for 90 minutes with serum opsonized S.aureus at an MOI of 5. The cells were washed, lysed, and the surviving bacteria were quantitated by plating serial dilutions of the lysates on Tryptic soy agar (D). In both panels, mean values and S.E.M. from n=6 and 8 experiments, respectively, are shown, normalized to the values obtained for the LucKD controls. In both panels, the observed differences between the LucKD and p40KD cells were statistically significant (p<0.005, Student's t-test, 2-tailed).
Differentiated lucKD cells and p40KD cells were activated using PMA and ROS production was measured using luminol-dependent chemiluminescence. As shown in Figure 1B, the p40KD cells showed a statistically significant defect in PMA-dependent ROS generation compared to lucKD cells at both 10 and 50 ng/ml. Next, we investigated the ability of p40KD PLB-985 cells to produce ROS when challenged with serum-opsonized S. aureus. In these experiments we observed a 43% decrease in total ROS production in the p40phox-deficient cells compared to the controls (Figure 1C). We also observed in each of 8 separate experiments that the p40KD cells were profoundly defective in their ability to kill opsonized microbes (Figure 1D), indicating that endogenous p40phox plays a physiologically important role in NADPH oxidase-dependent bacterial killing.
p40phox is necessary for optimal superoxide production by the NADPH oxidase in permeabilized PMNs
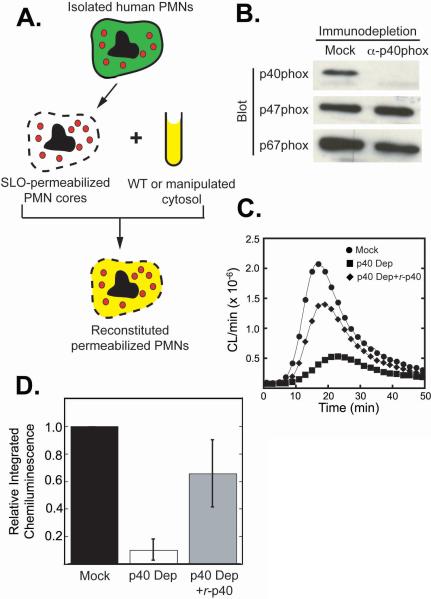
To further investigate the mechanism by which p40phox activates the neutrophil NADPH oxidase, we utilized a permeabilized neutrophil system previously developed in our laboratory that recapitulates intracellular ROS production (26). In this system, the plasma membranes of human neutrophils are permeabilized using Streptolysin-O (SLO), resulting in the production of cytosol-free neutrophil ‘cores’ containing intact intracellular granules and vesicles (Figure 2A). Robust PMA-stimulated activation of the NADPH oxidase is obtained in these neutrophil ‘cores’ when they are supplemented with purified cytosol, ATP, GTPγS, and NADPH, as measured by luminol-dependent chemiluminescence (26). In contrast to traditional cell-free NADPH oxidase assay systems, the permeabilized ‘core’ system preserves much of the complex intracellular structure of the neutrophil, and permits intracellular oxidase activation on granules and vesicles, where much of the initial oxidase activation is believed to occur in intact neutrophils (33,34). Importantly, the cytosol used in the reconstitution step can be manipulated prior to re-addition, allowing removal and/or replacement of specific cytoplasmic constituents.
Figure 2. p40phox is required for optimal ROS production in a permeabilized neutrophil reconstitution system.
(A) Schematic of PMN permeabilization and NADPH oxidase reconstitution procedure.
(B) Purified neutrophil cytosol was immunodepleted using non-specific rabbit IgG (Mock) or anti-p40phox antibodies and analyzed for remaining levels of p40phox, p47phox and p67phox by immunoblotting. Each lane contains 20 μg of total cytosolic protein.
(C, D) SLO-permeabilized PMN cores (8×104/100 μl) were reconstituted with mock depleted (●), p40phox-depleted (■) or p40phox-depleted cytosol re-supplemented with recombinant p40phox (◆), stimulated with PMA, and ROS production measured using luminol-dependent chemiluminescence. (C) A typical trace from n=6 experiments, where CL/min denotes the recorded number of chemiluminescence events per minute. (D) Mean values and standard deviations of total integrated chemiluminescence over 60 minutes, normalized to the mock-depleted controls (n=6).
To specifically address the biochemical role of p40phox in NADPH-oxidase-dependent ROS production, we reconstituted PMN cores with neutrophil cytosol that was immuno-depleted of p40phox or mock-depleted with non-specific rabbit IgG (Figure 2B). Depletion of all of the p40phox detectable by immuno-blotting dramatically reduced the activity of the NADPH oxidase, resulting in a 72 ± 0.8% decrease in the peak rate of ROS production, a kinetic delay of 7.6 ± 3.6 min in the time until the peak rate was reached (Figure 2C), and a 90 ± 7.1% reduction in the total amount of ROS produced (total integrated chemiluminescence over 60 min; Figure 2D). Immunodepletion of p40phox also resulted in co-depletion of 43 ± 13% of the p67phox present in the cytosol, with no change in p47phox levels (Figure 2B). Thus, the decrease in ROS production that we observed in the p40phox-depleted reactions could have been due either to the loss of p40phox itself, or to the partial depletion of p67phox.
To distinguish between these possibilities, we supplemented the p40phox-depleted reactions with purified bacterially-produced recombinant p40phox (r-p40phox). As shown in Figure 2C, the observed decrease in ROS production due to p40phox depletion, together with the kinetic delay, was largely reversed by re-addition of r-p40phox, suggesting that p40phox functions as a direct positive regulator of NADPH oxidase activity in this system. In contrast to the ∼600% increase in ROS production when r-p40phox was added to p40phox-depleted cytosols, we observed only minimal increases in ROS production (< 20%) when r-p40phox was added to the mock-depleted cytosols. Furthermore, addition of recombinant p67phox to levels 20-fold higher than those in the p40-depleted lysates failed to confer any increased ROS production in the absence of r-p40phox (data not shown).
p40phox enhances the initial enzymatic activity of the NADPH oxidase without affecting the apparent Km for NADPH
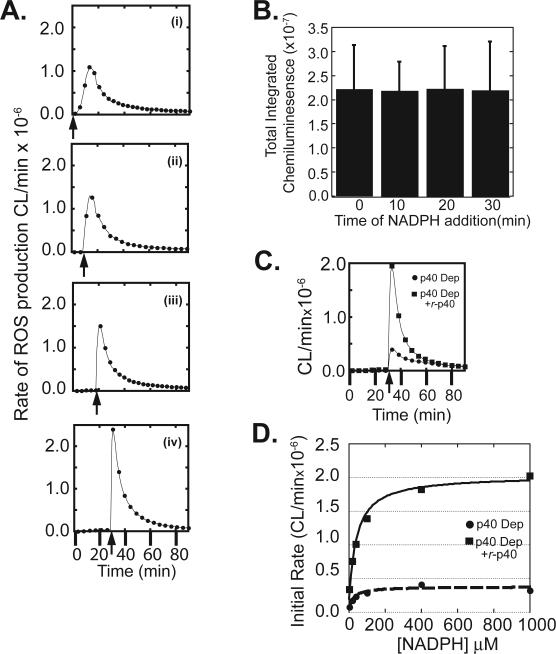
In intact neutrophils and reconstitution systems, superoxide production involves two interconnected events that overlap in time - a progressive assembly of the NADPH oxidase holoenzyme complex (c.f. (35-37), together with the catalysis of NADPH oxidation by those complexes that have already assembled. The coupling of these two events greatly complicates a kinetic analysis of the oxidase reaction. For example, in all of our permeabilized PMN reconstitution experiments we observed a slow steady rise in the rate of superoxide production that peaked at ∼15 min after PMA stimulation (Figure 2C), similar to that observed with intact neutrophils (27,38,39). In an effort to decouple these two events and measure the initial enzymatic activity of the oxidase, we made use of our previous observation that pre-incubation of the cores with cytosol, ATP, GTPγS and PMA prior to addition of NADPH results in assembly of the NADPH oxidase complex, without ROS production (27). As shown in panels i-iv of Figure 3A, addition of NADPH to reconstituted core reactions following 0, 10, 20, or 30 minutes of incubation progressively accelerated the onset of peak reaction velocity and dramatically increased the peak initial rate of ROS production. No further decrease in time to peak onset or increase in initial velocity was observed beyond 30 minutes of pre-incubation (data not shown). We therefore interpret this initial spike of oxidase activity when NADPH is added at 30 minutes to reflect the initial velocity of the pre-assembled oxidase complex. Interestingly, the total integrated chemiluminescence measured over 90 minutes remained the same regardless of the duration of pre-incubation (Figure 3B), indicating that total ROS production was unchanged.
Figure 3. Delayed addition of NADPH to reconstituted PMN cores allows measurement of initial rates of enzymatic activity; p40phox affects the maximal velocity of the NADPH oxidase but not the apparent KM for NADPH.
(A) Reconstitution reactions were performed using permeabilized PMN cores and mock-depleted cytosol. PMA was added at the beginning of the reaction, and NADPH was added 0 (i), 10 (ii), 20 (iii) or 30 (iv) minutes later (arrow). Luminol-dependent chemiluminescence was recorded over 90 min.
(B) Total integrated chemiluminescence over 90 minutes for reactions performed as in panels A (i)-(iv). Mean and standard deviations for n=3 experiments are shown.
(C) PMN cores were reconstituted using p40phox-depleted cytosol in the absence (●) or presence (■) of recombinant wild-type p40phox. NADPH was added to the reactions 30 min following PMA, and NADPH oxidase activity measured using luminol-dependent chemiluminescence.
(D) Reconstitution reactions were performed as in panel A using varying concentrations of NADPH. Initial rates of NADPH oxidase activity were measured as a function of NADPH concentration. Results are typical of 5 independent experiments
This pre-incubation assay, with addition of NADPH at 30 minutes, was then used to investigate the contribution of p40phox to the kinetics of ROS production by comparison of reactions with p40phox-depleted cytosol, with or without r-p40phox added prior to NADPH addition. As shown in Figure 3C, the initial reaction velocity was much lower in the absence, than in the presence of r-p40phox, however, the absence of p40phox did not significantly affect the time to peak ROS production.
p67phox, the constitutive binding partner of p40phox, has been implicated in enzymatic regulation of NADPH oxidase activity through direct participation in electron flow from NADPH (2,6,10,40). As such, we reasoned that the decrease in the initial rate of ROS production observed in p40phox-depleted reactions might result from interference with this process, manifesting from an alteration in the apparent KM of the enzyme for NADPH. Pre-incubation assays with NADPH added at 30 minutes were therefore performed both in the presence and in the absence of r-p40phox over a broad range of NADPH concentrations (Figure 3D). Analysis of the initial rates of reaction revealed little difference in the apparent KM of the reaction for NADPH in the absence or presence of r-p40phox, (15 ± 6.7 μM versus 24 ± 7.7 μM, respectively) despite the > 500% increase in the maximal velocity that was observed when p40phox was present. Our measured values compare favorably with the KM value of 34 ± 3.9 μM for NADPH in a traditional cell-free NADPH oxidase system following stimulation by the amphiphile SDS reported by Curnutte et al. (41). These findings suggest that p40phox enhances NADPH oxidase activity through a route other than a direct affect on the apparent Km for NADPH.
The PX, SH3 and PB1 domains of p40phox are all required for p40 function
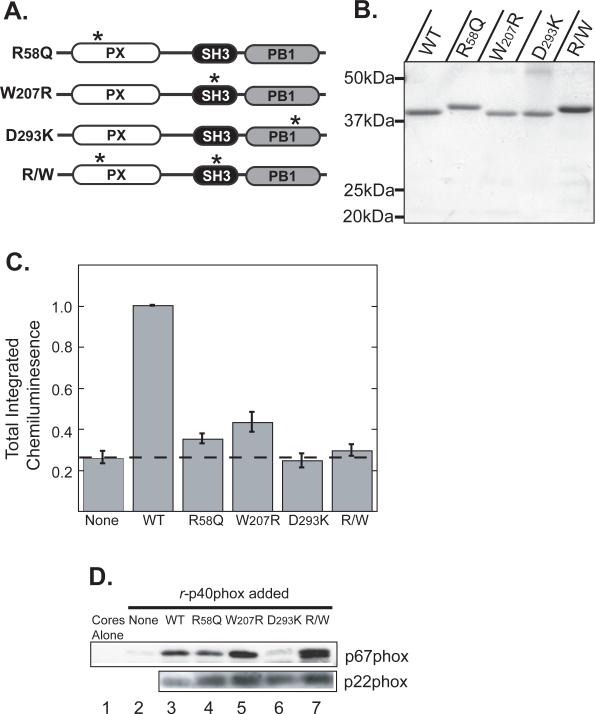
The p40phox protein has three modular domains: a PX domain, an SH3 domain and a PB1 domain. To investigate what roles these domains play in the p40phox-dependent stimulation of the NADPH oxidase, we made single inactivating point mutations in each domain (Figure 4A). We and others have previously shown that the p40phox PX domain is a phosphoinositide binding domain that specifically recognizes PtdIns(3)P (19,20). We therefore made an R58Q mutation in the PX domain which is sufficient to disrupt PtdIns(3)P binding (42). Although the physiological target(s) of the SH3 domain of p40phox remain unclear, this domain is known to recognize Pro-rich peptides in vitro (21). We therefore mutated Trp-207 to Arg (W207R), since this residue makes critical contacts with a synthetic Pro-peptide ligand in the p40phox SH3 crystal structure (21), and the equivalent Trp mutations in other SH3 domains are known to disrupt their interactions with cognate ligands (43,44). We also mutated Asp-293 in the p40phox PB1 domain to Lys (D293K), since this mutation is known to disrupt the constitutive interaction between p40phox and p67phox (45). Finally, we made a double mutant with both the R58Q mutation and the W207R mutation (R/W).
Figure 4. All three modular signaling domains within p40phox are required for reconstitution of the NADPH oxidase in permeabilized neutrophil cores.
(A) Schematic of p40phox mutants used in this figure. Asterisks denote the approximate location of point mutations within each domain.
(B) Recombinant wild-type and mutant p40phox proteins with an N-terminal His6 tag were produced in bacteria and batch purified on Ni-NTA agarose. Proteins were analyzed by SDS-PAGE and stained with Coomassie Blue. The R58Q mutants consistently displayed retarded migration. Positions of molecular weight markers are indicated.
(C) Reconstitution reactions were performed using permeabilized PMN cores and p40phox-depleted cytosol supplemented with the indicated recombinant p40phox proteins. Total integrated chemiluminescence was measured over 30 min after PMA stimulation and normalized to reactions supplemented with wild-type p40phox. Mean and standard deviations from n=4 experiments are shown.
(D) Differential effects of PX, SH3 and PB1 mutations on association of p67phox with neutrophil cores. Reconstitution reactions were performed as in panel C in the presence of PMA for 10 min. Cores were re-isolated and analyzed for p67phox by SDS-PAGE and immunoblotting. An immunoblot for p22phox is shown as a loading control. Each lane corresponds to 9×105 cores. Results are representative of 3 separate experiments.
Recombinant wild-type and mutant p40phox proteins were purified from bacteria (Figure 4B), added to p40phox-depleted cytosol, and ROS production then measured in reconstituted neutrophil cores. As shown in Figure 4C, mutations within any of the three domains of p40phox, as well as the double R/W mutant, dramatically reduced the ability of r-p40phox to stimulate NADPH oxidase activity in response to PMA. The least competent mutant was the D293K PB1 domain mutant, which produced amounts of total ROS approximately equivalent to cytosol lacking p40phox altogether. This finding is consistent with an obligatory requirement for interaction between p40phox and p67phox for p40phox-dependent stimulation of the NADPH oxidase.
One postulated function of p40phox is to facilitate the translocation of p67phox to intracellular membranes containing the b558 cytochrome (14). If the point mutants described above reduced the amount of p67phox that was associated with the cytochrome, this could explain why these mutants are unable to rescue ROS production in p40phox-depleted cores. To examine this possibility, we monitored the association of p67phox with cores following PMA stimulation by re-isolated the cores by centrifugation and washing at the end of the reconstitution experiments (Figure 4D). Control reactions using mock depleted cytosol revealed that 5−10% of the total p67phox present in the reactions translocated to the cores after PMA stimulation, consistent with what we had observed previously with unperturbed cytosol (26). These values compare favorably with the amount of p67phox (10−18%) reported by Quinn et al. to become membrane associated in PMA-stimulated neutrophils (46). Reactions with p40phox-depleted cytosol demonstrated very little, or no, core-associated p67phox after stimulation with PMA, whereas re-addition of wild-type r-p40phox to p40-depleted cytosol resulted in substantial translocation of p67phox to the cores (compare Figure 4D lanes 2 and 3). When cores reconstituted with p40phox-depleted cytosol were supplemented with r-p40phoxD293K, little if any p67phox was core-associated (Figure 4D lane 6), demonstrating the requirement of a direct PB1:PB1 interaction with p67phox to allow this translocation to occur. Interestingly, the R58Q PX domain mutant, the W207R SH3 domain mutant and the R/W double mutant, none of which were able to rescue significant ROS production in p40phox-depleted core reactions, caused core-association of p67phox to levels at least equivalent to those observed with wild-type r-p40phox (compare Figure 4D lanes 4, 5, and 7 to lane 3). The observation that p40phox-dependent translocation of p67phox did not strictly parallel reconstitution of ROS production demonstrates that, while p40phox does indeed translocate and localize p67phox, this is not sufficient for stimulation. Additional events must occur (through at least the PX and SH3 domains) to promote NADPH oxidase activation.
Addition of p40phox to ongoing oxidase reactions results in the instantaneous acceleration of ROS production
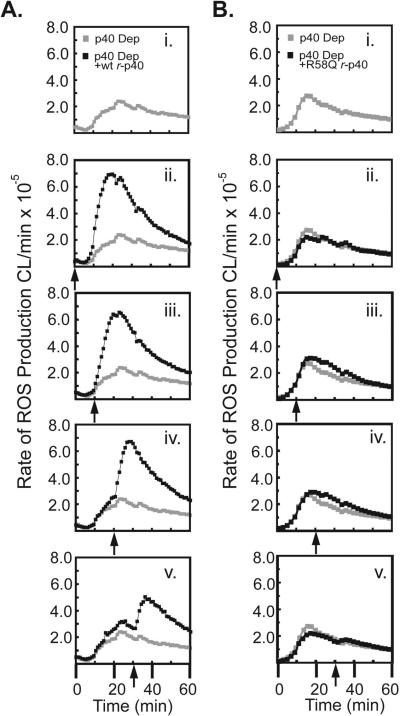
To further examine the parameters of p40phox's ability to stimulate NADPH oxidase activity, we investigated the effect of delayed addition of r-p40phox to ongoing oxidase reactions. In experiments analogous to those described in Figure 3, reactions containing permeabilized PMN cores, p40phox-depleted cytosol, ATP, GTPγS and PMA were incubated with NADPH at t=0 min, and then supplemented with recombinant p40phox protein 0,10, 20, or 30 min later (Figure 5). The addition of wild-type r-p40phox at 10, 20, or 30 min resulted in the instantaneous enhancement of the rate of oxidase activity at all subsequent time points (Figure 5A panels i-v). In contrast, little to no enhancement in rate was observed upon addition of the R58Q or W207R r-p40phox mutants (Figure 5B panels i-v and data not shown), despite the ability of these mutants to localize p67phox to the cores (Figure 4D). These data demonstrate that even when NADPH oxidase activity is decreasing, re-addition of p40phox induces a rapid and significant increase in ROS production, through a mechanism not solely reliant on simple co-localization of p67phox, requiring input from both the PX and SH3 domains.
Figure 5. Delayed addition of p40phox instantaneously accelerates the rate of ROS production in cores reconstituted with p40phox-depleted cytosol.
(A, B) Permeabilized PMN cores were reconstituted using p40phox-depleted cytosol and activated with PMA at t=0 min. Recombinant wild-type p40phox (A) or the R58Q p40phox mutant proteins (B) were either omitted entirely (i) or added back to the reactions at 0 min (ii), 10 min (iii), 20 min (iv), or 30 min (v) after PMA stimulation, as indicated by the arrows. ROS production was monitored using luminol-dependent chemiluminescence. The grey trace in panels ii-v is the minus p40phox control from panel i, shown for comparison.
The PX domain of p40phox plays both a PtdIns3P-dependent and a PtdIns3P-independent role in the activation of the NADPH oxidase
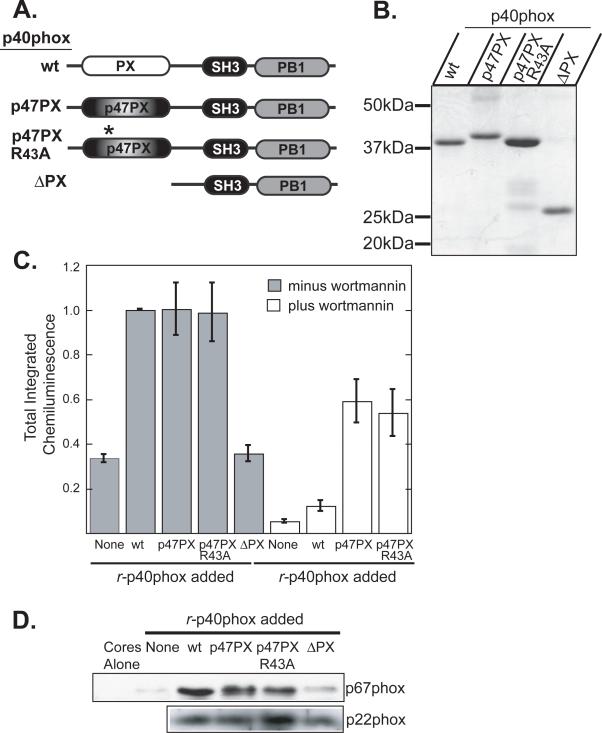
To better understand the role of the PX domain of p40phox in p40phox-dependent ROS production, we made a variety of additional PX domain mutants. We were particularly interested in whether the p47phox PX domain could functionally substitute for the p40phox PX domain. The p47phox PX domain has an almost identical structure to the p40phox PX domain (42), but each has different lipid-binding specificities. The p47phox PX domain shows most specific binding to PtdIns(3,4)P2 while the p40phox PX domain recognizes only PtdIns(3)P (19,20). We made a chimeric protein which swapped the PX domain of p40phox for the PX domain of p47phox (p47PX-p40phox). We also made this same p47PX-p40phox fusion protein with an R43A point mutation in the lipid-binding pocket of the p47phox PX domain, which eliminates PtdIns(3,4)P2 binding, and is the functionally equivalent mutation to R58Q in the p40phox PX domain. Finally we also generated a PX domain truncation of p40phox (ΔPX) (Figure 6A and B) which retains its ability to bind to p67phox (Supplemental Figure 1).
Figure 6. A chimeric p47phox PX domain-p40phox fusion protein activates the NADPH oxidase independently of binding to PI 3-kinase lipids.
(A) Schematic of p40phox chimeras and mutants studied.
(B) Recombinant wild-type and mutant p40phox proteins containing an N-terminal His6 tag were produced in bacteria and batch purified on Ni-NTA agarose. Proteins were analyzed by SDS-PAGE and stained with Coomassie Blue. Positions of molecular weight markers are indicated.
(C) Reconstitution reactions were performed using permeabilized PMN cores and p40phox-depleted cytosol supplemented with the indicated recombinant p40phox proteins in the presence (white bars) or absence (grey bars) of 100nM wortmannin. Total integrated chemiluminescence was measured over 30 min following PMA stimulation and normalized to reactions supplemented with wild-type r-p40phox. Mean and standard deviation from n=3 experiments are shown.
(D) Differential effects of p47phox PX domain-p40phox chimeric proteins and p40phox PX domain truncation on association of p67phox with neutrophil cores. Reconstitution reactions were performed as in panel C in the presence of PMA for 10 min. Cores were then re-isolated and analyzed for p67phox by SDS-PAGE and immunoblotting. An immunoblot for p22phox is shown as a loading control. Each lane corresponds to 9×105 cores.
Consistent with the inability of the R58Q point mutant to rescue oxidase activity of p40phox-depleted cores, the r-p40phox ΔPX truncation protein was also unable to rescue activity (Figure 6C). However, in contrast to the R58Q point mutant, which was able to localize equivalent amounts of p67phox to cores membranes as wild type p40phox (Figure 4C), the ΔPX truncation was unable to localize p67phox to the cores (Figure 6D). This demonstrates that co-translocation of p67phox is PX-domain-dependent, but PtdIns(3)P-independent, whereas reconstitution of oxidase activity requires both the PX domain and PtdIns(3)P binding.
The p47PX-p40phox chimeric mutant was able to activate the oxidase in p40phox-depleted cores at comparable levels to wild-type p40phox (Figure 6C), suggesting that in the context of the fusion protein, the PX domain of p47phox can functionally substitute for that of p40phox. However, the R43A p47PX-p40phox chimeric fusion protein also performed equivalently to the wild-type p47PX-p40phox fusion protein, demonstrating that, unlike the p40phox PX domain, the p47phox PX domain does not require contact with 3-phosphoinositides to confer activity on the system in the specific context of the p40phox fusion protein. This idea was further reinforced by the observation that while the NADPH oxidase activity of cores reconstituted with wild-type r-p40phox was highly sensitive to the PI3K inhibitor wortmannin, the fusion proteins were substantially less sensitive (Figure 6C), with the R43A mutant being no more sensitive than the wild-type p47PX-p40phox chimera. This again suggests that the p47PX-p40phox fusion protein is activating the oxidase in a manner that is not dependant on the interaction of the fusion protein with PI3-K-generated phosphoinositides. Upon examination of the ability of these p47PX-p40phox chimeric fusion proteins to translocate p67phox, we found that both were able to localize p67phox in amounts equivalent to that achieved by wild-type p40phox (Figure 6D).
Together, these data suggest that the PX domain of p40phox makes two distinct contributions to NADPH oxidase activity. The first is to translocate p67phox in a PX domain-dependent, but PtdIns(3)P-independent, manner, and this can be functionally substituted by the PX domain of p47phox. The second is a role in switching the oxidase to an activated state, which for wild-type p40phox, requires PtdIns(3)P binding.
“Opening” of p40phox causes enhanced p40phox-dependent ROS production by the NADPH oxidase
Recently the crystal structure of full-length p40phox was solved (24). Full length p40phox was found to exist in a “closed” state in which the N-terminal PX domain and the C-terminal PB1 domain interact with each other, allowing p67phox binding but preventing interaction of the PX domain with PtdIns(3)P (Figure 7A) (24,25). Mutating residues at the interface between the PX and PB1 domains was found to “open” full length p40phox, and permit the PX domain to bind to PtdIns(3)P (24). Masking of the lipid-binding pocket of the PX domain in the resting state clearly has implications for the current study in which we observed a PtdIns(3)P-independent role for the PX domain in translocation, but a PtdIns(3)P-dependent role on oxidase activation.
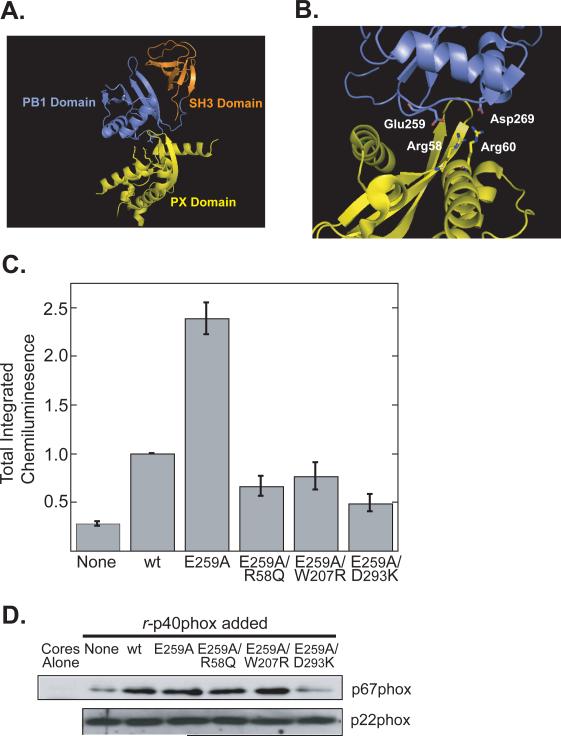
Figure 7. Relief of structural autoinhibition of p40phox enhances NADPH oxidase activity but still requires PX and SH3 Domain function.
(A) Ribbon diagram of full-length p40phox (PDB ID: 2DYB).
(B) Close up of the PX-PB1 domain interface showing some of the critical interacting residues that maintain the “closed” auto-inhibited conformation of p40phox.
(C) Reconstitution reactions were performed using permeabilized PMN cores and p40phox-depleted cytosol supplemented with the indicated recombinant p40phox proteins. Total integrated chemiluminescence was measured over 30 min and normalized to reactions supplemented with wild-type p40phox. Mean and standard deviations from n=3 experiments are shown.
(D) Differential effects of single and compound mutants that disrupt p40phox autoinhibition on association of p67phox with neutrophil cores. Reconstitution reactions were performed as in panel C in the presence of PMA for 10 min. Cores were then re-isolated and analyzed for p67phox by immunoblotting. Each lane corresponds to 9×105 cores. An immunoblot for p22phox is shown as a loading control. Results are representative of 3 separate experiments.
In order to determine the effect of the “open” or “closed” state of p40phox on the activation of the oxidase, we mutated Glu-259 of p40phox to Ala. Glu-259 is one of the key residues responsible for maintaining p40phox in a “closed” state (Figure 7B). Mutation of Glu-259 to Ala was reported to convert p40phox to an ‘open’ form and permit the PX domain to bind to PtdIns(3)P (24). When we added the r-p40phox E259A mutant to p40phox-depleted cores, we observed a dramatic increase in the amount of ROS produced by the cores relative to reactions reconstituted with wild-type r-p40phox (Figure 7C). Phe-320 of p40phox was found to have a similarly important role in keeping full-length p40phox “closed” (24). As observed with the E259A mutant, mutation of Phe-320 to Ala in r-p40phox also resulted in increased superoxide production when added to p40phox-depleted cores, compared to reactions containing wild-type r-p40phox (data not shown). Importantly, however, when we made mutations in the PX, SH3 or PB1 domains (R58Q, W207R or D293K respectively) in the background of an E259A mutant, we found that none of these double mutants were hyperactive (Figure 7C), suggesting that “opening” p40phox does not overcome the requirement for the PX, SH3 and PB1 domain functions.
To investigate whether the increased NADPH oxidase activity caused by the E259A mutant was due to the ability of this mutant protein to recruit more p67phox to the core membranes than wild-type p40phox, immunoblotting of the reconstituted PMA-stimulated cores for translocated p67phox was performed (Figure 7D). We observed that the E259A mutant p40phox protein translocated equal, but not larger amounts of p67phox to the cores as the wild type r-p40phox protein. Furthermore both the E259A/R58Q (PX domain) and E259A/W207R (SH3 domain) double mutants also delivered p67phox to the cores membranes to an extent comparable to wild-type r-p40phox, although consistent with results obtained with the p40phoxD293K (PB1 domain) single mutant (Figure 4C), the E259A/D293K double mutant translocated very little p67phox (Figure 7D). These data demonstrate that, while opening p40phox allows greater stimulation of the oxidase, the PX, SH3 and PB1 domains are all still required, and further establish that p40phox activates the oxidase in a manner that is not simply dependent on its ability to translocate p67phox.
Discussion
We used a combination of in cellulo RNAi experiments and ex vivo biochemical studies to probe the function and mechanism of p40phox function in assembly and activation of the NADPH oxidase. In cellulo studies were performed in PLB-985 cells, which can differentiate into mature neutrophils (32) that express all of the endogenous phox proteins, including p40phox, and display robust NADPH oxidase activity. Depletion of endogenous p40phox in these cells using lentiviral RNAi delivery revealed a pronounced decrease in PMA-dependent ROS production, and a marked defect in bacterial killing. These results are similar to those observed in bone-marrow derived neutrophils p40−/− mice (12). However, in contrast to p40phox−/− neutrophils, which demonstrated an ∼60% decrease in p67phox levels, the levels of p67phox in the PLB-985-derived knock-down neutrophils were unchanged, demonstrating a direct effect of endogenous p40phox on oxidase activity in a functionally relevant cell type.
An in depth biochemical analysis of p40phox function has been difficult until now since classical cell-free recombinant systems used for studying NADPH oxidase activity either do not show any enhancement of activity by p40phox (16,17), or demonstrate only a slight enhancement of activity (10−25%) if the system is modified to use crude neutrophil membranes as the source of the cytochrome (18). To overcome this limitation, we took advantage of a Streptolysin-O permeabilized neutrophil assay system previously described by our laboratory that maintains the ultrastructural features of neutrophils such as granules, compartments, etc, and allows intracellular oxidase activation in a PI 3-kinase dependent manner, mirroring the behavior of intact neutrophils (26). This permeabilized neutrophil ‘cores’ system permits biochemical manipulation and analysis, and allowed a series of structure-based mutations in p40phox to be studied in detail. Immunodepletion of p40phox from this system resulted in a profound decrease in PMA-stimulated intracellular ROS production that could be rescued by complementation with recombinant wild-type p40phox, but not by p40phox containing mutations in either the PX, PB1 or SH3 domains, illustrating the p40phox dependence of this system and indicating that all three domains are required for its biochemical activity.
p40phox is constitutively bound to p67phox through PB1-PB1 domain-mediated interactions, and p67phox has been shown to both bind to NADPH (40) and to facilitate electron flow from NADPH to FAD in the cytochrome (6,10). As such, we investigated whether p40phox might function as a molecular ‘chaperone’ for p67phox to facilitate interactions of NADPH with the oxidase. However, kinetic analysis of the reconstituted neutrophil cores revealed a similar KM for NADPH regardless of the presence or absence of p40phox, indicating that p40phox functions to stimulate the oxidase through its PX, PB1 and SH3 domains in some other manner. Even after the system has been assembled and activated in the absence of p40phox, the addition of p40phox still increases oxidase activity. We suggest this effect could be through either allosteric regulation of the pre-assembled holoenzyme complex, or by increased recruitment of p67phox to the region of the cytochrome, perhaps by enhancing the affinity of p67phox for p47phox during oxidase reconstitution as proposed by Cross (18). Indeed, we observed that p40phox-dependent translocation of p67phox was prevented by disruption of the PB1:PB1 domain interaction (D293K p40phox). However, contrary to the hypothesis that p40phox-dependent recruitment of p67phox would require the PX domain to interact with PtdIns(3)P-containing membranes, the R58Q p40phox mutant was fully competent to translocate p67phox compared to wild-type, as was an SH3 domain mutant (W207R p40phox). These data show that p40phox-dependent translocation of p67phox is necessary but not sufficient for oxidase activation in this system.
Very recently, the X-ray crystal structure of full-length p40phox was solved, revealing a ‘closed’ conformation via an intramolecular association of the PB1 domain with the PX domain that permits p40phoxp67phox binding but prohibits the PX domain from binding to PtdIns(3)P-containing membranes. Since mutations that abrogate PtdIns(3)P binding by the PX domain have significant impact on p40phox function, yet the full-length protein is in a ‘closed’ conformation such that the PX domain cannot contact PtdIns(3)P, then at some point during physiological activation of the oxidase, p40phox must transition to an ‘open’ state. This suggests that p40phox may itself be subject to allosteric regulation, perhaps via one or more interdomain interactions.
To investigate this, we first individually mutated key residues responsible for the intramolecular PX-PB1 domain interaction that have been shown to relieve the masking of the PX domain, E259A and F320A (24,25). These mutants displayed approximately 2.5-fold more activity than WT p40phox in our system, without increasing the amount of p67phox translocation, suggesting that opening of p40phox may be a limiting event in its mode of action. Furthermore, this result indicates that the stimulatory effect of p40phox on NADPH oxidase activity is functionally auto-inhibited in the resting state, and relieved upon disruption of the PX-PB1 interaction.
One function of the p40phox SH3 domain and/or the PX domain might be to assist in conversion of p40phox from its closed to its open conformation during oxidase activation (i.e. breathing movements in p40phox could permit transient association of the PX domain with PtdIns(3)P to prevent re-closure and stabilize the open form). Individual point mutations in the three modular domains (R58Q, W207R and D293K) were therefore generated in the background of the constitutively open E259A mutant. All three double mutants reversed the ability of the open form to activate the oxidase. These data demonstrate that although p40phox is opened during activation, none of the three domains are solely responsible for this event, such that constitutively opening the protein can circumvent the requirement for any of them. Instead both the PX domain and the SH3 domain directly participate in triggering and/or maintaining oxidase activity.
A mutant lacking the PX domain altogether (ΔPXp40phox) was incapable of rescuing the p40phox-dependent stimulation of the system, indicating that the presence of a PX domain is essential for the stimulatory function of p40phox. Surprisingly, however, we found that swapping the p40phox PX domain with the PX domain of p47phox resulted in production of equivalent amounts of ROS by the reconstituted oxidase, despite having very different phosphoinositide binding specificities (19). An R43A version of this p47PXp40phox fusion protein, rendering the PX domain of p47phox incapable of binding 3-phosphoinositides, displayed the same level of stimulatory capacity as the phosphoinositide-binding-competent version. Furthermore, compared to WT p40phox, the p47PX fusion proteins were much less sensitive to inhibition by wortmannin, underscoring the differential reliance of the two PX domains on interaction with 3-phosphoinositides, suggested by the R43A mutant chimera, for their positive influence on oxidase activity. We interpret the residual effect of wortmannin on inhibition of the p47PX chimeras to other PI3K-dependent processes necessary for oxidase activation, independent of signaling through p40phox. These data indicate a bifurcation in function of the two PX domains in the context of the rest of the p40phox protein: the wild-type PX domain of p40phox requires phosphoinositide binding to confer activity on the system, whereas the PX domain of p47phox does not. The p47phox PX domain has also been reported to bind to phoshatidic acid (PA) in vitro through a separate lipid binding pocket (47). We therefore cannot exclude the possibility that PA-binding to the p47phox PX domain in the p47PXp40phox fusion might be involved in the translocation and oxidase activation process.
Overlaying the structures of the two PX domains shows that the critical residues responsible for interaction of the p40 PX domain with the PB1 domain are not conserved in the PX domain of p47phox, suggesting that the p47PXp40phox constructs are in an open conformation. However, the p47PXp40phox construct displays equivalent activity as wild-type p40phox, whereas the p40phox E259A and F320A open mutants are 2.5-fold more active. This observation again suggests that PtdIns(3)P-binding to the p40phox PX domain has some type of specific and direct stimulatory effect on its ability to activate the enzyme.
When the ability of the p40phox constructs to translocate p67phox was examined, we found a stark lack of positive correlation between the ability of specific p40phox mutants to support oxidase activation and their ability to translocate p67phox. As expected, mutants unable to interact with p67phox did not translocate p67phox to the cores, but of the battery of other mutants (many of which had little or no activity), only ΔPXp40phox failed to localize p67phox. This disparity between the ability to activate the oxidase and the ability to localize p67phox suggests that: (a) p40phox-dependent translocation of p67phox is required, but not sufficient to activate the oxidase; (b) this localization is PX-domain-dependent but is independent of phosphoinositide binding of the PX domain; and (c) localization is independent of ligand binding by the SH3 domain. We do not know if the observed translocation of p67phox to the cores reflects its interaction with the membrane-bound cytochrome b558, or with the membrane-associated cytoskeleton. p40phox has been reported to bind to both coronin and moesin (48,49), two actin-binding proteins associated with the cytoskeleton. Intriguingly, the interaction of p40phox with moesin appears to involve the PX domain (49). Thus, it is tempting to speculate that this PX domain-moesin interaction is responsible for the p67phox translocation events that we observe, although additional experiments will be necessary to definitively support or refute this hypothesis.
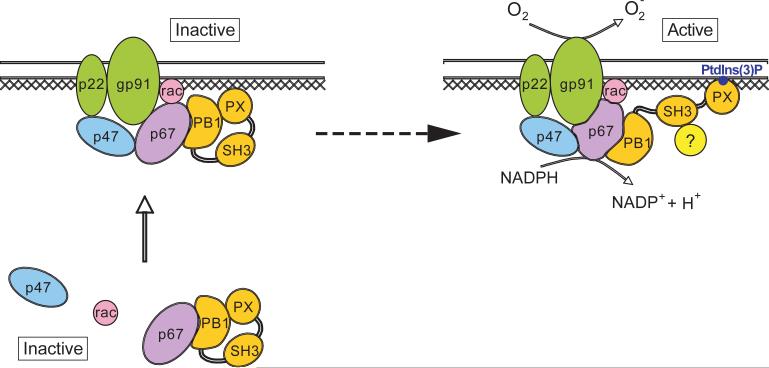
Taken together, our data would be consistent with the following model for the role of p40phox in the activation of the neutrophil NADPH oxidase (Fig. 8): in resting neutrophils, p40phox exists in an autoinhibited conformation. Upon neutrophil activation, p40phox is involved in the localization of p67phox to the cytochrome in a manner that requires the PX domain, but not its lipid-binding capacity. This localization is not sufficient to activate the oxidase, since mutants in the PX and SH3 domains are unable to activate the oxidase although they are competent to localize p67phox. Binding of PtdIns(3)P by the PX domain, and engagement of the SH3 domain, then functions as a molecular ‘switch’ that converts the holoenzyme from its inactive to its active form. Although molecular details of this activation process remain obscure, it might involve stimulating the ability of p67phox to act in a catalytic manner with multiple cytochome molecules, as proposed by Cross et al. (16). This model provides a succinct mechanism how the PX domain together with PtdIns(3)P could be involved in continual assembly of the oxidase holoenzyme during phaogocytosis followed by an acute burst of activation following phagosomal closure, since production of PtdIns(3)P appears to occur only following phagosomal closure via activation of a specific PI 3-K isoform, Vps34 (50). Definitive proof of this model, however, will likely require detailed X-ray structural studies of the holoenzyme complex in the presence and absence of PI 3-kinase derived lipid products.
Figure 8. Model for the role of p40phox in the activation of the neutrophil NADPH oxidase.
p40phox is shaded orange, with the PX, PB1 and SH3 domains indicated. Cross-hatching indicates a potential role for the cytoskeleton. See discussion for details.
Supplementary Material
A ΔPX domain truncation of p40phox retains the ability to bind to p67phox. Recombinant GST-p67phox was expressed in E. coli and bound to GSH beads. Following washing, the beads (20 μls) were incubated with 20μgs of His6-tagged wild-type or D293K forms of full-length p40phox, or 10μgs of the p40phox ΔPX truncation mutant for 2 hr at 4°C. Following 4 washes in TNE buffer containing 0.5% NP-40, the beads were eluted in Laemmli sample buffer, and analyzed by SDS-PAGE and immunoblotting using anti-p67phox and anti-His antibodies. The D293K mutant, which disrupts the PB1 domain function of p40phox known to bind to the PB1 domain of p67phox,serves as a negative control.
Acknowledgments
We gratefully acknowledge gifts of lentiviral plasmids from Drs. Christopher Dillon and Patrick Stern, and a p22phox antibody from Dr. Katrin Rittinger. We thank Drs. Mary Dinauer and Sergio Grinstein for helpful discussions. C.D.E. was funded by a fellowship from the Charles A. King Trust, Bank of America, Co-Trustee. This work was funded by NIH grant GM59281.
References
- 1.Nathan C. Nat Rev Immunol. 2006;6(3):173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 2.Babior BM, Lambeth JD, Nauseef W. Arch Biochem Biophys. 2002;397(2):342–344. doi: 10.1006/abbi.2001.2642. [DOI] [PubMed] [Google Scholar]
- 3.Meischl C, Roos D. Springer Semin Immunopathol. 1998;19(4):417–434. doi: 10.1007/BF00792600. [DOI] [PubMed] [Google Scholar]
- 4.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Medicine (Baltimore) 2000;79(3):170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Heyworth PG, Cross AR, Curnutte JT. Curr Opin Immunol. 2003;15(5):578–584. doi: 10.1016/s0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 6.Diebold BA, Bokoch GM. Nat Immunol. 2001;2(3):211–215. doi: 10.1038/85259. [DOI] [PubMed] [Google Scholar]
- 7.Clark RA. J Infect Dis. 1999;179(Suppl 2):S309–317. doi: 10.1086/513849. [DOI] [PubMed] [Google Scholar]
- 8.Heyworth PG, Curnutte JT, Nauseef WM, Volpp BD, Pearson DW, Rosen H, Clark RA. J Clin Invest. 1991;87(1):352–356. doi: 10.1172/JCI114993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith RM, Connor JA, Chen LM, Babior BM. J Clin Invest. 1996;98(4):977–983. doi: 10.1172/JCI118882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang PM, Babior BM, Smith RM. Biochemistry. 1999;38(18):5746–5753. doi: 10.1021/bi982750f. [DOI] [PubMed] [Google Scholar]
- 11.Vignais PV. Cell Mol Life Sci. 2002;59(9):1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellson CD, Davidson K, Ferguson GJ, O'Connor R, Stephens LR, Hawkins PT. J Exp Med. 2006;203(8):1927–1937. doi: 10.1084/jem.20052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh CI, Stull ND, Li XJ, Tian W, Price MO, Grinstein S, Yaffe MB, Atkinson S, Dinauer MC. J Exp Med. 2006;203(8):1915–1925. doi: 10.1084/jem.20052085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuribayashi F, Nunoi H, Wakamatsu K, Tsunawaki S, Sato K, Ito T, Sumimoto H. Embo J. 2002;21(23):6312–6320. doi: 10.1093/emboj/cdf642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sathyamoorthy M, de Mendez I, Adams AG, Leto TL. J Biol Chem. 1997;272(14):9141–9146. doi: 10.1074/jbc.272.14.9141. [DOI] [PubMed] [Google Scholar]
- 16.Cross AR, Erickson RW, Curnutte JT. Biochem J. 1999;341(Pt 2):251–255. [PMC free article] [PubMed] [Google Scholar]
- 17.Dagher MC, Pick E. Biochimie. 2007 doi: 10.1016/j.biochi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Cross AR. Biochem J. 2000;349(Pt 1):113–117. doi: 10.1042/0264-6021:3490113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. Nat Cell Biol. 2001;3(7):675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 20.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. Nat Cell Biol. 2001;3(7):679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 21.Massenet C, Chenavas S, Cohen-Addad C, Dagher MC, Brandolin G, Pebay-Peyroula E, Fieschi F. J Biol Chem. 2005;280(14):13752–13761. doi: 10.1074/jbc.M412897200. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs A, Dagher MC, Faure J, Vignais PV. Biochim Biophys Acta. 1996;1312(1):39–47. doi: 10.1016/0167-4889(96)00020-1. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Matsui Y, Ago T, Ota K, Sumimoto H. Embo J. 2001;20(15):3938–3946. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honbou K, Minakami R, Yuzawa S, Takeya R, Suzuki NN, Kamakura S, Sumimoto H, Inagaki F. Embo J. 2007;26(4):1176–1186. doi: 10.1038/sj.emboj.7601561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueyama T, Tatsuno T, Kawasaki T, Tsujibe S, Shirai Y, Sumimoto H, Leto TL, Saito N. Mol Biol Cell. 2007;18(2):441–454. doi: 10.1091/mbc.E06-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown GE, Stewart MQ, Liu H, Ha VL, Yaffe MB. Mol Cell. 2003;11(1):35–47. doi: 10.1016/s1097-2765(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 27.Brown GE, Stewart MQ, Bissonnette SA, Elia AE, Wilker E, Yaffe MB. J Biol Chem. 2004 doi: 10.1074/jbc.M314258200. [DOI] [PubMed] [Google Scholar]
- 28.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. Nat Genet. 2003;33(3):401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Karlin A, Loike JD, Silverstein SC. Proc Natl Acad Sci U S A. 2002;99(12):8289–8294. doi: 10.1073/pnas.122244799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N, Peitsch MC. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls A, Sharp KA, Honig B. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 32.Pedruzzi E, Fay M, Elbim C, Gaudry M, Gougerot-Pocidalo MA. Br J Haematol. 2002;117(3):719–726. doi: 10.1046/j.1365-2141.2002.03521.x. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T, Robinson JM, Seguchi H. J Cell Sci. 1998;111(Pt 1):81–91. doi: 10.1242/jcs.111.1.81. [DOI] [PubMed] [Google Scholar]
- 34.Vaissiere C, Le Cabec V, Maridonneau-Parini I. J Leukoc Biol. 1999;65(5):629–634. doi: 10.1002/jlb.65.5.629. [DOI] [PubMed] [Google Scholar]
- 35.Park JW, Hoyal CR, Benna JE, Babior BM. J Biol Chem. 1997;272(17):11035–11043. doi: 10.1074/jbc.272.17.11035. [DOI] [PubMed] [Google Scholar]
- 36.Rotrosen D, Leto TL. J Biol Chem. 1990;265(32):19910–19915. [PubMed] [Google Scholar]
- 37.van Bruggen R, Anthony E, Fernandez-Borja M, Roos D. J Biol Chem. 2004;279(10):9097–9102. doi: 10.1074/jbc.M309284200. [DOI] [PubMed] [Google Scholar]
- 38.DeLeo FR, Allen LA, Apicella M, Nauseef WM. J Immunol. 1999;163(12):6732–6740. [PubMed] [Google Scholar]
- 39.Karlsson A, Nixon JB, McPhail LC. J Leukoc Biol. 2000;67(3):396–404. doi: 10.1002/jlb.67.3.396. [DOI] [PubMed] [Google Scholar]
- 40.Dang PM, Johnson JL, Babior BM. Biochemistry. 2000;39(11):3069–3075. doi: 10.1021/bi9919807. [DOI] [PubMed] [Google Scholar]
- 41.Curnutte JT, Kuver R, Scott PJ. J Biol Chem. 1987;262(12):5563–5569. [PubMed] [Google Scholar]
- 42.Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. Mol Cell. 2001;8(4):829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 43.Hiroaki H, Ago T, Ito T, Sumimoto H, Kohda D. Nat Struct Biol. 2001;8(6):526–530. doi: 10.1038/88591. [DOI] [PubMed] [Google Scholar]
- 44.Tu Y, Liang L, Frank SJ, Wu C. Biochem J. 2001;354(Pt 2):315–322. doi: 10.1042/0264-6021:3540315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura R, Sumimoto H, Mizuki K, Hata K, Ago T, Kitajima S, Takeshige K, Sakaki Y, Ito T. Eur J Biochem. 1998;251(3):583–589. doi: 10.1046/j.1432-1327.1998.2510583.x. [DOI] [PubMed] [Google Scholar]
- 46.Quinn MT, Evans T, Loetterle LR, Jesaitis AJ, Bokoch GM. J Biol Chem. 1993;268(28):20983–20987. [PubMed] [Google Scholar]
- 47.Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL. Embo J. 2002;21(19):5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grogan A, Reeves E, Keep N, Wientjes F, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. J Cell Sci. 1997;110(Pt 24):3071–3081. doi: 10.1242/jcs.110.24.3071. [DOI] [PubMed] [Google Scholar]
- 49.Wientjes FB, Reeves EP, Soskic V, Furthmayr H, Segal AW. Biochem Biophys Res Commun. 2001;289(2):382–388. doi: 10.1006/bbrc.2001.5982. [DOI] [PubMed] [Google Scholar]
- 50.Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A ΔPX domain truncation of p40phox retains the ability to bind to p67phox. Recombinant GST-p67phox was expressed in E. coli and bound to GSH beads. Following washing, the beads (20 μls) were incubated with 20μgs of His6-tagged wild-type or D293K forms of full-length p40phox, or 10μgs of the p40phox ΔPX truncation mutant for 2 hr at 4°C. Following 4 washes in TNE buffer containing 0.5% NP-40, the beads were eluted in Laemmli sample buffer, and analyzed by SDS-PAGE and immunoblotting using anti-p67phox and anti-His antibodies. The D293K mutant, which disrupts the PB1 domain function of p40phox known to bind to the PB1 domain of p67phox,serves as a negative control.