Abstract
The structure–pharmacological activity relationships generally accepted for antitumor platinum compounds stressed the necessity for the cis-[PtX2(amine)2] structure while the trans-[PtX2(amine)2] structure was considered inactive. However, more recently, several trans-platinum complexes have been identified which are potently toxic, antitumor-active and demonstrate activity distinct from that of conventional cisplatin (cis-[PtCl2(NH3)2]). We have shown in the previous report that the replacement of ammine ligands by iminoether in transplatin (trans-[PtCl2(NH3)2]) results in a marked enhancement of its cytotoxicity so that it is more cytotoxic than its cis congener and exhibits significant antitumor activity, including activity in cisplatin-resistant tumor cells. In addition, we have also shown previously that this new trans compound (trans-[PtCl2(E-iminoether)2]) forms mainly monofunctional adducts at guanine residues on DNA, which is generally accepted to be the cellular target of platinum drugs. In order to shed light on the mechanism underlying the antitumor activity of trans-[PtCl2(E-iminoether)2] we examined oligodeoxyribonucleotide duplexes containing a single, site-specific, monofunctional adduct of this transplatin analog by the methods of molecular biophysics. The results indicate that major monofunctional adducts of trans-[PtCl2(E-iminoether)2] locally distort DNA, bend the DNA axis by 21° toward the minor groove, are not recognized by HMGB1 proteins and are readily removed from DNA by nucleotide excision repair (NER). In addition, the monofunctional adducts of trans-[PtCl2(E-iminoether)2] readily cross-link proteins, which markedly enhances the efficiency of this adduct to terminate DNA polymerization by DNA polymerases in vitro and to inhibit removal of this adduct from DNA by NER. It is suggested that DNA–protein ternary cross-links produced by trans-[PtCl2(E-iminoether)2] could persist considerably longer than the non-cross-linked monofunctional adducts, which would potentiate toxicity of this antitumor platinum compound toward tumor cells sensitive to this drug. Thus, trans-[PtCl2(E-iminoether)2] represents a quite new class of platinum antitumor drugs in which activation of trans geometry is associated with an increased efficiency to form DNA–protein ternary cross-links thereby acting by a different mechanism from ‘classical’ cisplatin and its analogs.
INTRODUCTION
cis-diamminedichloroplatinum(II) (cisplatin) (Fig. 1) is an efficient anticancer drug for the treatment of testicular and other germ-cell tumors (1). Since the discovery of its antitumor activity, the search continues for an improved platinum antitumor agent. The search is motivated by the desire to improve platinum chemotherapy since clinical use of cisplatin and some of its direct analogs is associated with diminished activity against a number of cancers, the acquired resistance developed by many tumors and severe side effects. In this search the hypothesis that platinum drugs which bind to DNA in a fundamentally different manner to that of cisplatin will have altered pharmacological properties has been tested (2). This concept has already led to the synthesis of several new unconventional platinum antitumor compounds that violate the original structure–activity relationships (3–5). The clinical inactivity of trans-diamminedichloro platinum(II) (transplatin) is considered a paradigm for the classical structure–activity relationships of platinum drugs (6), but to this end several new analogs of transplatin which exhibit a different spectrum of cytostatic activity including activity in tumor cells resistant to cisplatin have been identified (reviewed in 5,7,8). Examples of these antitumor trans-platinum complexes are the analogs of transplatin in which one ammine group is replaced by ligands such as thiazole, piperidine, piperazine, 4-picoline and cyclohexylamine; the analogs with branched asymmetric aliphatic amines; and the analogs containing iminoether groups of general formula trans-[PtCl2(E-iminoether)2] (trans-EE, Fig. 1) (5,7–9).
Figure 1.
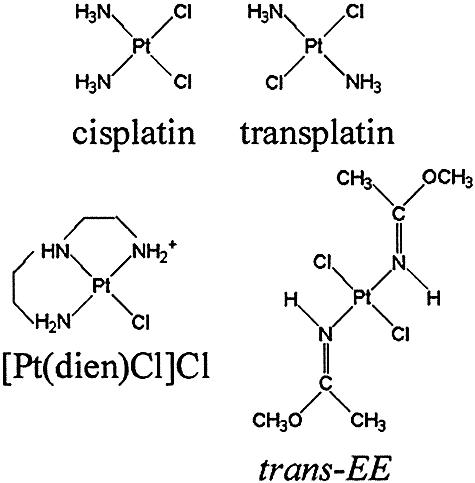
Structures of the platinum complexes.
trans-EE is not only more cytotoxic than its cis congener, but is also endowed with significant antitumor activity, including activity in cisplatin-resistant tumor cells (10–12). These results strongly imply a new mechanism of action for trans-EE. By analogy with the diamminedichloroplatinum(II) compounds, the inhibition of DNA synthesis by trans-EE (10,11) implies a role for DNA binding in the mechanism of action.
Bifunctional trans-EE preferentially forms stable monofunctional adducts at guanine residues in double-helical DNA (∼90% of monofunctional adducts are formed after 48 h at 37°C in 10 mM NaClO4) (13). The random modification of natural DNA in cell-free media results in non-denaturational alterations in the conformation of DNA (14), similar to the modification by cisplatin, but different from the modification by transplatin [which produces denaturational distortions in DNA (15)]. The most striking feature of the lesions of trans-EE is that they prematurely terminate RNA synthesis at similar sites and with a similar efficiency to major DNA adducts of cisplatin. This is a very intriguing finding since the prevalent lesions formed on DNA by trans-EE are monofunctional adducts at guanine residues and monofunctional DNA adducts of other platinum(II) complexes {such as those of [PtCl(NH3)3]Cl, chlorodiethylenetriamineplatinum(II) chloride ([PtCl(dien)]Cl), cisplatin and transplatin} do not terminate RNA synthesis (16,17). In addition, it is generally accepted that monofunctional DNA adducts of cisplatin are not relevant to its cytostatic effects.
Recently, a short duplex containing a single, monofunctional adduct of trans-EE at a central guanine residue has been analyzed by NMR spectroscopy (18). This analysis has yielded a model in which the bending induced by the monofunctional adduct of trans-EE was ∼45° towards the minor groove.
Since other structural details of the monofunctional DNA adducts formed by this antitumor transplatin analog are not yet available, it remains uncertain how these lesions affect conformation of DNA and how these alterations are further processed in the cells. Therefore, in order to shed light on the mechanism underlying activity of trans-EE, we examined in detail in the present work short oligodeoxyribonucleotide duplexes containing a single, site-specific, monofunctional adduct formed by this drug at a central guanine residue. We investigated how this adduct affects the local conformation of DNA (in particular bending and unwinding) and how this adduct is further processed by some cellular components in cell-free media.
MATERIALS AND METHODS
Chemicals
trans-EE (Fig. 1) and its mononitrato trans-[Pt(NO3)Cl(E-iminoether)2] analog was prepared by the methods described in detail previously (13,19). Cisplatin (Fig. 1A) was obtained from Sigma (Prague, Czech Republic). [PtCl(dien)]Cl was from Lachema a.s. (Brno, Czech Republic). The stock solutions of platinum compounds were prepared at concentrations of 5 × 10–4 M in 10 mM NaClO4 and stored at 4°C in the dark. The synthetic oligodeoxyribonucleotides were synthesized and purified as described previously (20). Expression and purification of domains A (residues 1–84) and B (residues 85–180) of the HMGB1 proteins (HMGB1a and HMGB1b, respectively) (HMG = high-mobility-group) were carried out as described (21,22). T4 DNA ligase, the Klenow fragment from DNA polymerase I (exonuclease minus, mutated to remove the 3′→5′ proofreading domain) (KF–), restriction endonuclease EcoRI and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). Reverse transcriptase from human immunodeficiency virus type 1 (RT HIV-1) was from Calbiochem (San Diego, CA). Histone H1 and deoxyribonucleoside 5′-triphosphates were from Roche Diagnostics, GmbH (Mannheim, Germany). Acrylamide, bis(acrylamide), urea and NaCN were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate (DMS), KMnO4, diethyl pyrocarbonate (DEPC), KBr and KHSO5 were from Sigma (Prague, Czech Republic). Nonidet P-30 was from Fluka (Prague, Czech Republic). Radioactive products were from Amersham (Arlington Heights, IL). Proteinase K and ATP were from Boehringer (Mannheim, Germany).
Platinations of oligonucleotides
The single-stranded oligonucleotides (the top, pyrimidine-rich, strands containing a single central G of the 19–23 bp duplexes) were reacted in stoichiometric amounts with either [PtCl(dien)]Cl or the mononitrato analog of trans-EE. The platinated oligonucleotides were purified by ion-exchange fast protein liquid chromatography (FPLC). It was verified by platinum flameless atomic absorption spectrophotometry (FAAS) and by optical density measurements that the modified oligonucleotides contained one platinum atom. It was also verified using DMS footprinting of platinum on DNA (23) that one trans-EE or [PtCl(dien)]Cl molecule was coordinated to the N7 atom of the single G in the top strands of each duplex. FPLC purification and FAAS measurements were carried out on a Pharmacia Biotech FPLC System with MonoQ HR 5/5 column and a Unicam 939 AA spectrometer equipped with a graphite furnace, respectively. The duplexes containing single, central 1,2-GG intrastrand cross-links (CL) of cisplatin in the pyrimidine-rich top strand were prepared as described (20). The unmodified or platinated duplexes used in the studies of recognition by HMGB1 domain proteins were still purified by electrophoresis on native 15% polyacrylamide (PAA) gel [mono:bis(acrylamide) ratio = 29:1]. Other details have been described previously (20,24).
Ligation and electrophoresis of oligonucleotides
Unplatinated 19–23mer single strands (bottom strands in Fig. S1A in Supplementary Material) were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. They were then annealed with their phosphorylated complementary strands (unplatinated or containing monofunctional adduct of trans-EE at the G residue). The duplexes were allowed to react with T4 DNA ligase. The resulting samples along with ligated unplatinated duplexes were subsequently examined on 8% native PAA [mono:bis(acrylamide) ratio = 29:1] electrophoresis gels. Other details of these experiments were as described in previous papers (25–27).
Chemical modifications
The modifications by KMnO4, DEPC and KBr/KHSO5 were performed as described previously (24,28–30). The strands of the duplexes were 5′-end-labeled with [γ-32P]ATP. In the case of the platinated oligonucleotides, the platinum complex was removed after reaction of the DNA with the probe by incubation with 0.2 M NaCN (pH 11) at 45°C for 10 h in the dark.
Gel-mobility-shift assay
The 5′-end labeled 20 bp oligonucleotide duplexes either unplatinated (controls) or containing the central platinum adduct in their top strands were used and their reaction with HMG-domain proteins was performed and analyzed as described previously (31).
Inhibition of DNA polymerization
We investigated in the present work DNA polymerization using the templates site-specifically modified by trans-EE or cisplatin by two DNA polymerases, which differ in processivity and fidelity. The DNA polymerase I class of enzymes has served as the prototype for studies on structural and biochemical mechanisms of DNA replication (32,33). In addition, as the most extensive genetic, biochemical and structural studies have been carried out on Klenow fragment of DNA polymerase I (including its exonuclease-deficient analog) this enzyme appears to be an ideal model system for investigating the molecular mechanisms associated with template-directed DNA synthesis (32,33). The other DNA polymerase used in these studies was RT HIV-1, showing a different mechanism underlying its catalytic activity and relatively low processivity and fidelity (34).
The 23-, 30- or 40mer templates (Figs 2 and 3) containing a single monofunctional adduct of trans-EE or [PtCl(dien)]Cl or 1,2-GG intrastrand CL of cisplatin were prepared in the same way as described above. Eight- or 17mer DNA primers whose sequences are also shown in Figures 2 and 3 were complementary to the 3′ termini of the 23, 40 or 30mer templates, respectively. The DNA substrates were formed by annealing templates and 5′-end-labeled primers at a molar ratio of 3:1. All experiments using KF– and RT HIV-1 were performed at 25°C in a volume of 50 µl in a buffer containing 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2, 0.1 mM dithiothreitol, 50 µg/ml BSA, 0.1% Nonidet P-30, 25 µM dATP, 25 µM dCTP, 25 µM dGTP, 25 µM TTP and 0.5 U KF–. The experiments with RT HIV-1 were performed at 37°C using the same conditions except that the nucleoside triphosphates were at a concentration of 100 µM and 1.0 U of the enzyme was used. Reactions were terminated by the addition of EDTA so that its resulting concentration was 20 mM and by heating at 100°C for 30 s. Products were resolved by denaturing 24 or 15% PAA/8 M urea gel and then visualized and quantified by using the FUJIFILM bio-imaging analyzer and AIDA image analyzer software.
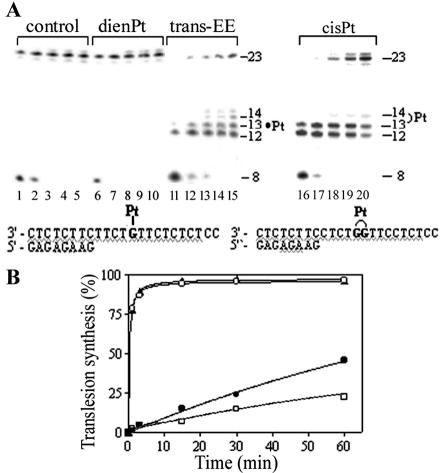
Figure 2.
Primer extension activity of exonuclease-deficient Klenow fragment of DNA polymerase I (KF–). (A) Experiments were conducted using the 8mer/23mer primer/template duplex for various times using undamaged template (lanes 1–5), the template containing monofunctional adduct of [Pt(dien)Cl]Cl (lanes 6–10), monofunctional adduct of trans-EE (lanes 11–15) or 1,2-GG intrastrand CL of cisplatin (lanes 16–20). Timings were as follows: 1 min, lanes 1, 6, 11 and 16; 3 min, lanes 2, 7, 12 and 17; 15 min, lanes 3, 8, 13 and 18; 30 min, lanes 4, 9, 14 and 19; 60 min, lanes 5, 10, 15 and 20. The pause sites opposite the platinated guanines and flanking residues are marked 12, 13 and 14 (the sites opposite the platinated residues are still marked ‘Pt’). The nucleotide sequences of the templates and the primer are shown beneath the gels. (B) The time dependence of the inhibition of DNA synthesis on undamaged (control) template (open circles), DNA containing monofunctional adduct of [Pt(dien)Cl]Cl (closed triangles), DNA containing monofunctional adduct of trans-EE (open squares) or DNA containing 1,2-GG intrastrand CL of cisplatin (closed circles). Data are means (±SE) from three different experiments with two independent template preparations.
Figure 3.
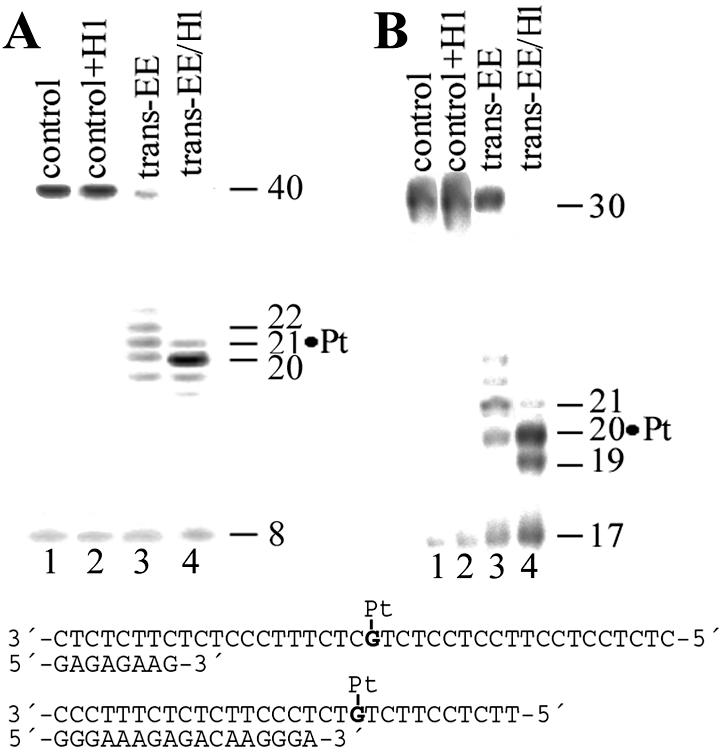
Primer extension activity of exonuclease-deficient Klenow fragment of DNA polymerase I (KF–) (A) and RT HIV-1 (B) using the 8mer/40mer and 17mer/30mer primer/template duplexes, respectively. The experiments were conducted for 30 min using undamaged templates (lanes 1), undamaged templates to which histone H1 was added at a molar ratio of 4:1 (lanes 2), the templates containing monofunctional adduct of trans-EE (lanes 3) and monofunctional adduct of trans-EE cross-linked to histone H1 (lanes 4). The pause sites opposite the platinated guanines and flanking residues are marked 19, 20, 21 and 22 (the sites opposite the platinated residue are still marked ‘Pt’). The nucleotide sequences of the templates and the primers are shown beneath the gels.
Nucleotide excision assay
The 148 bp substrates containing single, central, monofunctional adducts of trans-EE, [PtCl(dien)]Cl or 1,2-GG intrastrand CL of cisplatin were assembled from three oligonucleotide duplexes as described previously (35,36).
Oligonucleotide excision reactions were performed in cell-free extracts (CFEs) prepared from the HeLa S3 and Chinese hamster ovary (CHO) AA8 cell lines as described (37,38). In vitro repair was measured with excision assay using these CFEs and 148 bp linear DNA substrates in the same way as described previously (38). Reaction products were still treated overnight with 0.4 M NaCN, pH 10–11, at 45°C to remove platinum from excised fragments. The NaCN treatment was included to eliminate both the effect of the positively charged platinum complex bound to the excised fragments and the protein cross-linked to the excised fragments on their migration in the gel.
Mapping of incision sites was performed as described in the preceding reports (38). The major excision product (gel-purified) was further incubated for 10 min at 30°C with T4 DNA polymerase (0.25 U) in 10 µl of buffer composed of 50 mM Tris–HCl (pH 8.8), 15 mM (NH4)2SO4, 7 mM MgCl2, 0.1 mM EDTA, 50 mM β-mercaptoethanol and 20 µg BSA/ml, supplemented with 0.5 µg SmaI-digested pBluescript DNA and visualized by autoradiography following resolution in 10% denaturing PAA gel. Similar analyses using radiolabeled, platinated 20mers (used in the nucleotide excision assays) were also used to identify the nucleotide(s) at which the exonuclease activity of T4 DNA polymerase is blocked 3′ to the lesion. The location of the 5′ incision site made by the excinuclease was determined by comparison with the length of excision products observed in the absence of T4 DNA polymerase digestion.
RESULTS
Bending, unwinding and chemical probes of DNA conformation
Important structural motifs induced in DNA by antitumor platinum compounds that play a significant role in the mechanism underlying their antitumor activity are the unwinding and bending of the helix axis (5,39). For DNA adducts of cisplatin, the structural details of the bending and unwinding have been elucidated (26,40,41). Given the recent advances in our understanding of the structural basis of the bending and unwinding of DNA caused by cisplatin, it is of considerable interest to examine how the major monofunctional adduct of trans-EE affects these conformational properties of DNA. In this work we performed studies on the bending and unwinding afforded by a single, site-specific, monofunctional adduct formed by trans-EE at guanine residues using electrophoretic retardation as a quantitative measure of the extent of planar curvature (25,26).
Oligodeoxyribonucleotide duplexes (19–23 bp) (see Fig. S1A in Supplementary Material for their sequences) containing, in the top pyrimidine-rich strand, a central TGT sequence (42) were used for the bending and unwinding studies of the present work. The ligation products of these duplexes unplatinated or containing a single, site-specific, monofunctional adduct of trans-EE at the central guanine residue in the top strand were analyzed on native PAA electrophoresis gel (see Fig. S1B in Supplementary Material). Experimental details of these studies are given in our recent reports (23,27,43).
The K factor is defined as the ratio of calculated to actual length. The calculated length is based on a multimer’s mobility, and is obtained from a calibration curve constructed from the mobilities of unplatinated multimers. The variation of the K factor versus sequence length obtained for multimers of the 19–23 bp duplexes and containing the monofunctional adduct of trans-EE is shown in Figure S1C (Supplementary Material). Maximum retardation was observed for the 21 bp duplex. This observation suggests that the natural 10.5 bp repeat of B-DNA was not markedly changed. The exact helical repeat of the duplex containing the adduct of trans-EE, and from it the unwinding angle, were calculated by interpolation with the use of the K versus interadduct distance curve as described in the previous papers for other platinum adducts (23,40). The maximum of these curves constructed for the duplexes modified by trans-EE with a total length of 130 bp (see Fig. S1D in Supplementary Material) was determined to be 21.10 ± 0.01 bp. Total sequence lengths other than 130 bp were examined and gave identical results. To convert the interadduct distance in base pairs corresponding to the curve maximum into a duplex unwinding angle in degrees, the value is compared with that of the helical repeat of B-DNA, which is 10.5 ± 0.05 bp (44). The difference between the helical repeat of B-DNA and the DNA-containing monofunctional adduct of trans-EE, therefore, is [(21.10 ± 0.01) – 2(10.5 ± 0.05)] = 0.10 ± 0.06 bp. There are 360°/10.5 bp, so the DNA unwinding due to one monofunctional adduct of trans-EE is 3 ± 2°.
The quantitation of the bend angle of the monofunctional adduct of trans-EE was performed in the way described previously (24,26,41,45), utilizing the empirical equation
K – 1 = (9.6 × 10–5L2 – 0.47)(RC)2 1
where L represents the length of a particular oligomer with relative mobility K, and RC the curvature relative to a DNA bending induced at the tract of six adenines (A6 tract) (45). Application of 1 to the 120, 130 or 140 bp multimers of the 21 bp oligomers containing the single, monofunctional adduct of trans-EE (see Fig. S1C in Supplementary Material) leads to a mean curvature of 0.53, relative to an A6 tract. The average bend angle per helix turn can be calculated by multiplying the relative curvature by the absolute value of an A6 tract bend [20° (26)]. The results indicate that the bend induced by the monofunctional adduct of trans-EE is ∼21°. That this bend was oriented towards the minor groove of DNA was verified in the same way as previously using the duplex [TGT+(A/T)5](32) (see Fig. S1A in Supplementary Material for its sequence) (41,46).
Further studies of the present work were focused on analysis of the distortion induced by the monofunctional adducts of trans-EE by chemical probes of DNA conformation. The 20 bp duplex [the duplex TGT(20) in Fig. S1A in Supplementary Material] containing the single, site-specific adduct of trans-EE was treated with several chemical agents used as tools for monitoring the existence of conformations other than canon-ical B-DNA. These agents included KMnO4, DEPC and bromine. They react preferentially with single-stranded DNA and distorted double-stranded DNA (24,47). We used for this analysis the same methodology described in detail in our recent papers dealing with DNA adducts of various antitumor platinum drugs (43,48). The results (see Fig. S2 in Supplementary Material) indicate that the adduct of trans-EE induces in DNA a distortion that extends over at least 2 bp and is localized mainly at the base pair containing the platinated G residue and the adjacent base pairs on the 5′ side.
Recognition by the domains A and B of HMGB1
An important feature of the mechanism that underlies the antitumor activity of cisplatin and its direct analogs in a number of tumor cells is that the major adducts of these drugs (1,2-GG intrastrand CLs) are recognized by proteins containing HMG domains (5,39,49). Importantly, DNA modified by transplatin or monodentate platinum(II) compounds, such as [PtCl(dien)]Cl or [PtCl(NH3)3]Cl, is not recognized by these cellular proteins. Since the monofunctional adducts of trans-EE distort DNA in a different way to the monofunctional adducts of transplatin, [PtCl(dien)]Cl or [PtCl(NH3)3]Cl, we examined whether the monofunctional adducts of trans-EE enhance affinity of HMG-box proteins to DNA. The interactions of the rat HMGB1 domain A (HMGB1a) and HMGB1 domain B (HMGB1b) with DNA modified by trans-EE were investigated using gel-mobility-shift assay (31,50,51) (Fig. S3A and B in Supplementary Material). In these experiments, the 20 bp duplex (see Fig. S3C in Supplementary Material for its sequence) was modified so that it contained a single, site-specific, monofunctional adduct of trans-EE. The binding of HMGB1a and HMGB1b to these DNA probes was detected by retardation of the migration of the radiolabeled 20 bp probes through the gel (Fig. S3A and B in Supplementary Material) under identical conditions described in detail in our recent papers (42,52).
As indicated by the presence of a shifted band whose intensity increases with growing protein concentration, both HMGB1a and HMGB1b recognized the duplex containing the 1,2-GG intrastrand CL of cisplatin (Fig. S3A and B in Supplementary Material), consistent with earlier observations (49,53,54). These proteins exhibited, under the same experimental conditions, negligible binding to the 20 bp duplex unplatinated or containing the monofunctional adduct of trans-EE (see Fig. S3A and B in Supplementary Material). These data indicate that HMGB1 proteins do not bind the probe containing the major adducts of trans-EE.
Probing trans-EE adducts by DNA polymerases
It has been demonstrated that various DNA secondary structures have significant effects on processivity of a number of prokaryotic, eukaryotic and viral DNA polymerases (55–57). Interestingly, with DNA templates containing site-specifically placed adducts of various platinum compounds, a number of prokaryotic and eukaryotic DNA polymerases were blocked but could also traverse through platinum adducts depending on their character and conformational alterations induced in DNA. Inhibition of prokaryotic DNA and RNA polymerases by the adducts on DNA globally modified by trans-EE has already been demonstrated in in vitro replication or transcription mapping experiments (10,14). Similarly, the inhibition of DNA synthesis in human tumor cells treated with trans-EE has been demonstrated and found to be greater than that with the cis isomer (11). Monofunctional adducts of cisplatin or transplatin and those of the monodentate compounds such as [PtCl(dien)]Cl or [PtCl(NH3)3]Cl terminate DNA synthesis by DNA polymerases in vitro markedly less efficiently than major 1,2-GG intrastrand CLs of cisplatin (55). It is, therefore, interesting to examine whether DNA polymerases, processing DNA substrates containing either the major monofunctional adduct of trans-EE or bifunctional 1,2-GG intrastrand CL of cisplatin, could reveal potential differences in conformational alterations imposed on DNA by these two adducts.
We constructed the 8mer/23mer primer/template duplexes unplatinated or containing the monofunctional adduct of trans-EE or [PtCl(dien)]Cl in the central TGT sequence or the 1,2-GG intrastrand CL of cisplatin in the central TGGT sequence (for their sequences, see Fig. 2A). The first eight nucleotides on the 3′ terminus of the 23mer template strand were complementary to the nucleotides of the 8mer primer and the guanine involved in the monofunctional adduct of trans-EE, [PtCl(dien)]Cl or the 3′ guanine in the 1,2-GG CL of cisplatin on the template strand were located at the 13th position from the 3′ terminus (Fig. 2A). After annealing the 8 nt primer to the 3′ terminus of the unplatinated or platinated template strand positioning the 3′-end of the primer five bases before the adduct in the template strand, we examined DNA polymerization through the single, monofunctional adduct of trans-EE or [PtCl(dien)]Cl and the 1,2-intrastrand CL of cisplatin by KF– in the presence of all four deoxyribonucleoside 5′-triphosphates. The reaction was stopped at various time intervals, and the products were analyzed using a sequencing 24% PAA/8 M urea gel (Fig. 2A). Polymerization using the 23mer template containing the CL of cisplatin proceeded rapidly up to the nucleotide preceding and at the sites opposite the CL, such that the 12 and 13 nt products accumulated to a significant extent (shown in Fig. 2A, lanes 16–20). The larger DNA intermediates were not observed in a considerable extent, whereas no intermediate products were seen with the 23mer control template or the template containing the monofunctional adduct of [PtCl(dien)]Cl as the full-length products were formed (shown in Fig. 2A, lanes 1–10). The full-length products were also noticed with the 23mer template containing the CL of cisplatin, although in a significantly smaller amount (Fig. 2A, lanes 16–20). This result is in agreement with a previously published work (58) in which T7 DNA polymerase and RT HIV-1 were used and confirms that 1,2-GG intrastrand CL of cisplatin inhibits DNA synthesis (55), but translesion synthesis may occur. Under the same experimental conditions, DNA polymerization by KF– using the template containing the monofunctional adduct of trans-EE proceeded up to the nucleotide preceding the site opposite the platinated G involved in the adduct and to the following nucleotide residue (Fig. 2A, lanes 11–15). There was almost no accumulation of shorter and larger DNA intermediates and, importantly, the full-length products were also noticed. The amount of the full-length products increased with reaction time, but with a noticeably lower rate compared to polymerization using the template containing the CL of cisplatin (Fig. 2B). This result suggests that the monofunctional adducts of trans-EE are even more efficient inhibitors of DNA polymerization than the major adducts of cisplatin.
In order to further support the latter conclusion, we have also examined the effects of the monofunctional adduct of trans-EE on DNA polymerization by RT HIV-1. In these studies elongation of the 17mer/30mer primer/template duplexes was tested by rapidly mixing a solution of RT HIV-1 and DNA with a solution containing all four deoxyribonucleoside 5′-triphosphates. The 30mer template was non-modified or contained the monofunctional adduct of trans-EE located at the 20th position from the 3′ terminus (for its sequence, see the bottom sequence in Fig. 3). The reaction was stopped at various times, and the products were analyzed using a sequencing 15% PAA/8 M urea gel. Polymerization using the trans-EE template proceeded rapidly up to the nucleotide at the site opposite the platinated and following residues, such that 20 and 21 nt intermediates accumulated to a significant extent (shown in Fig. 3B for the incubation time of 30 min). Nevertheless, the synthesis by the RT HIV-1 across the monofunctional adduct of trans-EE was still possible, as in the case of the polymerization by KF–. Hence, we confirmed also by using DNA polymerase showing a different mechanism underlying its catalytic activity than KF– that the monofunctional adduct of trans-EE constitutes a fairly strong block to DNA synthesis catalyzed by KF– and RT HIV-1 but not absolute, thus permitting translesion DNA synthesis with a limited efficiency. Since there is a high degree of structural and sequence conservation of the domains among eukaryotic, prokaryotic and viral polymerases (59) insights gleaned from studies of the KF– and RT HIV-1 also should be applicable to other DNA polymerases (32,60,61). Hence, the observation that DNA polymerization is inhibited by trans-EE adducts more strongly than by the adducts of other simple monofunctional platinum(II) compounds and even by the major CL of cisplatin add a new dimension to the impact of the activation of the trans geometry in platinum compounds by iminoether ligands on processes in tumor cells, possibly including replication or DNA repair.
Nucleotide excision repair (NER)
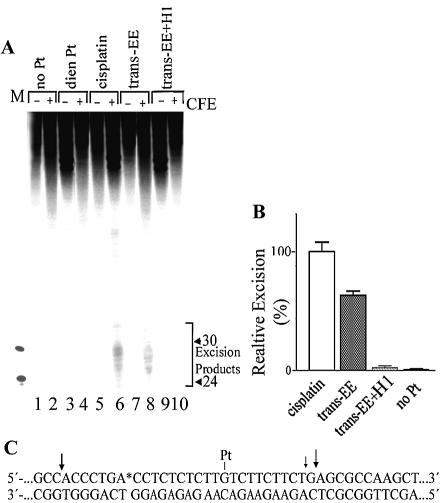
NER is a pathway used by human cells for the removal of damaged nucleotides from DNA (62,63). In mammalian cells, this repair pathway is an important mechanism for the removal of bulky, helix-distorting DNA adducts, such as those generated by various chemotherapeutics including cisplatin (64). Efficient repair of 1,2-GG or 1,3-GNG intrastrand CL of cisplatin has been reported by various NER systems including human and rodent excinucleases (38,65–69). The result presented in Figure 4A, lane 6 is consistent with these reports. The major excision fragment contains 28 nt and other primary excision fragments are 24–29 nt in length (38,70). In contrast, consistent with previous reports (42,71,72), no excision fragments were noticed if the monofunctional adduct of [PtCl(dien)]Cl was used as a substrate for human and rodent excinucleases (shown in Fig. 4A, lane 4 for rodent excinuclease). Importantly, the monofunctional adduct of trans-EE was also repaired by both human and rodent excinucleases, although with a somewhat lower efficiency than the major intrastrand CLs of cisplatin [shown in Fig. 4A (lane 8) and B for the adduct removed by rodent excinuclease].
Figure 4.
Excision of the adducts of platinum complexes by rodent excinuclease. (A) The 148 bp substrates were incubated with CHO AA8 CFE and subsequently treated overnight with NaCN prior to analysis in 10% PAA/8 M urea denaturing gel. Lanes 1 and 2, control, unplatinated substrate; lanes 3 and 4, the substrate containing the monofunctional adduct of [Pt(dien)Cl]Cl; lanes 5 and 6, 1,2-GG intrastrand CL of cisplatin; lanes 7 and 8, the monofunctional adduct of trans-EE; lanes 9 and 10, the monofunctional adduct of trans-EE cross-linked to histone H1. Lanes 1, 3, 5, 7 and 9, no extract added; lanes 2, 4, 6, 8 and 10, the substrates were incubated with CHO AA8 CFE for 40 min at 30°C. Lane M, the 20 and 30 nt markers. (B) Quantitative analysis of removal of the adducts. The columns marked cisplatin, trans-EE, trans-EE+H1 and noPt represent 1,2-GG intrastrand CL of cisplatin, monofunctional adduct of trans-EE, monofunctional adduct of trans-EE cross-linked to histone H1 and unplatinated substrate, respectively. The radioactivity associated with the fragments excised from the duplex containing the 1,2-GG intrastrand CL of cisplatin was taken as 100%. Data are the average of two independent experiments performed under the same conditions; bars indicate range of excision. (C) The central sequence of the 148 bp substrate. The site of the monofunctional adduct of trans-EE or [Pt(dien)Cl]Cl is marked ‘Pt’ and arrows indicate the major incision sites.
T4 DNA polymerase 3′→5′ exonuclease activity was used to map the primary sites of incision in the same way as described in recent papers (73). The major 3′ incision site is 10 nt (or at the 11th phosphodiester bond) 3′ to the adduct (the minor 3′ incision site is at the 10th phosphodiester bond) and the other major incision site at the 18th phosphodiester bond 5′ to the adduct (Fig. 4C).
Ternary DNA–protein complex formation
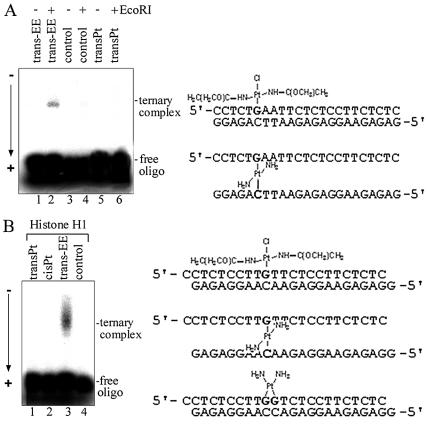
The monofunctional DNA adducts of trans-EE were investigated for their ability to specifically cross-link proteins. The 1,2-GG intrastrand CL of cisplatin and the interstrand CL of transplatin formed between G and complementary C were used as controls. The 22 or 23 bp duplexes 5′-end-labeled at their top, pyrimidine-rich strands (the sequences of which are given in Fig. 5) were modified by trans-EE so that they contained a single, site-specific, monofunctional adduct at the G residue. Ternary DNA–protein cross-linking efficiency was assessed by gel mobility shift assays. The 23 bp duplex containing the monofunctional adduct of trans-EE was mixed with EcoRI restriction endonuclease, a sequence-specific DNA-binding protein. A fraction was detected by denaturing gel electrophoresis with significantly retarded mobility (Fig. 5A, lane 2) compared with that of the free probe. This fraction was eliminated after treatment with sodium cyanide or proteinase K converting it to that of the unmodified probe (not shown). These results suggest that the species is a protein–DNA CL tethered by platinum–guanine and platinum–protein covalent bonds. While the proteinase K and NaCN experiments clearly indicate that protein is the species cross-linked to DNA, the amino acids participating in the cross-linking reaction have not been determined.
Figure 5.
Ternary complex formation of unmodified and platinated oligodeoxyribonucleotide duplexes containing single, site-specific platinum adduct with EcoRI (A) or histone H1 (B) assessed by SDS–PAA gel electrophoresis. (A) Lanes 1 and 2, the 23 bp duplex containing the monofunctional adduct of trans-EE; lanes 3 and 4, non-modified duplex; lanes 5 and 6, the duplex containing the interstrand CL of transplatin. Lanes 1, 3 and 5, no enzyme added; lanes 2, 4 and 6, EcoRI added. (B) Lane 1, the 22 bp duplex containing the interstrand CL of transplatin; lane 2, the duplex containing the 1,2-GG intrastrand CL of cisplatin; lane 3, the duplex containing the monofunctional adduct of trans-EE; lane 4, non-modified duplex. The nucleotide sequences of the duplexes with the platinum adducts are shown to the right of each panel.
Representative, non-sequence-specific DNA-binding protein, the linker histone H1 (which is an abundant nuclear protein) also effectively cross-links the 22 bp duplex containing the monofunctional adduct of trans-EE, as monitored by standard gel shift assays (Fig. 5B, lane 3) and confirmed by the treatment with NaCN and proteinase K. No ternary complexes containing EcoRI or histone H1 were formed under the same experimental conditions between the duplexes containing the CL of cisplatin or transplatin [Fig. 5A (lane 6) and B (lanes 1 and 2)]. Thus, in all cases studied it is clear that formation of the ternary DNA–platinum–protein complex is efficient for the major monofunctional adducts of trans-EE.
The 40- or 30mer templates cross-linked to histone H1 by trans-EE were isolated from the gel, purified, hybridized with 8- or 17mer primers, respectively and used as substrates to investigate the translesion synthesis across the monofunctional adduct of trans-EE. As shown in lanes 4 of Figure 3, polymerization by KF– or RT HIV-1 using the template cross-linked to histone H1 mainly proceeded to the sites close to the platinated G. However, in contrast to the polymerization using the same templates but containing the adduct of trans-EE not cross-linked to a protein, no full-length products were noticed and the polymerization was terminated mainly at the platinated site. Similarly, no removal of the monofunctional adducts of trans-EE cross-linked to histone H1 from the 148 bp substrates containing single, central, monofunctional adduct of trans-EE by human or rodent excinuclease was observed under conditions when the same adduct not cross-linked to a protein was readily excised [shown for rodent excinuclease in Fig. 4A (lane 10) and B].
DISCUSSION
It is generally accepted that the key intracellular target for antitumor platinum drugs is DNA on which these compounds form various types of adducts (74). The adducts of conventional cisplatin distort DNA conformation, inhibit replication and transcription (but they are also bypassed by DNA or RNA polymerases), and trigger apoptosis or necrosis (75). In addition, cisplatin adducts are removed from DNA mainly by NER. They are, however, also recognized by a number of proteins, such as, for instance, HMG-domain proteins. The details of how the binding of HMG-domain proteins to cisplatin-modified DNA sensitizes tumor cells to cisplatin are still not completely resolved, but possibilities such as shielding cisplatin–DNA adducts from excision repair or that these proteins could be titrated away from their transcriptional regulatory function, have been suggested as clues for how they are involved in antitumor activity. Experimental support of these aspects of the mechanism underlying antitumor activity of cisplatin or resistance of some tumors to this drug has recently been thoroughly reviewed (5,75–78). On the other hand, it has also been demonstrated (79) that the ability of HMGB1 protein, and probably other cisplatin–DNA-binding proteins, to influence the efficacy of the drug may be dependent on tumor cell type.
Lack of activity of transplatin against tumors has been proposed to be associated with selective recognition of its DNA adducts by cellular repair systems resulting in their removal (80,81). Moreover, the monofunctional transplatin–DNA intermediates are much longer lived than their cisplatin analogs (81). The transplatin intermediates may react readily with sulfur-containing nucleophiles, such as glutathione or metallothioneins, preventing closure to the bifunctional cytotoxic lesion. This feature of the monofunctional adducts of transplatin and the fact that the free transplatin molecules can readily react with sulfur-containing nucleophiles (81,82) may be at least partly responsible for clinical inefficiency of transplatin (8,81,83–85). trans-EE forms on DNA stable monofunctional adducts preferentially at guanine residues (13). These lesions distort DNA, although differently from the adducts of cisplatin or transplatin (11,13,14,86). These adducts can inhibit DNA polymerization to a limited extent so that they can also be bypassed by DNA polymerases (Figs 2 and 3). In contrast to the adducts of cisplatin, the monofunctional adducts of trans-EE are not recognized by HMG-domain proteins. An important structural motif recognized by HMG-domain proteins on DNA containing the major 1,2-GG intrastrand CL of cisplatin is a stable, directional bend of the helix axis toward the major groove (87). As demonstrated in the present work the major monofunctional adduct of trans-EE also bends the helix axis of DNA (by 21°), but towards the minor groove, and no recognition of these adducts by HMGB1 proteins was observed. Plausible explanation of this observation may be that the pre-bending due to the monofunctional adduct of trans-EE is too small and/or in an incorrect direction to be recognized by HMG-domain proteins. Thus, from these considerations we could conclude that the mechanism of antitumor activity of trans-EE does not involve recognition by HMG-domain proteins as a crucial step, in contrast to the proposals for cisplatin and its direct analogs in certain types of cells.
Several reports have demonstrated (38,66) that NER is a major mechanism contributing to cisplatin resistance. The examinations of excision of monofunctional adducts of trans-EE have revealed that these adducts can also be efficiently removed by NER (Fig. 4). Hence, by analogy to the mechanism proposed for antitumor effects of cisplatin, the monofunctional adducts of trans-EE should be shielded by damaged-DNA recognition proteins, such as those containing HMG-domains, to prevent their repair. However, we demonstrate in the present work that, in contrast to cisplatin CLs, the adducts of trans-EE are not recognized by HMG-domain proteins. Hence, it is reasonable to assume that the monofunctional adducts of trans-EE would be removed from DNA too early to trigger downstream processes leading to toxicity of trans-EE toward tumor cells sensitive to this drug, unless other factors than affinity of the HMG-domain proteins to the adducts protect them from being removed from DNA. In addition, lack of affinity of HMG-domain proteins for the adducts of trans-EE implies that these proteins will not be titrated away from their transcriptional regulatory function as they can be in certain types of cells in the presence of cisplatin CLs (88). Thus, a reasonable alternative for the mechanism underlying antitumor effects of trans-EE may also be that proteins other than HMG-domain proteins bind to DNA adducts of this drug and by other mechanisms.
The results of the present work demonstrate for the first time that the monofunctional adducts of trans-EE readily cross-link proteins (Fig. 5). Interestingly, earlier observations have demonstrated that cisplatin and transplatin also form DNA–protein ternary CLs (89–97). However, the monofunctional adducts of cisplatin formed in the first step of the reaction with DNA close to bifunctional CLs with a relatively fast rate so that these adducts do not persist for long enough to allow their extensive cross-linking to proteins to occur (82). Consistent with this conclusion is the relatively very low frequency of the DNA–protein ternary CLs produced in cells treated with cisplatin (<1%) (98). Transplatin forms more monofuntional adducts on DNA than cisplatin, and the rate of rearrangement of monofunctional to bifunctional adducts is considerably slower compared to cisplatin [24 h are required for ∼50% rearrangement of monofunctional adducts of transplatin (81)], but still considerably faster than that of the monofunctional adducts of trans-EE (13). In addition, in contrast to trans-EE, free transplatin and its monofunctional DNA adducts readily react with sulfur-containing nucleophiles, such as glutathione or thiourea, which may translabilize transplatin from DNA (81). On the other hand, free trans-EE and its DNA monofunctional adducts react with sulfur-containing nucleophiles, such as thiourea (13), glutathione or Zn-MT2 metallothionein (unpublished results) much less readily than free cisplatin, transplatin and their monofunctional DNA adducts. Hence, the formation of the DNA–protein ternary CLs in cells treated with transplatin is much less likely than in the cells treated with trans-EE. Taken together, the capacity of the monofunctional DNA adducts of trans-EE to cross-link proteins supports the idea that the capability of trans-EE monofunctional DNA adducts to cross-link proteins represents an important feature of the mechanism underlying antitumor effects of this platinum compound.
The results of the present work demonstrate that cross-linking proteins to monofunctional DNA adducts of trans-EE markedly enhances the efficiency of this adduct to terminate DNA polymerization by DNA polymerases in vitro (Fig. 3) and to inhibit removal of this adduct from DNA by NER (Fig. 4). Hence, it is reasonable to suggest that DNA–protein ternary CLs produced by trans-EE could persist considerably longer than their non-cross-linked monofunctional adducts, potentiating the toxicity of trans-EE toward tumor cells sensitive to this drug. In other words, covalent cross-linking of DNA and proteins by trans-EE represents a potential novel mechanism through which this compound could exert its antitumor activity.
To date several strategies on ways to activate trans geometry in bifunctional platinum(II) compounds, including circumvention of resistance to cisplatin, have been proposed. One strategy consists of chemical modification of the ineffective transplatin which results in an increased efficiency to form in DNA interstrand CLs and/or in an increased stability of its 1,3-intrastrand CLs in double-helical DNA (99). Examples of such compounds are the analogs of transplatin in which at least one ammine ligand is replaced by a heterocyclic amine ligand [such as quinoline, thiazole or pyridine (7,100) or piperidine, piperazine or 4-picoline (99)]. Another strategy consists of a chemical modification of transplatin resulting in the capability of the new complex to form DNA adducts that mimic the structure of the adducts of cisplatin (42,101). For instance, antitumor trans-[PtCl2(NH3)(thiazole)] forms on DNA monofunctional adducts which mimic 1,2-intrastrand CLs of cisplatin including their recognition by HMG-domain proteins and NER (42). In addition, this trans compound also forms interstrand CLs which are similar to those formed in DNA by cisplatin (101).
Efforts to investigate the generality of the DNA–protein ternary CLs formed by trans-EE are under study in these laboratories and will be reported in due course. Given that cellular DNA is intimately associated with proteins, DNA–protein ternary CLs formed by trans-EE stresses the importance of this type of DNA damage for antitumor effects of the new platinum compound. Although formation of DNA–protein CLs seems to be limited to proteins that are able to bind to DNA, the potential in vivo CL formation with other DNA-binding proteins, including those directly related to neoplastically transformed cells, is not excluded. The capacity of trans-EE to cross-link the oncoproteins or other functional proteins with nuclear DNA in vivo would represent a potentially novel mechanism that might contribute to the antitumor efficacy of trans-EE by abrogating the functional integrity of these proteins.
In conclusion, trans-EE represents a quite new class of platinum antitumor drugs in which activation of the trans geometry is associated with an increased efficiency to form DNA–protein ternary CLs. In addition, our results provide additional strong support for the hypothesis that platinum drugs which bind to DNA in a fundamentally different manner to that of cisplatin have altered pharmacological properties. Hence, trans-EE and its analogs may represent a novel class of platinum anticancer drugs acting by a different mechanism to cisplatin and its analogs.
SUPPLEMENTARY MATERAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. T. Reardon and A. Sancar for HeLa and CHO cells. We also acknowledge participation in the EC COST Chemistry Actions D20 and D21, which enabled us to regularly exchange the most recent ideas in the field of platinum anticancer drugs with several European colleagues. This research was supported by the Grant Agency of the Czech Republic (grant no. 305/02/1552A), the Grant Agency of the Academy of Sciences of the Czech Republic (grant no. KJB5004301), the Wellcome Trust (to J.K. and V.B.) and by the Italian Ministry for Education, University and Research.
REFERENCES
- 1.O'Dwyer P.J., Stevenson,J.P. and Johnson,S.W. (1999) Clinical status of cisplatin, carboplatin and other platinum-based antitumor drugs. In Lippert,B. (ed.), Cisplatin. Chemistry and Biochemistry of a Leading Anticancer Drug. VHCA, WILEY-VCH, Zürich, Weinheim, pp. 31–72. [Google Scholar]
- 2.Farrell N., Kelland,L.R., Roberts,J.D. and Van Beusichem,M. (1992) Activation of the trans geometry in platinum antitumor complexes: a survey of the cytotoxicity of trans complexes containing planar ligands in murine-L1210 and human tumor panels and studies on their mechanism of action. Cancer Res., 52, 5065–5072. [PubMed] [Google Scholar]
- 3.Wong E. and Giandomenico,C.M. (1999) Current status of platinum-based antitumor drugs. Chem. Rev., 99, 2451–2466. [DOI] [PubMed] [Google Scholar]
- 4.Judson I. and Kelland,L.R. (2000) New developments and approaches in the platinum arena. Drugs, 59, 29–36. [DOI] [PubMed] [Google Scholar]
- 5.Brabec V. (2002) DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog. Nucleic Acids Res. Mol. Biol., 71, 1–68. [DOI] [PubMed] [Google Scholar]
- 6.Reedijk J. (1996) Improved understanding in platinum antitumour chemistry. Chem. Comm., 801–806. [Google Scholar]
- 7.Farrell N. (1996) Current status of structure-activity relationships of platinum anticancer drugs: Activation of the trans geometry. In Sigel,A. and Sigel,H. (eds), Metal Ions in Biological Systems. Marcel Dekker, Inc., New York, Basel, Hong Kong, Vol. 32, pp. 603–639. [PubMed] [Google Scholar]
- 8.Perez J.-M., Fuertes,M.A., Alonso,C. and Navarro-Ranninger,C. (2000) Current status of the development of trans-platinum antitumor drugs. Crit. Rev. Oncol. Hematol., 35, 109–120. [DOI] [PubMed] [Google Scholar]
- 9.Natile G. and Coluccia,M. (1999) trans-Platinum compounds in cancer therapy: a largely unexplored strategy for identifying novel antitumor platinum drugs. In Clarke,M.J. and Sadler,P.J. (eds), Metallopharmaceuticals. Springer, Berlin, Germany, Vol. 1, pp. 73–98. [Google Scholar]
- 10.Coluccia M., Nassii,F., Loseto,F., Boccarelli,A., Marigió,M.A., Giordano,D., Intini,F.P., Caputo,P. and Natile,G. (1993) A trans-platinum complex showing higher antitumor activity than the cis congeners. J. Med. Chem., 36, 510–512. [DOI] [PubMed] [Google Scholar]
- 11.Coluccia M., Boccarelli,A., Mariggio,M.A., Cardellicchio,N., Caputo,P., Intini,F.P. and Natile,G. (1995) Platinum(II) complexes containing iminoethers: A trans platinum antitumour agent. Chem. Biol. Interact., 98, 251–266. [DOI] [PubMed] [Google Scholar]
- 12.Coluccia M., Nassi,A., Boccarelli,A., Giordano,D., Cardellicchio,N., Intini,F.P., Natile,G., Barletta,A. and Paradiso,A. (1999) In vitro antitumour activity and cellular pharmacological properties of the platinum-iminoether complex trans-[PtCl2{E-HN=C(OMe)Me}2]. Int. J. Oncol., 15, 1039–1044. [DOI] [PubMed] [Google Scholar]
- 13.Brabec V., Vrana,O., Novakova,O., Kleinwachter,V., Intini,F.P., Coluccia,M. and Natile,G. (1996) DNA adducts of antitumor trans-[PtCl2(E-imino ether)2]. Nucleic Acids Res., 24, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaludova R., Zakovska,A., Kasparkova,J., Balcarova,Z., Vrana,O., Coluccia,M., Natile,G. and Brabec,V. (1997) DNA modifications by antitumor trans-[PtCl2(E-iminoether)2]. Mol. Pharmacol., 52, 354–361. [DOI] [PubMed] [Google Scholar]
- 15.Brabec V., Kleinwächter,V., Butour,J.L. and Johnson,N.P. (1990) Biophysical studies of the modification of DNA by antitumour platinum coordination complexes. Biophys. Chem., 35, 129–141. [DOI] [PubMed] [Google Scholar]
- 16.Lemaire M.A., Schwartz,A., Rahmouni,A.R. and Leng,M. (1991) Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum(II) and DNA. Proc. Natl Acad. Sci. USA, 88, 1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brabec V. and Leng,M. (1993) DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl Acad. Sci. USA, 90, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen B., Margiotta,N., Coluccia,M., Natile,G. and Sletten,E. (2000) Antitumor trans platinum DNA adducts: NMR and HPLC study of the interaction between a trans-Pt iminoether complex and the deoxy decamer d(CCTCGCTCTC).d(GAGAGCGAGG). Metal-Based Drugs, 7, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cini R., Caputo,P.A., Initini,F.P. and Natile,G. (1995) Mechanistic and stereochemical investigation of iminoethers formed by alcoholysis of coordinated nitrils: X-ray crystal structure of cis- and trans-[bis(1-imino-1-methoxyethane) dichloroplatinum(II)]. Inorg. Chem., 34, 1130–1137. [Google Scholar]
- 20.Brabec V., Reedijk,J. and Leng,M. (1992) Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding. Biochemistry, 31, 12397–12402. [DOI] [PubMed] [Google Scholar]
- 21.Stros M. (1998) DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem., 273, 10355–10361. [PubMed] [Google Scholar]
- 22.Stros M. (2001) Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry, 40, 4769–4779. [DOI] [PubMed] [Google Scholar]
- 23.Kasparkova J., Mellish,K.J., Qu,Y., Brabec,V. and Farrell,N. (1996) Site-specific d(GpG) intrastrand cross-links formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry, 35, 16705–16713. [DOI] [PubMed] [Google Scholar]
- 24.Brabec V., Sip,M. and Leng,M. (1993) DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II). Biochemistry, 32, 11676–11681. [DOI] [PubMed] [Google Scholar]
- 25.Koo H.S., Wu,H.M. and Crothers,D.M. (1986) DNA bending at adenine·thymine tracts. Nature, 320, 501–506. [DOI] [PubMed] [Google Scholar]
- 26.Bellon S.F. and Lippard,S.J. (1990) Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-Pt(NH3)2(N3-cytosine)Cl+. Biophys. Chem., 35, 179–188. [DOI] [PubMed] [Google Scholar]
- 27.Kasparkova J., Farrell,N. and Brabec,V. (2000) Sequence specificity, conformation and recognition by HMG1 protein of major DNA interstrand cross-links of antitumor dinuclear platinum complexes. J. Biol. Chem., 275, 15789–15798. [DOI] [PubMed] [Google Scholar]
- 28.Bailly C., Gentle,D., Hamy,F., Purcell,M. and Waring,M.J. (1994) Localized chemical reactivity in DNA associated with the sequence-specific bisintercalation of echinomycin. Biochem. J., 300, 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross S.A. and Burrows,C.J. (1996) Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate. Nucleic Acids Res., 24, 5062–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailly C. and Waring,M.J. (1997) Diethylpyrocarbonate and osmium tetroxide as probes for drug-induced changes in DNA conformation in vitro. In Fox,K.R. (ed.), Drug–DNA Interaction Protocols. Humana Press Inc, Totowa, NJ, pp. 51–79. [DOI] [PubMed] [Google Scholar]
- 31.He Q., Ohndorf,U.-A. and Lippard,S.J. (2000) Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry, 39, 14426–14435. [DOI] [PubMed] [Google Scholar]
- 32.Lam W.C., VanderSchans,E.J.C., Sowers,L.C. and Millar,D.P. (1999) Interaction of DNA polymerase I (Klenow fragment) with DNA substrates containing extrahelical bases: Implications for proofreading of frameshift errors during DNA synthesis. Biochemistry, 38, 2661–2668. [DOI] [PubMed] [Google Scholar]
- 33.Patel P.H., Suzuki,M., Adman,E., Shinkai,A. and Loeb,L.A. (2001) Prokaryotic DNA polymerase I: Evolution, structure and ‘base flipping’ mechanism for nucleotide selection. J. Mol. Biol., 308, 823–837. [DOI] [PubMed] [Google Scholar]
- 34.Johnson K.A. (1993) Conformational coupling in DNA-polymerase fidelity. Annu. Rev. Biochem., 62, 685–713. [DOI] [PubMed] [Google Scholar]
- 35.Matsunaga T., Mu,D., Park,C.-H., Reardon,J.T. and Sancar,A. (1995) Human DNA repair excision nuclease. J. Biol. Chem., 270, 20862–20869. [DOI] [PubMed] [Google Scholar]
- 36.Buschta-Hedayat N., Buterin,T., Hess,M.T., Missura,M. and Naegeli,H. (1999) Recognition of non-hybridizing base pairs during nucleotide excision repair of DNA. Proc. Natl Acad. Sci. USA, 96, 6090–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manley J.L., Fire,A., Cano,A., Sharp,P.A. and Gefter,M.L. (1980) DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl Acad. Sci. USA, 77, 3855–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reardon J.T., Vaisman,A., Chaney,S.G. and Sancar,A. (1999) Efficient nucleotide excision repair of cisplatin, oxaliplatin and bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res., 59, 3968–3971. [PubMed] [Google Scholar]
- 39.Jamieson E.R. and Lippard,S.J. (1999) Structure, recognition and processing of cisplatin-DNA adducts. Chem. Rev., 99, 2467–2498. [DOI] [PubMed] [Google Scholar]
- 40.Bellon S.F., Coleman,J.H. and Lippard,S.J. (1991) DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry, 30, 8026–8035. [DOI] [PubMed] [Google Scholar]
- 41.Huang H.F., Zhu,L.M., Reid,B.R., Drobny,G.P. and Hopkins,P.B. (1995) Solution structure of a cisplatin-induced DNA interstrand cross-link. Science, 270, 1842–1845. [DOI] [PubMed] [Google Scholar]
- 42.Kasparkova J., Novakova,O., Farrell,N. and Brabec,V. (2003) DNA binding by antitumor trans-[PtCl2(NH3)(thiazole)]. Protein recognition and nucleotide excision repair of monofunctional adducts. Biochemistry, 42, 792–800. [DOI] [PubMed] [Google Scholar]
- 43.Zehnulova J., Kasparkova,J., Farrell,N. and Brabec,V. (2001) Conformation, recognition by high mobility group domain proteins and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464. J. Biol. Chem., 276, 22191–22199. [DOI] [PubMed] [Google Scholar]
- 44.Rhodes D. and Klug,A. (1980) Helical periodicity of DNA determined by enzyme digestion. Nature, 286, 573–578. [DOI] [PubMed] [Google Scholar]
- 45.Rice J.A., Crothers,D.M., Pinto,A.L. and Lippard,S.J. (1988) The major adduct of the antitumor drug cis-diamminedichloroplatinum(II) with DNA bends the duplex by 40o toward the major groove. Proc. Natl Acad. Sci. USA, 85, 4158–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kostrhunova H. and Brabec,V. (2000) Conformational analysis of site-specific DNA cross-links of cisplatin-distamycin conjugates. Biochemistry, 39, 12639–12649. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen P.E. (1990) Chemical and photochemical probing of DNA complexes. J. Mol. Recogit., 3, 1–24. [DOI] [PubMed] [Google Scholar]
- 48.Malina J., Hofr,C., Maresca,L., Natile,G. and Brabec,V. (2000) DNA interactions of antitumor cisplatin analogs containing enantiomeric amine ligands. Biophys. J., 78, 2008–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei M., Cohen,S.M., Silverman,A.P. and Lippard,S.J. (2001) Effects of spectator ligands on the specific recognition of intrastrand platinum-DNA cross-links by high mobility group box and TATA-binding proteins. J. Biol. Chem., 276, 38774–38780. [DOI] [PubMed] [Google Scholar]
- 50.Ohndorf U.M., Rould,M.A., He,Q., Pabo,C.O. and Lippard,S.J. (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature, 399, 708–712. [DOI] [PubMed] [Google Scholar]
- 51.Cohen S.M., Mikata,Y., He,Q. and Lippard,S.J. (2000) HMG-domain protein recognition of cisplatin 1,2-intrastrand d(GpG) cross-links in purine-rich sequence contexts. Biochemistry, 39, 11771–11776. [DOI] [PubMed] [Google Scholar]
- 52.Kasparkova J., Zehnulova,J., Farrell,N. and Brabec,V. (2002) DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by high mobility group domain proteins and nucleotide excision repair. J. Biol. Chem., 277, 48076–48086. [DOI] [PubMed] [Google Scholar]
- 53.Stehlikova K., Kostrhunova,H., Kasparkova,J. and Brabec,V. (2002) DNA bending and unwinding due to the major 1,2-GG intrastrand cross-link formed by antitumor cis-diamminedichloroplatinum(II) are flanking-base independent. Nucleic Acids Res., 30, 2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasparkova J., Delalande,O., Stros,M., Elizondo-Riojas,M.A., Vojtiskova,M., Kozelka,J. and Brabec,V. (2003) Recognition of DNA interstrand cross-link of antitumor cisplatin by HMGB1 protein. Biochemistry, 42, 1234–1244. [DOI] [PubMed] [Google Scholar]
- 55.Comess K.M., Burstyn,J.N., Essigmann,J.M. and Lippard,S.J. (1992) Replication inhibition and translesion synthesis on templates containing site-specifically placed cis-diamminedichloroplatinum(II) DNA adducts. Biochemistry, 31, 3975–3990. [DOI] [PubMed] [Google Scholar]
- 56.Suo Z. and Johnson,K. (1998) DNA secondary structure effects on DNA synthesis catalyzed by HIV-1 reverse transcriptase. J. Biol. Chem., 273, 27259–27267. [DOI] [PubMed] [Google Scholar]
- 57.Vaisman A., Warren,M.W. and Chaney,S.G. (2001) The effect of DNA structure on the catalytic efficiency and fidelity of human DNA polymerase beta on templates with platinum-DNA adducts. J. Biol. Chem., 276, 18999–19005. [DOI] [PubMed] [Google Scholar]
- 58.Suo Z., Lippard,S. and Johnson,K. (1999) Single d(GpG)/cis-diammineplatinum(II) adduct-induced inhibition of DNA polymerization. Biochemistry, 38, 715–726. [DOI] [PubMed] [Google Scholar]
- 59.Hubscher U., Nasheuer,H.P. and Syvaoja,J.E. (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci., 25, 143–147. [DOI] [PubMed] [Google Scholar]
- 60.Steitz T.A. (1999) DNA polymerases: Structural diversity and common mechanisms. J. Biol. Chem., 274, 17395–17398. [DOI] [PubMed] [Google Scholar]
- 61.Lone S. and Romano,L.J. (2003) Mechanistic insights into replication across from bulky DNA adducts: A mutant polymerase I allows an N-acetyl-2-aminofluorene adduct to be accommodated during DNA synthesis. Biochemistry, 42, 3826–3834. [DOI] [PubMed] [Google Scholar]
- 62.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 63.Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- 64.Reardon J.T. and Sancar,A. (1998) Molecular mechanism of nucleotide excision repair in mammalian cells. In Dizdaroglu,M. and Karakaya,A. (eds), Advances in DNA Damage and Repair. Plenum Publishing Corp., New York, NY, pp. 377–393. [Google Scholar]
- 65.Jones S.L., Hickson,I.D., Harris,A.L. and Harnett,P.R. (1994) Repair of cisplatin–DNA adducts by protein extracts from human ovarian carcinoma. Int. J. Cancer, 59, 388–393. [DOI] [PubMed] [Google Scholar]
- 66.Zamble D.B., Mu,D., Reardon,J.T., Sancar,A. and Lippard,S.J. (1996) Repair of cisplatin–DNA adducts by the mammalian excision nuclease. Biochemistry, 35, 10004–10013. [DOI] [PubMed] [Google Scholar]
- 67.Moggs J.G., Szymkowski,D.E., Yamada,M., Karran,P. and Wood,R.D. (1997) Differential human nucleotide excision repair of paired and mispaired cisplatin–DNA adducts. Nucleic Acids Res., 25, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koberle B., Masters,J.R.W., Hartley,J.A. and Wood,R.D. (1999) Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr. Biol., 9, 273–276. [DOI] [PubMed] [Google Scholar]
- 69.Li M.J. and Yang,L.Y. (1999) Use of novel plasmid constructs to demonstrate fludarabine triphosphate inhibition of nucleotide excision repair of a site-specific 1,2-d(GpG) intrastrand cisplatin adduct. Int. J. Oncol., 15, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 70.Huang J.-C., Svoboda,D.L., Reardon,J.T. and Sancar,A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calsou P., Frit,P. and Salles,B. (1992) Repair synthesis by human cell extracts in cisplatin-damaged DNA is prefentially determined by minor adducts. Nucleic Acids Res., 20, 6363–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buterin T., Hess,M.T., Gunz,D., Geacintov,N.E., Mullenders,L.H. and Naegeli,H. (2002) Trapping of DNA nucleotide excision repair factors by non-repairable carcinogen adducts. Cancer Res., 62, 4229–4235. [PubMed] [Google Scholar]
- 73.Malina J., Kasparkova,J., Natile,G. and Brabec,V. (2002) Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem. Biol., 9, 629–638. [DOI] [PubMed] [Google Scholar]
- 74.Johnson N.P., Butour,J.-L., Villani,G., Wimmer,F.L., Defais,M., Pierson,V. and Brabec,V. (1989) Metal antitumor compounds: The mechanism of action of platinum complexes. Prog. Clin. Biochem. Med., 10, 1–24. [Google Scholar]
- 75.Fuertes M.A., Alonso,C. and Perez,J.M. (2003) Biochemical modulation of cisplatin mechanisms of action: Enhancement of antitumor activity and circumvention of drug resistance. Chem. Rev., 103, 645–662. [DOI] [PubMed] [Google Scholar]
- 76.Guo Z.J. and Sadler,P.J. (1999) Metals in medicine. Angew. Chem. Int. Ed., 38, 1513–1531. [DOI] [PubMed] [Google Scholar]
- 77.Jordan P. and Carmo-Fonseca,M. (2000) Molecular mechanisms involved in cisplatin cytotoxicity. Cell. Mol. Life Sci., 57, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohen S.M. and Lippard,S.J. (2001) Cisplatin: From DNA damage to cancer chemotherapy. Prog. Nucleic Acid Res. Mol. Biol., 67, 93–130. [DOI] [PubMed] [Google Scholar]
- 79.Wei M., Burenkova,O. and Lippard,S.J. (2003) Cisplatin sensitivity in Hmgb1(–/–) and Hmgb1(+/+) mouse cells. J. Biol. Chem., 278, 1769–1773. [DOI] [PubMed] [Google Scholar]
- 80.Ciccarelli R.B., Solomon,M.J., Varshavsky,A. and Lippard,S.J. (1985) In vivo effects of cis- and trans-diamminedichloroplatinum(II) on SV 40 chromosomes: Differential repair, DNA–protein cross-linking and inhibition of replication. Biochemistry, 24, 7533–7540. [DOI] [PubMed] [Google Scholar]
- 81.Eastman A. and Barry,M.A. (1987) Interaction of trans-diamminedichloroplatinum(II) with DNA: Formation of monofunctional adducts and their reaction with glutathione. Biochemistry, 26, 3303–3307. [DOI] [PubMed] [Google Scholar]
- 82.Bancroft D.P., Lepre,C.A. and Lippard,S.J. (1990) Pt-195 NMR kinetic and mechanistic studies of cis-diamminedichloroplatinum and trans-diamminedichloroplatinum(II) binding to DNA. J. Am. Chem. Soc., 112, 6860–6871. [Google Scholar]
- 83.Lepre C.A. and Lippard,S.J. (1990) Interaction of platinum antitumor compounds with DNA. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, Heidelberg, Germany, Vol. 4, pp. 9–38. [Google Scholar]
- 84.Kelland L.R. (1993) New platinum antitumor complexes. Crit. Rev. Oncol. Hematol., 15, 191–219. [DOI] [PubMed] [Google Scholar]
- 85.Leng M., Schwartz,A. and Giraud-Panis,M.J. (2000) Transplatin-modified oligonucleotides as potential antitumor drugs. In Kelland,L.R. and Farrell,N.P. (eds), Platinum-Based Drugs in Cancer Therapy. Humana Press Inc, Totowa, NJ, pp. 63–85. [Google Scholar]
- 86.Zaludova R., Natile,G. and Brabec,V. (1997) The effect of antitumor trans-[PtCl2(E-iminoether)2] on B→Z transition in DNA. Anti-Cancer Drug Des., 12, 295–309. [PubMed] [Google Scholar]
- 87.Zamble D.B. and Lippard,S.J. (1999) The response of cellular proteins to cisplatin-damaged DNA. In Lippert,B. (ed.), Cisplatin. Chemistry and Biochemistry of a Leading Anticancer Drug. VHCA, WILEY-VCH, Zürich, Weinheim, pp. 73–110. [Google Scholar]
- 88.Kartalou M. and Essigmann,J.M. (2001) Recognition of cisplatin adducts by cellular proteins. Mutation Res., 478, 1–21. [DOI] [PubMed] [Google Scholar]
- 89.Zwelling L.A., Anderson,T. and Kohn,K.W. (1979) DNA–protein and DNA interstrand cross-linking by cis- and trans-platinum(II)diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res., 39, 365–369. [PubMed] [Google Scholar]
- 90.Banjar Z.M., Hnilica,L.S., Briggs,R.C., Stein,J. and Stein,G. (1984) cis- and trans-Diamminedichloroplatinum(II)-mediated cross-linking of chromosomal non-histone proteins to DNA in HeLa cells. Biochemistry, 23, 1921–1926. [DOI] [PubMed] [Google Scholar]
- 91.Olinski R., Wedrychovski,A., Schmidt,W.N., Briggs,R.C. and Hnilica,L.S. (1987) In vivo DNA-protein cross-linking by cis- and trans-diamminedichloroplatinum(II). Cancer Res., 47, 201–205. [PubMed] [Google Scholar]
- 92.Baudin F., Romby,P., Romaniuk,P.J., Ehresmann,B. and Ehresmann,C. (1989) Crosslinking of transcription factor TfIIa to ribosomal 5S RNA from X.laevis by trans-diamminedichloroplatinum(II). Nucleic Acids Res., 17, 10035–10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller C.A., Cohen,M.D. and Costa,M. (1991) Complexing of actin and other nuclear proteins to DNA by cis-diamminedichloroplatinum(II) and chromium compounds. Carcinogenesis, 12, 269–276. [DOI] [PubMed] [Google Scholar]
- 94.Olinski R.B.R.C. (1991) DNA–protein cross-linking in L1210 cells and resistant to cis-diamminedichloroplatinum(II). Mol. Biol. Rep., 15, 81–86. [DOI] [PubMed] [Google Scholar]
- 95.Comess K.M. and Lippard,S.J. (1993) Molecular aspects of platinum-DNA interactions. In Neidle,S. and Waring,M. (eds), Molecular Aspects of Anticancer Drug–DNA Interactions. The MacMillan Press Ltd, Houndmills, UK, Vol. 1, pp. 134–168. [Google Scholar]
- 96.Wozniak K. and Walter,Z. (2000) Induction of DNA–protein cross-links by platinum compounds. Z. Naturforsch. C, 55, 731–736. [DOI] [PubMed] [Google Scholar]
- 97.Chichiarelli S., Coppari,S., Turano,C., Eufemi,M., Altieri,F. and Ferraro,A. (2002) Immunoprecipitation of DNA–protein complexes cross-linked by cis-diamminedichloroplatinum. Anal. Biochem., 302, 224–229. [DOI] [PubMed] [Google Scholar]
- 98.Plooy A.C.M., Van Dijk,M. and Lohman,P.H.M. (1984) Induction and repair of DNA cross-links in Chinese hamster ovary cells treated with various platinum coordination compounds in relation to platinum binding to DNA, cytotoxicity, mutagenicity and antitumor activity. Cancer Res., 44, 2043–2051. [PubMed] [Google Scholar]
- 99.Kasparkova J., Marini,V., Najajreh,Y., Gibson,D. and Brabec,V. (2003) DNA binding mode of the cis and trans geometries of new antitumor non-classical platinum complexes containing piperidine, piperazine or 4-picoline ligand in cell-free media. Relations to their activity in cancer cell lines. Biochemistry, 42, 6321–6332. [DOI] [PubMed] [Google Scholar]
- 100.Zakovska A., Novakova,O., Balcarova,Z., Bierbach,U., Farrell,N. and Brabec,V. (1998) DNA interactions of antitumor trans-[PtCl2(NH3)(quinoline)]. Eur. J. Biochem., 254, 547–557. [DOI] [PubMed] [Google Scholar]
- 101.Brabec V., Neplechova,K., Kasparkova,J. and Farrell,N. (2000) Steric control of DNA interstrand cross-link sites of trans platinum complexes: specificity can be dictated by planar non-leaving groups. J. Biol. Inorg. Chem., 5, 364–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.