Abstract
Urinary tract infection (UTI) is the second most common infectious disease, and is caused predominantly by type 1-fimbriated uropathogenic E. coli (UPEC). UPEC initiates infection by attaching to uroplakin Ia, its urothelial surface receptor, via the FimH adhesins capping the distal end of its fimbriae. Uroplakin Ia, together with uroplakins Ib, II and IIIa, forms a 16 nm receptor complex that is assembled into hexagonally packed two-dimensional crystals (urothelial plaques) covering >90% of the urothelial apical surface. Recent studies indicate that FimH is the invasin of UPEC as its attachment to the urothelial surface can induce cellular signaling events including calcium elevation and the phosphorylation of the uroplakin IIIa cytoplasmic tail, leading to cytoskeletal rearrangements and bacterial invasion. However, it remains unknown how the binding of FimH to the uroplakin receptor triggers a signal that can be transmitted through the highly impermeable urothelial apical membrane. We show here by cryo-electron microscopy that FimH-binding to the extracellular domain of UPIa induces global conformational changes in the entire uroplakin receptor complex, including a coordinated movement of the tightly bundled transmembrane helices. This movement of the transmembrane helix bundles can cause a corresponding lateral translocation of the uroplakin cytoplasmic tails, which can be sufficient to trigger downstream signaling events. Our results suggest a novel pathogen-induced transmembrane signal transduction mechanism that plays a key role in the initial stages of UPEC invasion and receptor-mediated bacterial invasion in general.
Keywords: bacterial adhesin, tetraspanins, transmembrane signaling, urinary tract infections, uroplakins
INTRODUCTION
Bacterial infections of the urinary tract, caused predominantly by type 1-fimbriated uropathogenic E. coli (UPEC), are the second most common human bacterial infection.1-3 Initial binding of UPEC to the urothelium is mediated by the FimH adhesin, a lectin that caps the tip of the fimbria and binds with high affinity to mannose glycans exposed on the apical surface of the bladder.4 Over 90% of the luminal surface of the superficial umbrella cell layer of the bladder urothelium is covered by rigid-looking plaques (“urothelial plaques”), which are 0.3-1 μm in diameter and consist of two-dimensional crystals of hexagonally packed 16-nm particles5-8 composed of four integral membrane proteins, the uroplakins (UPs; reviewd in 9). We and others have shown that the 16-nm uroplakin particles have 6 inner and 6 outer structural subdomains,10-13 and that each of these subdomains contains a cylindrical 4-helix-bundled tetraspanin uroplakin (UP Ia for the inner subdomains and UP Ib for the outer ones) in association with a single-pass transmembrane uroplakin (UP II and UP IIIa, respectively).14,15 These 16-nm particles are packed hexagonally into 2D crystalline arrays that form the urothelial plaques, and contribute to the extraordinary permeability barrier function of the mammalian urothelium.15-19 The uroplakin complex is also a unique example of tetraspanin webs – membrane protein networks that play important signaling roles in a wide range of cells.20,21 Importantly, uroplakin Ia has been identified as the urothelial receptor for the FimH protein.22,23
FimH binding to the uroplakin receptor complex enables the bacterium to adhere to the urothelial surface, thus allowing it to establish a foothold for infection.24 Moreover, FimH plays a key role as the “invasin” that enables the UPEC to invade into the host cells, i.e., it is necessary and sufficient for triggering host cell signaling cascades leading to bacterial internalization by the so-called “zipper”-type invasion mechanism.25-27 UPEC invasion subsequently leads to the formation of intracellular bacterial communities in the superficial umbrella cells.28,29 Despite the critical roles of the uroplakin receptor complex in the establishment of UTI, the mechanism by which the bacterial binding-induced signal is transmitted through the lipid bilayer to initiate the downstream signaling cascade is unclear.
We speculated earlier that the binding of FimH to the uroplakin particle might induce the receptor complex to undergo conformational changes that can mediate transmembrane signal transduction.14,15 We report here a three-dimensional (3D) cryo-EM structure, at 8 Å resolution, of the mouse uroplakin particle in complex with the FimH adhesin. Comparison of this 3D structure with that of the native particle14 shows that FimH binding to UP Ia induces large conformational changes in the extracellular domains of the two single-pass transmembrane uroplakins, UPs II and IIIa, as well as a twisting movement of the transmembrane helices of all the uroplakin molecules of the 16-nm particle. The movements of these transmembrane helices can cause conformational changes in the cytoplasmic tail of UP IIIa, thus triggering the downstream signaling pathways and enabling bacterial invasion and host cell response.
RESULTS
FimH-binding induces major conformational changes in the extracellular domains of the uroplakin receptor complex
The structure of the 16-nm uroplakin particles bound to recombinant FimH (Fig. 1), in the form of a FimH/C complex, was determined by cryo-EM at 8 Å resolution (Table 1 and Fig. S1). The FimH/C complex was used as a ligand because FimC, a periplasmic chaperone, is required to stabilize the FimH adhesin.27,30-32 The FimH/C-bound 16-nm uroplakin particle retained the stellate-shape of the native particle,14,15 with six easily recognizable subunits, each consisting of an inner subdomain made of UPs Ia and II and an outer subdomain made of UPs Ib and IIIa (Figs. 2A and 2C). To compare the electron density maps of the FimH-bound particle with the previously-determined native one, we contoured their density maps to enclose similar volumes and segmented the maps into individual uroplakins.14
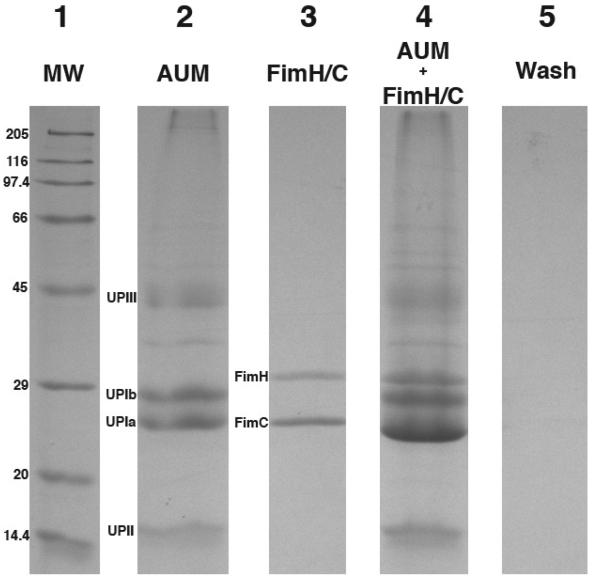
Figure 1. Gel electrophoresis (SDS-PAGE) analysis of FimH binding to mouse urothelial plaques.
Purified mouse urothelial plaques were incubated with FimH/C and washed excessively; the proteins in the complex were resolved by SDS-PAGE and stained with Coomassie blue. Lane 1: Molecular weight marks. Lane 2: Mouse urothelial plaques (AUM). The positions of the 4 uroplakins are indicated on the left. Lane 3: Recombinant FimH/C complex. Lane 4: Urothelial plaques incubated with FimH/C and washed 3 times with buffers showing FimH/C was complexed with urothelial plaques. Note that FimH is clearly visible while FimC co-migrates with the UP Ia band. Lane 5: Supernatant of the last wash.
Table 1.
Summary of the cryo-EM 3D data analysis
| Two-side plane group | P6 | |
| Unit cell dimension | a = b = 165 Å; γ = 120° | |
| IQ cut-off of data | ≤6 | |
| Number of observed amplitudes/phases | 16562 | |
| Overall R-factor | 34.5% | |
| Overall weighted R-factor | 34.3% | |
| Overall phase residual | 35.1° | |
| Overall weighted phase residual | 23.8° | |
| Phase residual in resolution zones | 21.8° (200-14 Å) | |
| 24.7° (14-10 Å) | ||
| 34.8° (10-8 Å) | ||
| In-plane resolution cutoff | 8.0 Å | |
| Vertical resolution cutoff | 19.2 Å | |
| Maximum tilt | 55° | |
| Number of images | 0° – 15° tilt | 19 |
| 15° – 25° tilt | 16 | |
| 25° – 35° tilt | 18 | |
| 35° – 55° tilt | 12 | |
| Total | 65 | |
Figure 2. Structures of native and FimH-bound mouse uroplakin particles at 8 Å resolution.
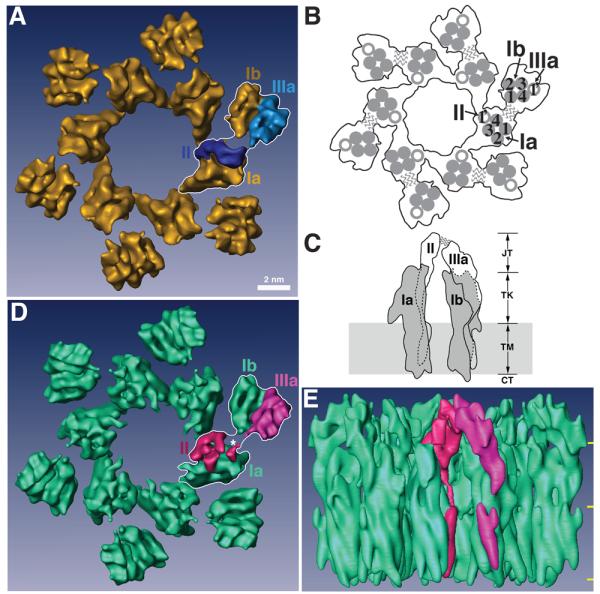
(A) Top view of the native 16-nm particle, presented as the reference structure. One of the 6 subunits is outlined by a white line stroke with its individual uroplakins labeled. Each subunit consists of an inner and an outer subdomain that contains the uroplakins pairs Ia/II and Ib/IIIa, respectively. Bar scale: 2 nm. This scale also applies to panels (D) and (E). (B) Schematic top view of the 16-nm particle showing the relative positions of the UP transmembrane helices. In one subunit, the transmembrane helices of the tetraspanin UPs Ia and Ib are numbered. The wavy lines indicate the weak connection between the joint regions of UPs II and IIIa that link the inner and outer subdomains within each subunit. (C) Schematic side view of a subunit of the 16-nm particle. The tetraspanin UPs Ia and Ib are shown in grey, while the single-pass UPs II and IIIa are shown in white. Dashed lines indicate the portions of the structures obscured by the molecules in front. The wavy lines again indicate the weak joint connection between the two subdomains. In the vertical dimension the particle can be divided into four regions, from top to bottom: the joint (JT), the trunk (TK), the transmembrane domain (TM), and the cytoplasmic tail (CT). Previous modeling depicted the UPs Ia and Ib as cylindrically shaped structures with a total height of ∼9 nm (CT+TM+TK).14,62 (D) Top view of the FimH-bound 16-nm particle. As in panel (A), one subunit is outlined by a white line stroke and its UPs II and IIIa are rendered in red and magenta, respectively. The asterisk points to the FimH binding site (see text). Note the considerable conformational differences in UPs II and IIIa when compared with the native structure in (A). (E) Side view of the FimH-bound 16-nm particle colored as in panel (D). The three horizontal yellow marks on the right demarcate the 4 regions explained in panel (C).
Major FimH-induced conformational changes could be readily detected in the top regions of the UPs II and IIIa extracellular domains – the so-called “joint” domains (“JT” in Fig. 2C) which bridge the inner and outer subdomains at the top of the uroplakin particle.14,15 In the native particle, the joint domain of UP II has an elongated globular shape connected by a long helical electron density anchored to the single transmembrane helix (Figs. 3A and 3C). After FimH binding to UP Ia, however, the globular joint domain of UP II opens up to form two smaller domains linked at their lower parts to form an approximately “Y” shaped structure (Fig. 3B), in which one of the arms swings ∼120° towards the opposite side (Fig. 3G, curved arrow). Thus, the joint domain of UP II undergoes a drastic change in both its shape and orientation (Figs. 3A-3G). The rotation of the UP II head resulting from FimH/UP Ia binding can be appreciated from Figures 2D and 3F. In addition to the changes in the joint domain, FimH binding also affected the conformation of the trunk domains of UP II (Fig. 3). In the unliganded native particle there is a discontinuity of density in the trunk regions that link the joint domains to the transmembrane helices14 of both uroplakins II and IIIa (Figs. 3A and 3H, dashed lines), indicating a greater degree of flexibility in these regions. However, in the FimH-bound structure of UP II, the joint and transmembrane regions are connected by an uninterrupted trunk region density (Figs. 3B and 3D), suggesting that this helical region has been stabilized by FimH binding.
Figure 3. Comparisons of the subdomain structures of native and FimH-bound uroplakin receptor complexes.
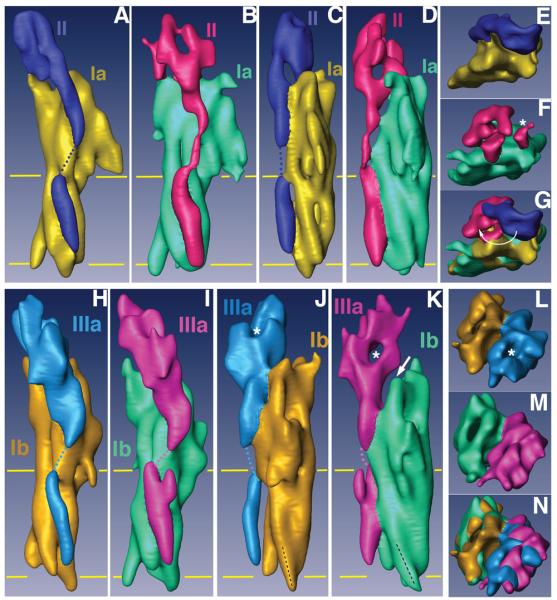
(A)-(G) represent different views of the UP Ia/II pair of the native and FimH-bound particles, and (H)-(N) the comparable views for UP Ib/IIIa pair. The individual uroplakins are segmented and labeled. The horizontal yellow lines mark the TM region. (A) and (B) Side views of the UP Ia/II pair in the inner subdomains of native (A) and FimH-bound (B) particles. (C) and (D) Comparison of the same UP Ia/II pair as in (A) and (B) but rotated ∼90°. Note the drastic FimH binding-induced shape change in the joint domain of UP II. Also note the connecting density in the TK of UP II (B and D) as a result of FimH-binding.14 (E) and (F) Top views of the UP Ia/II pair in the inner subdomains of native (E) and FimH-bound (F) particles. The asterisk in (F) indicates the FimH binding site of UP Ia. (G) Superimposed top views of the UP Ia/II structures shown in (E) and (F). Note the large swing of the joint domain of UP II indicated by a curved arrow. (H) and (I) Side views of the outer subdomain UP Ib/IIIa pair of the native (H) and FimH-bound (I) particles. (J) and (K) Comparison of the same UP Ib/IIIa pair as in (H) and (I) but rotated ∼90°. Note in (J) and (K) the orientation (black dashed line) of the cytoplasmic end of UP Ib helix 3 changed after FimH binding. Also note the separation (arrow in panel K) between UPs Ib and IIIa resulting from the FimH-induced conformational changes. (L) and (M) Top views of the outer subdomains of the native (L) and FimH-bound (M) particles. (N) Top view of the outer subdomains of the two structures superimposed.
Although less drastic than those observed for UP II, FimH binding also induced major conformational changes in UP III (Figs. 3H-3N). In its native structure, a density tunnel in the joint region of UP III runs approximately perpendicular to the plane of the membrane and is thus visible only from the top (Figs. 3J and 3L, asterisks). In the FimH-bound structure, however, this tunnel is re-oriented to run horizontally, thus becoming clearly visible from the side (Fig. 3K, asterisk). In addition, FimH binding forces the head of UP IIIa, i.e., its joint region, to lean away from its UP Ib partner, thus creating a separation between these two uroplakins (Fig. 3K, arrow). Interestingly, the large conformational changes of the joint domains of both UPs II and IIIa appeared to be centered on the FimH-binding site, which lies in a crevice between the two domains (Fig. 2D, asterisk). This suggests that the interactions between FimH and the uroplakins cause the receptor complex to open up its joint regions thus facilitating FimH binding to the mannose moiety in the shorter UP Ia receptor.
Although the affinity between FimH and uroplakins is sufficiently strong (Kd ∼ 100 nM)23 to allow a stable binding (Fig. 1), we did not observe densities that directly correspond to the whole FimH/C complex itself, an elongated mass with a total molecular weight of about 50-kDa.30 However, this is anticipated for the following reasons: (i) the high mannose moiety of UP Ia to which FimH binds is likely flexible, (ii) the sugars of the high mannose moiety of UP Ia do not have a unique number of branches and length35 and FimH only binds the terminal mannose, and (iii) the opening of the crevice that FimH has to reach in for binding the mannose is larger than the diameter of FimH. Hence, FimH may not occupy a crystallographically unique position when complexed with the 16-nm particle. In our previous molecular model of the native tetraspanin UP Ia,14 we predicted the location of its single glycosylation site. We now observed in the FimH-bound receptor complex some density at that site (Figs. 2D and 3F, asterisks; see also Fig. S2), possibly a residual density of the FimH/C complex after crystallographic average.33
FimH-binding induces movements of the transmembrane helices of the uroplakin receptor complex
To compare the transmembrane helices of the 4 uroplakins in native and FimH-bound 16-nm particles,14,15 we segmented the transmembrane helices by marking the center of the transmembrane helices in cross-sections of density maps parallel to the plane of the membrane (Fig. 4A). A similar method was used by Unwin and Ennis to detect conformational changes in the transmembrane helices of gap junctions induced by calcium ions.34 Superimposition of the segmented helices of the native and FimH-bound 16-nm particle showed that FimH-binding induced all the uroplakin transmembrane helices to move in a concerted fashion (Figs. 4B-4D). The top ends (near the luminal side) of the 5 helices of each subdomain underwent a clockwise rotation around the center of the subdomain (Fig. 4C). Since the swing of the UPII head was also clockwise (Fig. 3G, curved arrow), the rotation of the transmembrane helices most likely resulted from the conformational changes in the joint regions induced by FimH binding. Notably, the movement of the cytoplasmic ends of the helices differed from that of the top ends, in both direction and magnitude (Fig. 4D), indicating that a degree of twisting between helices takes place upon ligand binding (see also Fig. S3 in Supplementary Data). For example, the cytoplasmic end of transmembrane helix 3 of UP Ib tilted ∼10° more in the FimH-bound structure than that in the native structure (Figs. 3J and 3K, black dashed lines). A comparison of the cytoplasmic ends of the helices in the native and the FimH-bound uroplakins showed that FimH-binding causes displacements of up to ∼6 Å (Table 2).
Figure 4. FimH binding-induced movements of the transmembrane helices of uroplakins.
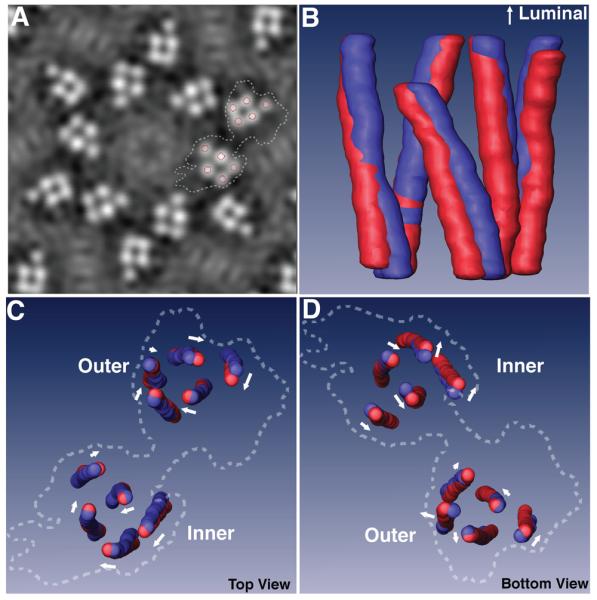
(A) A slice of the density map in the middle of the transmembrane domain of the FimH-bound 16-nm particle. Circles mark the center of the transmembrane helices in the segmentation process, and dashed lines outline a subunit. (B) Side view of the superimposed, segmented transmembrane helices of the inner subdomains of the native (blue) and FimH-bound (red) particles. (C) and (D) Superimpositions of the segmented transmembrane helices of the inner and outer subdomains viewed from the luminal (C) and the cytoplasmic (D) sides. The arrow near each helix approximates the direction and magnitude of the helix's movement induced by FimH-binding. Dashed lines again indicate the outlines of a whole subunit.
Table 2.
FimH-binding induced lateral translocations of the TM helices measured at their cytoplasmic ends (see Fig. 2B for the numbering of the helices).
| Subdomain | Inner | Outer | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Molecule | UP Ia | UP II | UP Ib | UPIIIa | ||||||
| Helix | 1 | 2 | 3 | 4 | 1′ | 1 | 2 | 3 | 4 | 1′ |
| Shift (Å) | 4.4 | 3.2 | 2.6 | 1.0 | 3.8 | 5.8 | 3.6 | 3.4 | 1.0 | 1.7 |
DISCUSSION
Our data showed that FimH binding to its UP Ia receptor induces major conformational changes in the extracellular domains as well as concerted movements of the transmembrane helices of all the uroplakins in the 16-nm uroplakin receptor complex (Figs. 2-4). The fact that FimH, which binds to the mannose moiety of UP Ia, can induce widespread conformational changes throughout the uroplakin complex is consistent with the unique architecture of the 16-nm particle. First, FimH has to reach into a crevice on the top of the uroplakin particle, because UP Ia, the only uroplakin harboring the high mannose moiety bound by FimH,23,35 is shorter than the single-pass UPs II and IIIa.14 The crevice formed by the two joints (connecting UPs II and IIIa between the inner and outer subdomains at the top) of the neighboring subunits of the 16 nm particle is ∼3 nm wide, large enough to accommodate the lectin domain (∼2.5 nm in diameter and 5 nm long) of the FimH molecule.14 Therefore the binding of FimH to UP Ia, located at the bottom of the crevice formed with the joint regions of UPs II and IIIa, favors the formation of protein-protein contacts between FimH with the UP II and IIIa joints. These contacts can then induce large conformational changes in the joint region. Second, we showed previously that the joint region is more flexible, indicated by the weaker electron density, than the rest of the 16-nm particle.14 Hence FimH binding can relatively easily induce conformational changes in this region (Fig. 3). Third, all uroplakins in the 16-nm particle are closely connected, thus enabling conformational changes in one region to be propagated to other regions.14 This high degree of connection occurs on 4 levels: (i) in each subdomain, the single-pass uroplakin is closely packed against the extracellular loop as well as the transmembrane helical bundle of its tetraspanin uroplakin partner; (ii) within each subunit, the inner domain is connected to the outer domain through the top joint; (iii) in the 16-nm particle, the 6 inner domains are interconnected through linkages at the juxtamembrane region;14 (iv) the 5 transmembrane helices of each subdomain form a tightly packed helix bundle;14 thus the helices within a subdomain have to move in a concerted manner. Collectively, these considerations suggest that FimH establishes its initial binding to the UP Ia receptor via its specific interaction with the mannose moiety, and then interacts with a broader area of the uroplakin protein complex. Such a secondary protein-protein interaction then induces global conformational changes in the entire 16-nm particle. Interestingly, we did not observe electron densities in the 3D construction that can be interpreted as the FimH/C complex used in our experiments, even though we have used enough FimH/C to saturate the binding sites on the urothelial plaques and the binding appears stoichiometric (Fig. 1). This suggests that FimH in the complex may be somewhat mobile and its density was averaged out.33
Recent data indicate that the uroplakin complex can transmit signals across the cell membrane. Klumpp and coworkers showed that FimH binding to the uroplakin receptor complex can induce the phosphorylation of a specific threonine residue located in the UP IIIa cytoplasmic tail, and that this binding is followed by an elevation of the intracellular calcium concentration.36 Pharmacological inhibition of these signaling events can block the invasion of the UPEC into the host urothelial cells. These data support the idea that the uroplakin receptor complex is a mediator of urothelial responses to UPEC attachment. In addition, uroplakins are known to play signaling roles in the fertilization of Xenopus eggs.37-39 Fertilization of Xenopus eggs requires the phosphorylation of a cytoplasmic tyrosine residue of UP IIIa by the egg tyrosine kinase Src, and can be blocked by antibodies to the extracellular domain of UPIIIa. These data indicate that uroplakins have multiple transmembrane signaling roles and regulate cellular functions in both mammals and amphibians.
Although the FimH-induced movements of the transmembrane helices of uroplakins are relatively small in comparison with the conformational changes of the extracellular domains, these movements may be sufficient to transmit signals recognizable by downstream molecules that interact with the cytoplasmic domains of the uroplakins. First, an excellent example is provided by G protein-coupled receptors that are known to be able to transmit signals via conformational changes of their transmembrane helices.40 Recent X-ray crystallographic studies have shown that ligand-induced conformational changes of the human β2-adrenergic receptor and A2A adenosine receptor include a movement of the transmembrane helices in the range of 2-7 Å.41,42 This is comparable with the magnitude of the FimH-induced movements of the uroplakin transmembrane helices, which amount to ∼6 Å at the cytoplasmic ends of these helices. Such a movement of the helices was shown to be sufficient to expose the G protein binding site on the cytoplasmic surface, thus allowing binding and the initiation of downstream signaling events. Second, the binding of FimH to the high-mannose glycans can exert a mechanical force of up to a hundred pico-Newtons,43,44 and it has been suggested that a force of this magnitude is strong enough to modify the interactions between the cytoplasmic tails of the transmembrane receptors complexes and the cytoskeleton.45 Finally, since the FimH/FimC complex alone, in the absence of other bacterial virulence factors, is sufficient to mediate bacterial uptake by the umbrella cells,27,46 the FimH-induced conformational changes in the uroplakin receptor complex most likely occur in vivo. In addition, EM studies have revealed that, after the bacteria attach to the host urothelial surface, their type 1 fimbriae undergo retraction.25,47 This retraction may further enhance the conformational changes of the uroplakin receptor complex beyond what we have observed in our cryo-EM study performed under static conditions.
The tetraspanin network nature of the urothelial plaques may further enhance the FimH binding-induced signal sensed by the host urothelial umbrella cells. UP Ia and UP Ib belong to the tetraspanin family of proteins that often form ‘tetraspanin-enriched microdomains’ (TEMs), or tetraspanin networks.48 TEMs are membrane domains similar to but distinct from lipid rafts; they serve as a signaling platform and play important roles in regulating cell migration, fusion and other signaling events.20,21,49 The urothelial plaque may be regarded as a unique TEM as thousands of 16-nm particles in each plaque aggregate into 2D membrane crystalline arrays.14,21 This crystalline network may enhance the propagation of the bacterial binding-signal to the cytoplasmic side through multiple cytoskeleton connections within each plaque. The almost complete coverage of the urothelial apical surface by the crystalline plaques may be particularly advantageous to UPEC, allowing this pathogen, which previously thought to be extracellular only, to become invasive in the urinary tract.50 UPEC is able to initiate invasion into the urothelial umbrella cells by the FimH-binding-induced conformational changes in its receptor complex because the structural features of the complex are particularly suitable to transmit this signal.
This is the first report to show that bacterial adhesin-binding can induce conformational changes in transmembrane domains of the host receptor leading to bacterial invasion using the zipper-mechanism. Bacterial pathogens invade the non-phagocytic host cells by two general mechanisms.25,26 In the much better understood trigger mechanism, bacteria such as Shigella and Salmonella use a specialized type III secretion system to inject bacterial proteins into the host cells in order to modulate the activity of the host cell's Rho GTPases, leading to the formation of actin-rich membrane ruffles promoting bacterial invasion. In the zipper mechanism, bacteria such as Listeria and Yersinia as well as UPEC invade the host cells by binding to specific receptors using adhesin molecules.25,51 This binding eventually leads to cytoskeletal rearrangements and bacterial internalization. However, there is a gap in our understanding of how the initial transmembrane signal in this receptor-mediated mechanism sets off the downstream signaling cascade leading to the cytoskeleton rearrangement.51 We showed here that the UPEC adhesin can induce conformational changes in the transmembrane domains of its uroplakin receptor complex, hence transmitting a signal to the cytoplasmic side. The urothelial plaque is known to be connected to the cytoskeleton,52 and the bacterial binding signal may propagate through these linkers to activate Rho-family GTPase pathways and cause cytoskeletal rearrangements and down stream events similar to those of the trigger mechanism.27,53 Hence, results from this study provide an improved understanding of how transmembrane signaling facilitates the early stages of the zipper mechanism of bacterial invasion.
MATERIALS AND METHODS
Preparation of mouse urothelial plaques and FimH binding to the plaques
Mouse urothelial plaques (AUM) were isolated by sucrose density gradient centrifugation and differential detergent wash;54,55 the quality of the purified urothelial plaques was assessed by negative staining and EM. FimH/C complex was expressed in E. coli HM 125 56 harboring pFimH-FimC and purified by ion-exchange chromatography as described.57 FimC, which does not affect the function of FimH, is the periplasmic chaperone required to stabilize FimH in its active mannose binding state.23,30-32,58 To form the FimH-bound 16-nm uroplakin particle, two times excess FimH/C was mixed with purified mouse AUM (0.1 mg/ml) and incubated on ice for 45 minutes to 1 hour. The formation of the FimH-uroplakin complexes was verified by pull-down assays and gel electrophoresis (Fig. 1). Judging from the intensities of the protein bands on the gels, FimH bound UP Ia in an approximately 1:1 stoichiometry. All experiments on animals received prior approval from the institutional animal care and use committee of New York University School of Medicine and conformed to the standards set by the appropriate regulatory agencies.
Cryo-EM Sample Preparation
Preparation of the frozen-hydrated samples for cryo-EM has been described previously.14 collecting un-tilted data, we used the back-injection method for grid preparation.59 Drops of ∼4.5μl of FimH-bound mouse urothelial plaques were absorbed onto newly prepared carbon films on a molybdenum grid (300 mesh) and transferred to a tannic acid solution (0.75%, pH 6.0). After the excess liquid was blotted with filter paper, the sample was quickly immersed into liquid nitrogen. For collecting tilted data, the carbon sandwich technique,60 for which the urothelial plaques were sandwiched between two sheets of carbon film, was adopted to prepare cryo-EM samples. The frozen samples were then loaded onto an Oxford cryo-holder and transferred to a Philips CM200 FEG electron microscope operated at 200 kV.
Image recording and processing
Data recorded from a tilted series of images of large urothelial plaques with a diameter up to ∼1 μm enabled us to reconstruct a 3D structure of the FimH-liganded particle at 8 Å resolution in the membrane plane and 19.2 Å in the vertical direction (Figs. 2, Table 1 and Fig. S1 in Supplementary Data). Electron Micrographs were taken at a magnification of 50,000× under low-dose mode and with 0.5-1.5 μm underfocus, at up to 55° tilt angles. Only one image was acquired from each plaque. The micrographs were selected using an optical diffractometer and digitized using a ZEISS SCAI scanner at a step of 14 μm, which corresponds to 2.8 Å in the crystal. A total of 65 images was selected from more than 1000 micrographs and processed using the MRC software suite.61 The 3D density maps were visualized and segmented using AMIRA (Mercury Computer System, Inc.). To compare the 3D map of the FimH-liganded 16-nm particle with that of the native one, we first contoured the two density maps, both calculated at 8 Å resolution, at compatible levels (1.2σ for the native and 1.47σ for the FimH bound maps, respectively) so that the electron density map-enclosed volumes are about the same for the two structures. We then segmented the two maps into individual uroplakins to compare the conformational changes in the two structures.14
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. George Georgiou (University of Texas at Austin) for the bacterial strain for overexpress FimH/C, Anchi Cheng for the program to calculate the resolution in the vertical direction of the 3D reconstruction. We thank Dr. David Klumpp for sharing a manuscript before publication, and Drs. Milton Adesnik, Gert Krebich and David Sabatini for critically reading the manuscript. This work was supported by National Institutes of Health grant DK52206 and the Schweizerische Nationalfonds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Litwin MS, Saigal CS, Yano EM, Avila C, Geschwind SA, Hanley JM, Joyce GF, Madison R, Pace J, Polich SM, Wang M. Urologic diseases in America Project: analytical methods and principal findings. J Urol. 2005;173:933–7. doi: 10.1097/01.ju.0000152365.43125.3b. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon. 2003;49:53–70. doi: 10.1067/mda.2003.7. [DOI] [PubMed] [Google Scholar]
- 3.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, Hultgren SJ. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–11. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 4.Abraham SN, Sun D, Dale JB, Beachey EH. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature. 1988;336:682–4. doi: 10.1038/336682a0. [DOI] [PubMed] [Google Scholar]
- 5.Kachar B, Liang F, Lins U, Ding M, Wu XR, Stoffler D, Aebi U, Sun TT. Three-dimensional analysis of the 16 nm urothelial plaque particle: luminal surface exposure, preferential head-to-head interaction, and hinge formation. J Mol Biol. 1999;285:595–608. doi: 10.1006/jmbi.1998.2304. [DOI] [PubMed] [Google Scholar]
- 6.Kreplak L, Wang H, Aebi U, Kong XP. Atomic force microscopy of Mammalian urothelial surface. J Mol Biol. 2007;374:365–73. doi: 10.1016/j.jmb.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks RM. The mammalian urinary bladder: an accommodating organ. Biol Rev Camb Philos Soc. 1975;50:215–46. doi: 10.1111/j.1469-185x.1975.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 8.Porter KR, Kenyon K, Badenhausen S. Specializations of the unit membrane. Protoplasma. 1967;63:262–74. [PubMed] [Google Scholar]
- 9.Sun TT. Altered phenotype of cultured urothelial and other stratified epithelial cells: implications for wound healing. Am J Physiol Renal Physiol. 2006;291:F9–21. doi: 10.1152/ajprenal.00035.2006. [DOI] [PubMed] [Google Scholar]
- 10.Hicks RM, Ketterer B. Hexagonal lattice of subunits in the thick luminal membrane of the rat urinary bladder. Nature. 1969;224:1304–5. doi: 10.1038/2241304a0. [DOI] [PubMed] [Google Scholar]
- 11.Vergara J, Longley W, Robertson JD. A hexagonal arrangement of subunits in membrane of mouse urinary bladder. J Mol Biol. 1969;46:593–6. doi: 10.1016/0022-2836(69)90200-9. [DOI] [PubMed] [Google Scholar]
- 12.Oostergetel GT, Keegstra W, Brisson A. Structure of the major membrane protein complex from urinary bladder epithelial cells by cryo-electron crystallography. J Mol Biol. 2001;314:245–52. doi: 10.1006/jmbi.2001.5128. [DOI] [PubMed] [Google Scholar]
- 13.Walz T, Haner M, Wu XR, Henn C, Engel A, Sun TT, Aebi U. Towards the molecular architecture of the asymmetric unit membrane of the mammalian urinary bladder epithelium: a closed “twisted ribbon” structure. J Mol Biol. 1995;248:887–900. doi: 10.1006/jmbi.1995.0269. [DOI] [PubMed] [Google Scholar]
- 14.Min G, Wang H, Sun TT, Kong XP. Structural basis for tetraspanin functions as revealed by the cryo-EM structure of uroplakin complexes at 6-A resolution. J Cell Biol. 2006;173:975–83. doi: 10.1083/jcb.200602086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min G, Zhou G, Schapira M, Sun TT, Kong XP. Structural basis of urothelial permeability barrier function as revealed by Cryo-EM studies of the 16 nm uroplakin particle. J Cell Sci. 2003;116:4087–94. doi: 10.1242/jcs.00811. [DOI] [PubMed] [Google Scholar]
- 16.Hu P, Deng FM, Liang FX, Hu CM, Auerbach A, Shapiro E, Wu XR, Kachar B, Sun TT. Ablation of uroplakin III gene results in small urothelial plaques, urothelial leakage, and vesicoureteral reflux. Urology. 2001;57:117. doi: 10.1016/s0090-4295(01)01062-7. [DOI] [PubMed] [Google Scholar]
- 17.Hu P, Meyers S, Liang FX, Deng FM, Kachar B, Zeidel ML, Sun TT. Role of membrane proteins in permeability barrier function: uroplakin ablation elevates urothelial permeability. Am J Physiol Renal Physiol. 2002;283:F1200–7. doi: 10.1152/ajprenal.00043.2002. [DOI] [PubMed] [Google Scholar]
- 18.Chang A, Hammond TG, Sun TT, Zeidel ML. Permeability properties of the mammalian bladder apical membrane. Am J Physiol. 1994;267:C1483–92. doi: 10.1152/ajpcell.1994.267.5.C1483. [DOI] [PubMed] [Google Scholar]
- 19.Negrete HO, Lavelle JP, Berg J, Lewis SA, Zeidel ML. Permeability properties of the intact mammalian bladder epithelium. Am J Physiol. 1996;271:F886–94. doi: 10.1152/ajprenal.1996.271.4.F886. [DOI] [PubMed] [Google Scholar]
- 20.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–48. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 21.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–11. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 22.Wu XR, Sun TT, Medina JJ. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc Natl Acad Sci U S A. 1996;93:9630–5. doi: 10.1073/pnas.93.18.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J Cell Sci. 2001;114:4095–103. doi: 10.1242/jcs.114.22.4095. [DOI] [PubMed] [Google Scholar]
- 24.Beachey EH. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J Infect Dis. 1981;143:325–45. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- 25.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–7. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 26.Finlay BB, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–25. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 27.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. Embo J. 2000;19:2803–12. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–7. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 29.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A. 2004;101:1333–8. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–6. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 31.Pellecchia M, Sebbel P, Hermanns U, Wuthrich K, Glockshuber R. Pilus chaperone FimC-adhesin FimH interactions mapped by TROSY-NMR. Nat Struct Biol. 1999;6:336–9. doi: 10.1038/7573. [DOI] [PubMed] [Google Scholar]
- 32.Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J Biol Chem. 2007;282:23437–46. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- 33.Buhle EL, Jr., Aebi U. Specific labeling of protein domains with antibody fragments. J Ultrastruct Res. 1984;89:165–78. doi: 10.1016/s0022-5320(84)80012-x. [DOI] [PubMed] [Google Scholar]
- 34.Unwin PN, Ennis PD. Two configurations of a channel-forming membrane protein. Nature. 1984;307:609–13. doi: 10.1038/307609a0. [DOI] [PubMed] [Google Scholar]
- 35.Xie B, Zhou G, Chan SY, Shapiro E, Kong XP, Wu XR, Sun TT, Costello CE. Distinct glycan structures of uroplakins Ia and Ib: structural basis for the selective binding of FimH adhesin to uroplakin Ia. J Biol Chem. 2006;281:14644–53. doi: 10.1074/jbc.M600877200. [DOI] [PubMed] [Google Scholar]
- 36.Thumbikat P, Berry RE, Zhou G, Billips BK, Yaggie RE, Zaichuk T, Sun TT, Schaeffer AJ, Klumpp DJ. Bacteria-induced uroplakin signaling mediates bladder response to infection. PLoS Pathog. 2009;5:e1000415. doi: 10.1371/journal.ppat.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahbub Hasan AK, Ou Z, Sakakibara K, Hirahara S, Iwasaki T, Sato K, Fukami Y. Characterization of Xenopus egg membrane microdomains containing uroplakin Ib/III complex: roles of their molecular interactions for subcellular localization and signal transduction. Genes Cells. 2007;12:251–67. doi: 10.1111/j.1365-2443.2007.01048.x. [DOI] [PubMed] [Google Scholar]
- 38.Mahbub Hasan AK, Sato K, Sakakibara K, Ou Z, Iwasaki T, Ueda Y, Fukami Y. Uroplakin III, a novel Src substrate in Xenopus egg rafts, is a target for sperm protease essential for fertilization. Dev Biol. 2005;286:483–92. doi: 10.1016/j.ydbio.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 39.Sakakibara K, Sato KI, Yoshino KI, Oshiro N, Hirahara S, Hasan AK, Iwasaki T, Ueda Y, Iwao Y, Yonezawa K, Fukami Y. Molecular identification and characterization of Xenopus egg uroplakin III, an egg raft-associated transmembrane protein that is tyrosine-phosphorylated upon fertilization. J Biol Chem. 2005 doi: 10.1074/jbc.M410538200. [DOI] [PubMed] [Google Scholar]
- 40.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 41.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang MN, Smith SP, Metallo SJ, Choi IS, Prentiss M, Whitesides GM. Measuring the forces involved in polyvalent adhesion of uropathogenic Escherichia coli to mannose-presenting surfaces. Proc Natl Acad Sci U S A. 2000;97:13092–6. doi: 10.1073/pnas.230451697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas W. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008 doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 46.Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc Natl Acad Sci U S A. 2000;97:8829–35. doi: 10.1073/pnas.97.16.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Liang FX, Kong XP. Characteristics of the phagocytic cup induced by uropathogenic Escherichia coli. J Histochem Cytochem. 2008;56:597–604. doi: 10.1369/jhc.2008.950923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 49.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 50.Bower JM, Eto DS, Mulvey MA. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic. 2005;6:18–31. doi: 10.1111/j.1600-0854.2004.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–27. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Staehelin LA, Chlapowski FJ, Bonneville MA. Lumenal plasma membrane of the urinary bladder. I. Three-dimensional reconstruction from freeze-etch images. J Cell Biol. 1972;53:73–91. doi: 10.1083/jcb.53.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez JJ, Hultgren SJ. Requirement of Rho-family GTPases in the invasion of Type 1-piliated uropathogenic Escherichia coli. Cell Microbiol. 2002;4:19–28. doi: 10.1046/j.1462-5822.2002.00166.x. [DOI] [PubMed] [Google Scholar]
- 54.Wu XR, Manabe M, Yu J, Sun TT. Large scale purification and immunolocalization of bovine uroplakins I, II, and III. Molecular markers of urothelial differentiation. J Biol Chem. 1990;265:19170–9. [PubMed] [Google Scholar]
- 55.Liang F, Kachar B, Ding M, Zhai Z, Wu XR, Sun TT. Urothelial hinge as a highly specialized membrane: detergent-insolubility, urohingin association, and in vitro formation. Differentiation. 1999;65:59–69. doi: 10.1046/j.1432-0436.1999.6510059.x. [DOI] [PubMed] [Google Scholar]
- 56.Meerman HJ, Georgiou G. Construction and characterization of a set of E. coli strains deficient in all known loci affecting the proteolytic stability of secreted recombinant proteins. Biotechnology (N Y) 1994;12:1107–10. doi: 10.1038/nbt1194-1107. [DOI] [PubMed] [Google Scholar]
- 57.Vetsch M, Sebbel P, Glockshuber R. Chaperone-independent folding of type 1 pilus domains. J Mol Biol. 2002;322:827–40. doi: 10.1016/s0022-2836(02)00845-8. [DOI] [PubMed] [Google Scholar]
- 58.Jones CH, Pinkner JS, Nicholes AV, Slonim LN, Abraham SN, Hultgren SJ. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci U S A. 1993;90:8397–401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang DN, Kuhlbrandt W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J Mol Biol. 1991;217:691–9. doi: 10.1016/0022-2836(91)90526-c. [DOI] [PubMed] [Google Scholar]
- 60.Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y. Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J Struct Biol. 2004;146:325–33. doi: 10.1016/j.jsb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 62.Seigneuret M. Complete Predicted Three-Dimensional Structure of the Facilitator Transmembrane Protein and Hepatitis C Virus Receptor CD81: Conserved and Variable Structural Domains in the Tetraspanin Superfamily. Biophys J. 2006;90:212–27. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.