Summary
Glucose-6-phosphatase-α (G6Pase-α or G6PC) catalyzes the hydrolysis of glucose-6-phosphate to glucose and is a key enzyme in interprandial glucose homeostasis. Mutations in the human G6PC gene, expressed primarily in the liver, kidney, and intestine, cause glycogen storage disease type Ia (GSD-Ia), an autosomal recessive disorder characterized by a disturbed glucose homeostasis. For better understanding of the roles of G6Pase-α in different tissues and in pathological conditions, we have generated mice harboring a conditional null allele for G6pc by flanking exon 3 of the G6pc gene with loxP sites. We confirmed the null phenotype by using the EIIa-Cre transgenic approach to generate mice lacking exon 3 of the G6pc gene. The resulting homozygous Cre-recombined null mice manifest a phenotype mimicking G6Pase-α-deficient mice and human GSD-Ia patients. This G6pc conditional null allele will be valuable to examine the consequence of tissue-specific G6Pase-α deficiency and the mechanisms of long-term complications in GSD-Ia.
Introduction
Glucose-6-phosphatase-α (G6Pase-α also known as G6PC) is a key enzyme in glucose homeostasis catalyzing the hydrolysis of glucose-6-phosphate to glucose and phosphate in the terminal steps in gluconeogenesis and glycogenolysis (Chou et al., 2002). The expression of G6Pase-α is tissue-specific, restricted primarily to the liver, kidney, and intestine (Pan et al., 1998). Deleterious mutations in the G6PC gene causes glycogen storage disease type Ia (GSD-Ia), an autosomal recessive metabolic disorder characterized by hypoglycemia, hepatomegaly, nephromegaly, hyperlipidemia, hyperuricemia, lactic acidemia, and growth retardation (Chou et al., 2002). Currently, there is no cure for GSD-Ia, but many of the disease symptoms can be managed or improved using dietary therapies (Greene et al., 1976; Chen et al., 1984) that maintain normoglycemia. This strategy enables patients to attain near normal growth and pubertal development, with fewer complications as they age. Despite intensive dietary therapy, the long-term complications of GSD-Ia - renal disease and hepatocellular adenomas with risk for malignant transformation - remain (Chou et al., 2002). Moreover, GSD-Ia patients suffer from intermittent diarrhea that worsens with age (Visser et al., 2002). Therefore, the metabolic disruption by G6Pase-α deficiency has tissue-specific consequences. We have previously generated a null allele for G6Pase-α (G6pc−/−) by knocking out exon 3 and shown that the G6pc−/−mice manifest a phenotype virtually identical to that of human GSD-Ia patients (Lei et al., 1996). Despite administering a glucose therapy to prevent hypoglycemic seizures, the G6pc−/− mice rarely live to 3 months of age. Consequently, we have been unable to study the molecular mechanisms leading to long-term complications of GSD-Ia. While liver and/or kidney transplantation have been proposed for the treatment of late stage liver and renal disease in GSD-Ia (Matern et al., 1999; Labrune, 2002), their effectiveness is not known. To understand whether such transplants are viable, and whether single or double transplant is necessary, we need to understand the importance of G6Pase-α expression in the liver and kidney, individually, and whether wild-type G6Pase-α expression at one site is sufficient to compensate for lack of expression at the other.
To address the role of G6Pase-α in maintaining normal liver, kidney, and intestinal functions, we report here the generation of mice harboring a conditional null allele for G6pc using DNA homologous recombination in embryonic stem cells and the Cre-loxP and FLPe-Frt strategies. These G6pc conditional null mice should facilitate the study of the individual consequence of G6Pase-α deficiency in the liver and kidney, and help clarify whether a deficiency in one can affect the other. The conditional mice could also be used to study the consequence of G6Pase-α deficiency in the intestine, an organ also capable of endogenous glucose production (Croset et al., 2001).
Generation of a Conditional Allele of G6pc
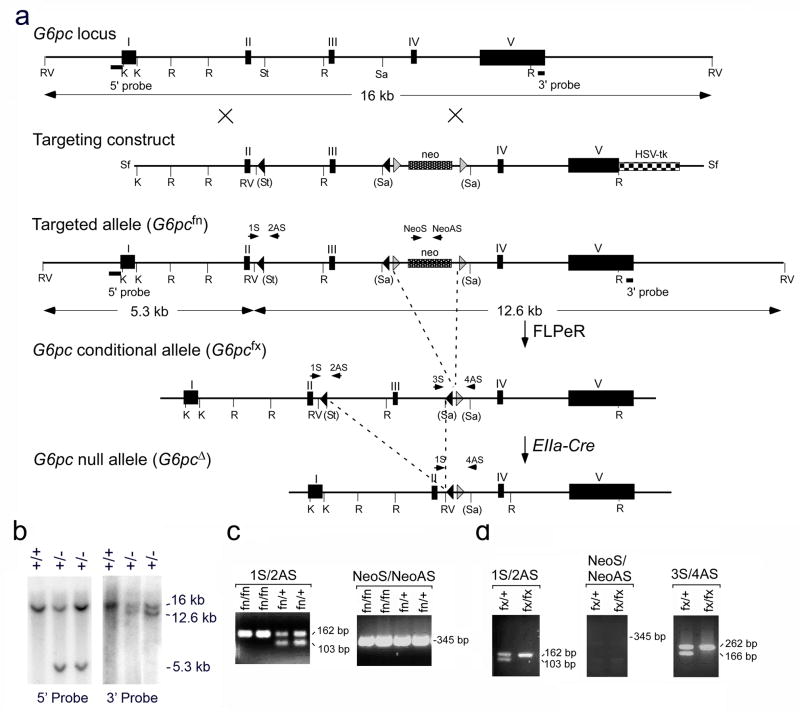
The murine G6pc gene consists of 5 exons spanning approximately 12 kb of mouse genomic DNA. To generate a conditional allele for G6pc, a targeting vector was constructed using plasmid pPNT4 which contains the neo gene flanked by Frt sites and one loxP site at its 5′ region, and the HSV-tk gene (Conrad et al., 2003). The neo gene provides a positive selection for insertion of the targeting sequences in ES cells in the presence of G418, and the HSV-tk gene for negative selection in the presence of gancyclovir. The neo gene, along with the flanking Frt and loxP sites, was inserted at the SacII site within intron 3 of the G6pc gene. An additional DNA sequence containing a loxP site and three diagnostic restriction sites (AvaI, SspI, and EcoRV) was inserted at the StuI site within intron 2 of the G6pc gene. The resulting targeting vector contained loxP sites to flox the G6pc exon 3, and Frt sites to flank the neo gene (Fig. 1a). The targeting vector was introduced into J1 ES cells and the cells, subjected to selection with the drugs G418 and gancyclovir. Homologous recombinants were identified by Southern hybridization of EcoRV digested genomic DNA using two probes corresponding to specific 5′ or 3′ fragment located outside of the targeted region (Fig. 1a and 1b). One of the G6pc-targeted ES clones was used for the production of chimeric animals. The chimeric mice were then mated with C57BL/6 mice and germline transmission of the G6pc targeted allele in F1 heterozygous (fn/+; fn denotes floxP-neo) mice was confirmed by PCR analysis (Fig. 1c). The fn/+ mice were mated and homozygous (fn/fn) progenies were identified by PCR analysis (Fig. 1c). The G6pcfn/fn mice were phenotypically indistinguishable from the wild-type mice.
FIG. 1.
Conditional disruption of the murine G6pc gene. (a) The G6pc locus is presented along with the targeting construct, the targeted allele (G6pcfn), the G6pc conditional allele (G6pcfx) and the anticipated outcome of the G6pc null allele (G6pcΔ). Introns are denoted as lines, exons as boxes. K, KpnI; R, EcoRI; RV, EcoRV; St. StuI; Sa, SacII; Sf, SfiI. The PCR primers used to verify the conditional alleles are denoted by arrows. (b) Southern-blot analysis of EcoRV digested mouse genomic DNA from wild-type (+/+) and targeted ES (+/−) clones using 5′ and 3′ external probes. The wild-type locus hybridizes with either probe to give a 16 kb band while the disrupted locus hybridizes with the 5′ probe to give a 5.3 kb band, and with the 3′ probe to give a 12.6 kb band. (c) PCR analysis of genomic DNA of the targeted allele (G6pcfn). The primer pair, 1S/2AS is expected to amplify a fragment of 103 bp in wild-type alleles and a fragment of 162 bp in the mutant allele. The primer pair Neo1/NeoAs is expected to amplify a fragment of 345 bp in the neo gene of the targeted allele. (d) PCR analysis of genomic DNA of the G6pc conditional allele (G6pcfx). The primer pairs, 1S/2AS and 3S/4AS are expected to amplify fragments of 103 and 166 bp, respectively, in wild-type alleles and fragments of 162 and 262 bp, respectively, in the mutant allele. Deletion of the neo gene is confirmed by the absence of a 345 bp fragment predicted to amplify the neo gene by the primer pair NeoS/NeoAS.
The Frt-flanked neo gene was deleted in vivo by breeding the G6pcfn/fn mice with the FLPeR mice which express the enhanced version of the site-specific recombinase FLPe ubiquitously (Farley et al., 2000). The resulting G6pcfx/+ (fx denotes floxP with neo-deleted) conditional mice which have loxP sites flanking G6pc exon 3 and one Frt site in the G6pc allele were confirmed by PCR analysis (Fig. 1d). The G6pcfx/+ mice were mated and the resulting homozygous G6pcfx/fx conditional mice were viable and fertile, and they did not display any recognizable phenotype. To demonstrate that the loxP flanked G6pc exon 3 could be deleted in vivo, the G6pcfx/fx mice were crossed with the EIIa-Cre transgenic mice, where Cre is expressed under the control of the adenoviral EIIa promoter that targets expression of the Cre recombinase during preimplanation development (Lakso et al., 1996). The exon 3- and neo-deleted (G6pcΔ/Δ) mice were confirmed by PCR analysis. There were no detectable G6Pase enzymatic activity in the liver and kidney of the G6pcΔ/Δ mice (data not shown), confirming that the targeted mutation in exon 3 had resulted in a null G6pc locus in vivo.
The G6pcΔ/Δ Mice Manifest Disturbed Glucose Homoestasis
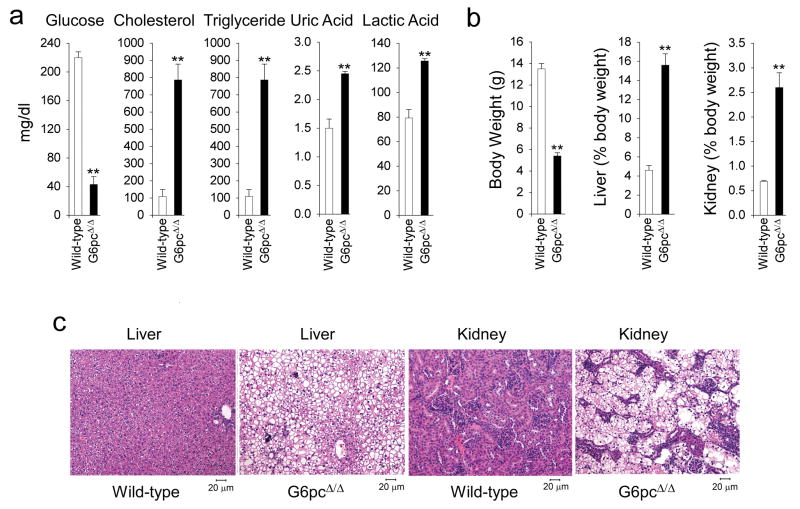
GSD-Ia patients (Chou et al., 2002) and G6pc−/− mice (Lei et al., 1996) manifest a phenotype of disturbed glucose homeostasis characterized by hypoglycemia, hepatomegaly, nephromegaly, hypercholesterolemia, hypertriglyceridemia, hyperuricemia, lactic acidemia, and growth retardation. The G6pcΔ/Δ mice exhibit similar characteristics. Metabolic analysis reveals a fasting hypoglycemia typical of GSD-Ia. The mean serum glucose level in G6pcΔ/Δ mice (43 ± 11 mg/dl) was significantly lower than those in wild-type (220 ± 8 mg/dl) littermates (Fig. 2a). The G6pcΔ/Δ mice suffered from hypoglycemic seizures requiring glucose therapy, like the G6pc−/−mice (Lei et al., 1996). Additionally, serum cholesterol, triglyceride, uric acid, and lactic acid levels in G6pcΔ/Δ mice averaged 7.2-, 15.9-, 1.6- and 1.6-fold higher, respectively, than that in the control littermates (Fig. 2a), like the G6pc−/− mice (Lei et al., 1996). The G6pcΔ/Δ mice are growth retarded, and exhibit hepatomegaly and nephromegaly (Fig. 2b) similar to that observed in GSD-Ia patients (Chou et al., 2002) and G6pc−/− mice (Lei et al., 1996). The average liver and kidney weights of 3-week-old G6pcΔ/Δ mice were elevated to 15.6% and 2.6% of body weight, respectively, compared to the wild-type value of 4.6% and 0.7%. Hematoxylin-eosin (H&E) staining revealed glycogen accumulation in the hepatocytes of the liver and tubular epithelial cells of the kidney in G6pcΔ/Δ mice (Fig. 2c), like the G6pc−/− mice (Lei et al., 1996).
FIG. 2.
The G6pcΔ/Δ mice manifest a phenotype of disturbed glucose homeostasis. Metabolic functions were analyzed in 3-week-old G6pcΔ/Δ and wild-type mice. (a) Serum glucose, cholesterol, triglyceride, uric acid, and lactic acid levels. Data are presented as mean ± SEM. **p < 0.005. (b) The body weight and the weights of the liver and kidney relative to total body weight. Data are presented as mean ± SEM. **p < 0.005. (c) H&E stained liver and kidney sections from G6pcΔ/Δ and wild-type mice with magnifications of ×200.
In conclusion, we have generated a conditional null G6pc allele and shown that homologous exon 3 deletion leads to a phenotype of disturbed glucose homeostasis mimicking that of G6pc−/−mice and human GSD-Ia patients. The mice carrying the floxed G6pc allele are now excellent candidates to study tissue-specific role of the G6pc gene and the mechanism of long term complications in GSD-Ia.
METHODS
Gene Targeting and Generation of a G6pc-Floxed Allele
A bacterial artificial chromosome (BAC) containing the murine G6pc gene was obtained by screening a BAC library in the 129/svJ mouse strain (Invitrogen, CA) with a G6pc cDNA probe. The targeting construct was generated in two steps from the neo- and HSV-tk-containing plasmid pPNT4 where the neo gene is flanked by Frt sites and one loxP site at its 5′ region (Conrad et al., 2003). First, the 3.5-kb SacII-EcoRI fragment of the murine G6pc gene containing 3′ region of intron 3, intron 4, exon 4 and 5′ region of exon 5 was inserted into the KpnI-EcoRI sites of pPNT4 downstream of the neo gene, to create the construct pPNT4-G6pc-SE. In a second step, the 5.8-kb G6pc KpnI-SacII fragment containing exons 2, 3, introns 1, 2, and 5′ region of intron 3, including an additional loxP site plus three diagnostic restriction sites, AvaI, SspI, and EcoRV at the StuI site within intron 2, was inserted into SalI and NotI sites of pPNT4-G6pc-SE plasmid upstream of the neo gene, yielding the targeting construct. The final targeting construct contained a neo gene flanked by two Frt sites, and exon 3 of the G6pc gene flanked by two loxP sites (fig. 1a).
The targeting-construct, linearized at the SfiI site, was introduced into J1 ES cells by electroporation and the cells subjected to selection with G418 and gancyclovir (Mansour et al., 1988). Homologous recombinants were identified by Southern blot hybridization of EcoRV digested genomic DNA using two probes corresponding to specific 5′ or 3′ fragment located outside of the targeting region. Twenty-one correctly targeted clones were obtained after screening 318 G418 resistant clones. The G6pc-targeted ES cells were microinjected into blastocysts from C57BL/6 mice for the production of chimeric embryos, which then was implanted into the uterine horn of pseudopregnant FVB/N foster mice for the production of chimeric animals. The chimeric mice were mated with C57BL/6 mice and germline transmission of the exon 3-floxed G6pc targeted allele in F1 heterozygous (G6pcfn/+) mice was confirmed by PCR analysis. The Frt-flanked neo gene was deleted in vivo by breeding the homozygous G6pcfn/fn mice with the FLPeR mice (Farley et al., 2000). Exon 3 of the G6pc gene was deleted by crossing the G6pcfx/fx mice with the EIIa-Cre transgenic mice (Lakso et al., 1996).
PCR, Genotype, and Phenotype Analyses
The introduced loxP site in intron 2 was identified using a primer pair, 1S (5′-CGGGCAATGGCTAGTTAGAG-3′) and 2AS (5′-CAGGCTGCTAGGAAGGACAC-3′), expected to amplify a fragment of 103 bp in wild-type alleles and a fragment of 162 bp in the mutant allele. The FLPe allele was identified using the primer pair, FS (5′-CCCATTCCATGCGGG GTATCG -3′) and FAS (5′-GCATCTGGG AGATCACTGAG -3′), expected to amplify a fragment of ~700 bp. A neo-specific primer pair, neoS (5′-GATCGGCCATTGAACAAGAT-3′) and neoAS (5′-ATACTTTCTCGGCAGGAGCA-3′) is expected to amplify a fragment of 345 bp in the neo gene of the targeted allele. The FLPeR-mediated deletion of the neo cassette was identified using the primer pair, 3S (5′-CAGTCGGGTGCTCTTACCTG-3′) and 4AS (5′-AGCCTGTTCTCTCTGCTTGG-3′), expected to amplify a fragment of 166 bp in wild-type alleles and a fragment of 262 bp in the neo-deleted mutant allele. The primer pair 1S/4AS, expected to amplify a fragment of 289 bp, was used to identify EIIa-Cre-mediated deletion of floxed exon 3.
Serum glucose, total cholesterol, and uric acid were analyzed using kits obtained from Thermo Electron (Louisville, CO), triglycerides using a kit from Sigma Diagnostics (St Louis, MO), and lactic acid using a kit from Trinity Biotech (St. Louis, MO). For H&E staining, tissues were preserved in 10% neutral buffered formalin, embedded in paraffin, and sectioned at 4–6 micron thickness.
Acknowledgments
We thank Dr. M. Conrad for gift of pPNT4 plasmid, Dr. S. M. Dymecki for the gift of FLPeR mice, and Syntex Research for the gift of gancylovir. This research was supported by the Intramural Research Program of the NICHD, NIH.
LITERATURE CITED
- Chen YT, Cornblath M, Sidbury JB. Cornstarch therapy in type I glycogen storage disease. N Engl J Med. 1984;310:171–175. doi: 10.1056/NEJM198401193100306. [DOI] [PubMed] [Google Scholar]
- Chou JY, Matern D, Mansfield BC, Chen YT. Type I glycogen storage diseases: disorders of the glucose-6-phosphatase complex. Curr Mol Med. 2002;2:121–143. doi: 10.2174/1566524024605798. [DOI] [PubMed] [Google Scholar]
- Conrad M, Brielmeier M, Wurst W, Bornkamm GW. Optimized vector for conditional gene targeting in mouse embryonic stem cells. Biotechniques. 2003;34:1136–1140. doi: 10.2144/03346bm03. [DOI] [PubMed] [Google Scholar]
- Croset M, Rajas F, Zitoun C, Hurot JM, Montano S, Mithieux G. Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes. 2001;50:740–746. doi: 10.2337/diabetes.50.4.740. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Greene HL, Slonim AE, O’Neill JA, Jr, Burr IM. Continuous nocturnal intragastric feeding for management of type 1 glycogen-storage disease. N Engl J Med. 1976;294:423–425. doi: 10.1056/NEJM197602192940805. [DOI] [PubMed] [Google Scholar]
- Labrune P. Glycogen storage disease type I: indications for liver and/or kidney transplantation. Eur J Pediatr. 2002;161 (Suppl 1):S53–S55. doi: 10.1007/s00431-002-1004-y. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei KJ, Chen H, Pan CJ, Ward JM, Mosinger B, Lee EJ, Westphal H, Mansfield BC, Chou JY. Glucose-6-phosphatase dependent substrate transport in the glycogen storage disease type 1a mouse. Nat Genet. 1996;13:203–209. doi: 10.1038/ng0696-203. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Matern D, Starzl TE, Arnaout W, Barnard J, Bynon JS, Dhawan A, Emond J, Haagsma EB, Hug G, Lachaux A, Smit GP, Chen YT. Liver transplantation for glycogen storage disease types I, III, and IV. Eur J Pediatr. 1999;158:S43–S48. doi: 10.1007/pl00014320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C-J, Lei K-J, Chen H, Ward JM, Chou JY. Ontogeny of the murine glucose-6-phosphatase system. Arch Biochem Biophy. 1998;358:17–24. doi: 10.1006/abbi.1998.0849. [DOI] [PubMed] [Google Scholar]
- Visser G, Rake JP, Kokke FT, Nikkels PG, Sauer PJ, Smit GP. Intestinal function in glycogen storage disease type I. J Inherit Metab Dis. 2002;25:261–267. doi: 10.1023/a:1016572706488. [DOI] [PubMed] [Google Scholar]