Abstract
The accumulation of cellular damage is a feature common to all aging cells and leads to a decreased ability of the organism to survive. The overall rate at which damage accumulates is influenced by conserved metabolic factors (longevity pathways and regulatory proteins), which control life span through adjusting mechanisms for maintenance and repair. Autophagy, the major catabolic process of eukaryotic cells, which has the capacity to degrade and recycle damaged macromolecules and organelles, is implicated in aging and in the incidence of diverse age-related pathologies. Recent evidence shows further that autophagic activity is required for life span extension in various long-lived mutant organisms, and that numerous autophagy-related genes or proteins serve as targets that longevity pathways regulate directly. These findings support the emerging view that autophagy acts as a central regulatory mechanism of aging in divergent eukaryotic species.
Implicating autophagy in the control of aging
In the 21st century, one of the major challenges human societies face is the aging of the population and the associated medical, social and economic implications. In several developed countries, life expectancy exceeds 80 years, and is projected to continue to increase during the next half century [1]. As the ratio of elderly people gradually increases, it becomes more important to understand the biological bases (i.e., mechanism and regulation) of aging and the molecular mechanisms that underlie various age-related pathologies. This knowledge may lead to future genetic and pharmacological interventions delaying many aspects of decline that occurs at advanced ages, thereby extending the number of active, healthy years that one can live.
Aging in diverse eukaryotic organisms is driven by the progressive accumulation of damaged macromolecules and organelles [1, 2]. Such types of damage appear as oxidized, misfolded, cross-linked or aggregated macromolecules, which have abnormal structure and, therefore, cannot function properly. In addition, damaged macromolecules and organelles, or their aggregates, can actively compromise cellular function, presumably by interfering with signaling pathways, acting as sinks for binding proteins or sending erroneous signals. Hence, the need to eliminate them is essential for the cells to operate. Cellular damage is generated continuously during the entire life of an organism. For example, ionizing radiation of biological molecules, endogenous factors including specific enzymes, and mitochondria produce reactive oxygen species (ROS), which in turn oxidize macromolecules, leading to their abnormal function [3]. The lifelong accumulation of aberrant cellular constituents leads to a decreased ability of the cells, and ultimately of the organism, to survive. Thus, aging is a natural decline in the fitness of an organism over time as the result of accumulation of unrepaired damage [2]. Cellular damage is also an important contributory factor in diverse age-related pathologies, including cancer, neurodegeneration, infection and muscle atrophy [1]. Oxidation of DNA, for instance, results in double- or single-strand breaks, the repair of which can lead to mutation causing cancer, whereas oxidized proteins tend to form toxic aggregates that are the primary cause of several age-related neurodegenerative diseases [4]. It is therefore not surprising that these pathologies acting as the major causes of human mortality generally occur at advanced age. The fact that the cells of aged organisms are particularly prone to undergo uncontrolled proliferation or premature death suggests a considerable overlap between the underlying mechanisms of these events and aging. Indeed, certain conserved signaling pathways and regulatory proteins, such as the insulin/IGF-1 (insulinlike growth factor-1) hormonal system or the tumor suppressor p53, are implicated in each of the processes of aging, cell division and death [5, 6].
Genetic studies in model organisms from yeast and invertebrates to mammals reveal that single gene mutations can extend the life span of the model organism dramatically [2]. For example, mutations reducing the activity of the insulin/IGF-1 receptor double natural life span in nematodes and insects, and increase longevity by 60% in female mice. Intriguingly, the genes that affect longevity do so by refining mechanisms for cellular maintenance and repair [1]. In agreement with this supposition, accumulating data indicate that molecular chaperones and cellular self-degradative systems, including the ubiquitin–proteasome system and especially lysosome-mediated autophagy, play a pivotal role in the aging process [7–10]. These systems maintain cellular homeostasis, a function known to be impaired during aging. Heat-shock and stress proteins referred to as chaperones preserve the conformational homeostasis of cytosolic proteins through either assisting in refolding or sequestering them for proteasome-mediated degradation. Indeed, forced expression of certain heat-shock proteins increases life span in invertebrates [11–12]. Proteasomes mediate the degradation of misfolded and short-lived proteins [4]. In the nematode Caenorhabditis elegans, proteasomal components are required for the extended life span of insulin/IGF-1 signaling-defective mutants [13]. Moreover, proteasome activity normally attenuates with the aging process in Drosophila, and, when its age-dependent decline is prevented, can result in life span lengthening [14, 15].
Autophagy is the major degradative process of eukaryotic cells [8, 16]. It is required for the bulk clearance of damaged macromolecules and the turnover of long-lived proteins, and is the only known intracellular degradative mechanism for the removal of superfluous or dysfunctional organelles. Autophagic degradation has two main cellular functions: it provides macromolecular components (the products of lysosomal degradation, including amino acids and fatty acids) as an alternative energy source for fuelling the cells during starvation, and eliminates damaged macromolecules and organelles, thereby contributing to cellular clearance. Autophagy is also implicated in the cellular response to various stress conditions such as hypoxia or elevated temperature, when rapid reorganization of cellular functions is necessary. Moreover, it is important in cell growth, proliferation and survival, as well as in defense against intracellular microorganisms; in humans, diverse age-related pathological conditions such as cancer, neurodegenerative diseases (e.g., Alzheimer, Parkinson and Huntington disease), stroke, sarcopenia, infections and heart attack are linked to compromised autophagy [8, 16].
During autophagy, parts of the cytoplasm are delivered to lysosomes for degradation. Depending on the mechanism by which cytosolic materials are transported into lysosomes, there are three general types of autophagy: microautophagy, chaperone-mediated autophagy (CMA) and macroautophagy (for the major forms of autophagy, see Box 1, and Refs 8 and 17).
Box 1. The major types of autophagy.
Microautophagy
in this process, cytoplasmic material is sequestered into lysosomes through direct invagination or protrusion of the lysosomal membrane. Although it occurs in yeast and mammals, this type of autophagy is the least characterized.
Chaperone-mediated autophagy (CMA)
it selectively degrades proteins bearing a particular consensus pentapeptide motif, KFERQ. These proteins are recognized by a chaperone complex, and translocated into the lysosome through a specific receptor called the lysosome-associated membrane protein (LAMP)-2A. CMA appears to be a secondary response that temporally follows macroautophagy.
Macroautophagy
this process involves the formation of subcellular typically double-membrane-bound structures called autophagosomes to sequester cytoplasmic materials and deliver them into lysosomes for breakdown. The process of macroautophagy starts with the initiation of the formation of the phagophore. The growth of the phagophore terminates in the completion of the autophagosome. Owing to the convergence of the endocytic and autophagic pathways, the autophagosome may fuse with an endosome to generate an amphisome. The subsequent fusion of the autophagosome or amphisome with a lysosome forms an autolysosome, in which the enclosed material is degraded by acidic hydrolases, including proteases, lipases, glycosidases and nucleases.
Figure I[RH1]. The major types of autophagy
The sizes of the organelles and proteins are not proportional.
Measuring the rate of degradation of long-lived proteins in rat liver shows an age-dependent decline in autophagic activity that correlates with the progressive accumulation of damaged proteins and energetic compromise of the aging cells [18, 19]. Decline in autophagic activity with age can be prevented by calorie restriction, which is an effective anti-aging intervention [20]. Decreased expression of some autophagy genes in aging human brain is also evident [21]. Genetic studies in invertebrates demonstrate further that autophagic activity is required for survival in wild-type animals maintained under both normal and starvation-induced stress conditions [22–24], and for life span extension in various long-lived mutant animals [reviewed in [10, 17]. In C. elegans, for example, mutations decreasing insulin/IGF-1 or TOR (target of rapamycin) kinase signaling, mitochondrial activity and food intake extend life span, and autophagy genes are required for the expression of the long-lived phenotype triggered by such genetic interventions [7, 9, 25–27]. As the life-shortening effect of compromised autophagy on long-lived mutant nematodes is more significant than on wild-type animals, autophagy may specifically function in an anti-aging process rather than effecting shortened life span through nonspecific toxicity to the animal. In Drosophila, a general decline in autophagic activity is a normal part of aging [24]. The effect of autophagy on aging has also been recently reported in yeast, plants and diverse mammalian species [28–30]. Together, these results strongly implicate autophagy in life span control in eukaryotic organisms. Furthermore, prolonging basal expression of the autophagy-related gene Atg8 in Drosophila extends life span by 50% [24]. In mice, forced expression of the CMA receptor LAMP-2A to maintain CMA activity also results in reduced levels of damaged proteins and subsequent improvement in liver function [31]. These data provide evidence that the autophagic machinery regulates life span and the rate at which morphological changes occur during aging (representative references are listed in Table 1). To date, among the different forms of autophagy only macroautophagy has been implicated in life span control, while both macroautophagy and CMA have been linked to age-related morphological alterations. Hereafter we use therefore the term of autophagy as the synonym of macroautophagy. Together, autophagy-related genes constitute an anti-aging pathway.
Table 1.
Organisms in which autophagy has been directly linked to the control of life span.
However, based upon accumulating data from yeast to mammals, we speculate that autophagy, as the main cellular degradation process, does not merely represent one of the mechanisms that control aging; rather, it appears as the mechanism that integrates signals from and mediates the effect of distinct longevity pathways and regulatory proteins on aging. Hereafter, we summarize these results and propose a model in which autophagy plays a central regulatory role in the aging process. In addition, we provide a comprehensive view of how distinct longevity pathways, which have other outputs unrelated to autophagy and life span extension (e.g., cell growth, proliferation and survival), are interconnected with the autophagic machinery to form the signaling network of aging.
Longevity pathways converge on autophagy genes to regulate life span
Aging in divergent animal species is influenced by multiple conserved signaling pathways, which in turn respond to various environmental cues, such as nutrient availability, temperature and oxygen level. Various intrinsic factors including mitogens, growth factors and cellular energy status also affect the rate at which the cells age. Similarly, molecular chaperones, sirtuin-type chromatin-remodeling factors, cellular degradative processes (i.e., proteasome-mediated and autophagic), mitochondrial activity and the protein translation machinery regulate aging too. Therefore, aging is a multi-factorial process. Autophagy, however, appears to be unique among these pro- and anti-aging factors as it genetically interacts with all the others to mediate the longevity response; genes underlying the autophagic process mediate the regulatory effect that distinct aging factors have on life span extension, and the activity of some autophagy genes is directly regulated by such aging factors. This raises the possibility that autophagy has a central role in aging control. We next survey those molecular links and genetic interactions that potentially place the autophagic machinery at the base of the signaling network of aging.
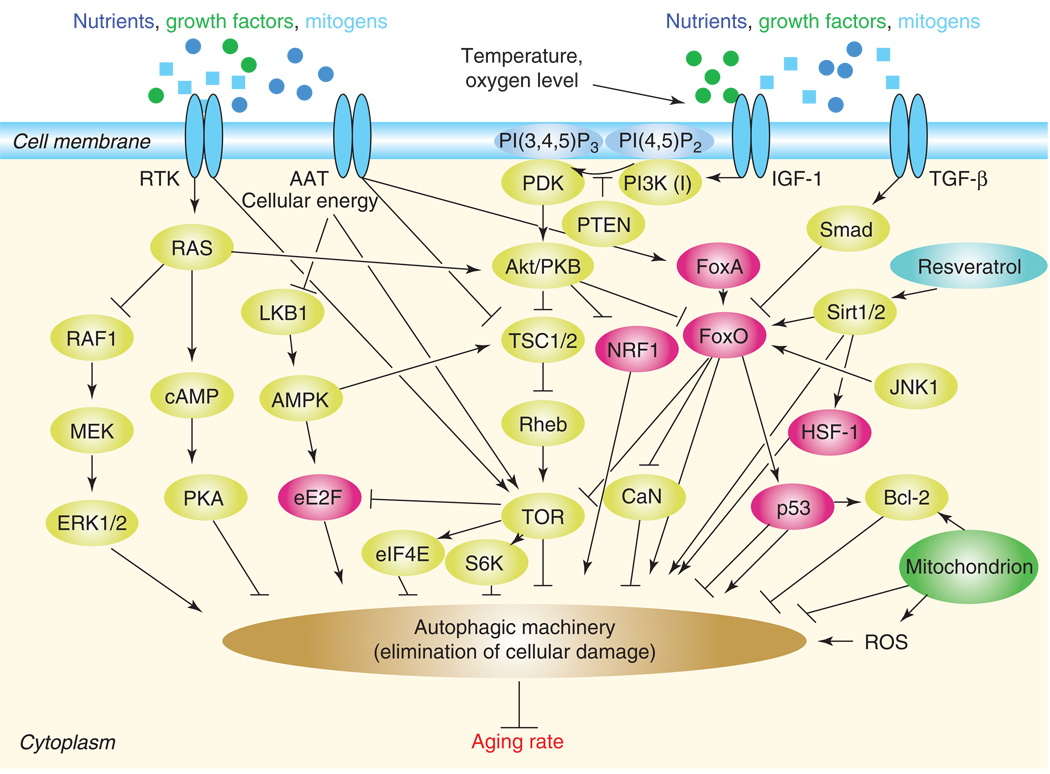
Insulin/IGF-1 signaling regulates aging in worms, flies and mammals [2]. The insulin/IGF-1 receptor, through activating a cascade of conserved kinases, inhibits the forkhead transcription factor FoxO. Longevity increases when FoxO becomes activated in response to reduced insulin/IGF-1 signaling (Fig. 1). In C. elegans, the insulin/IGF-1 receptor DAF-2 also influences both reproductive and cell growth, and these two functions of DAF-2 depend on the activity of certain autophagy genes [7, 32]. Thus, the autophagic machinery appears to function, at least in these processes, downstream of DAF-2. Consistently, autophagy genes are required for the expression of the long-lived phenotype of daf-2(-) mutants [7, 9, [25]. In mammals, the relationship between autophagy and insulin/IGF-1 signaling is remarkably intimate even at the molecular level. For instance, FoxO in mice promotes the transcription of several autophagy-related genes to induce protein degradation in various cell types. The regulatory effect of FoxO on Atg8 and Atg12 transcription seems to be direct as it is able to bind to their promoters in a sequence-specific manner [33]. Based upon these findings, one can conclude that insulin/IGF-1 signaling converges on the autophagic machinery to accelerate aging (Fig. 1).
Figure 1. Model for the signaling network of aging.
The overall rate at which cellular damage accumulates is regulated by multiple pathways (yellow components) and transcription factors (red) that adjust mechanisms for maintenance and repair. These transcription factors influence autophagy through modulating the activity of autophagy-related genes in the nucleus (for simplicity, the nucleus is not shown). Autophagy (brown) is the main cellular degradation process for eliminating or recycling damaged cellular constituents (the proteasomal system also plays an important role in eliminating damage and thereby in aging, but it is not shown here). Longevity pathways, which respond to diverse environmental cues such as nutrient availability, temperature and oxygen level, converge on the autophagic pathway to control aging. The chemical substance resveratrol is indicated by blue coloring, the mitochondrion is green. This figure illustrates the mammalian counterparts of signaling and metabolic components involved in aging. Several interrelationships among these components have only been demonstrated thus far in model organisms. Arrows (activations) and bars (inhibitions) indicate direct interactions that have been experimentally confirmed. However, not all the indirect interactions have as yet necessarily been demonstrated (e.g., it is unknown whether Sirt1/2 regulate eE2F activity). RTK: receptor tyrosine kinase; AAT: amino acid transporters; IGF-1: insulin-like growth factor-1 receptor; PI(4,5)P2; phosphatidylinositol (4,5)-bisphosphate; PI(3,4,5)P3: phosphatidylinositol (3,4,5)-trisphosphate; PI3K (I): class I phosphatidylinositol 3-kinase; PI3K (III): class III phosphatidylinositol 3-kinase; TGF-β: transforming growth factor-beta receptor; ROS: reactive oxygen species; Bcl-2: B cell lymphoma-2; JNK1: c-Jun N-terminal kinase-1; HSF-1: heat shock response transcription factor-1; NFR1: isoform 1 of nuclear factor erythroid 2-related factor 2; FoxO: Forkhead-family transcription factor; FoxA: FoxA transcription factor; Sirt1/2: NAD-dependent histone deacetylase-1/2; Smad: SMAD family member; PTEN: phosphatase and tensin homolog; Akt/PKB: AKT8 virus protooncogene/protein kinase B; PDK: 3-phosphoinositide-dependent protein kinase; CaN: calcineurin; TSC1/2: tuberous sclerosis complex-1/2; Rheb: Ras homologue enriched in brain; TOR: target of rapamycin; S6K: S6 kinase; eIFE4: eukaryotic translation initiation factor; eE2F: eukaryotic transcription factor; AMPK: AMP-activated protein kinase; LKB1: serine/threonine protein kinase; PKA: protein kinase A; ERK1/2: extracellular signal-related tyrosine kinase.
The JNK (c-Jun N-terminal kinase) and TGF-β (transforming growth factor-beta) signaling pathways also influence aging in flies and worms [34–36], and presumably do in mammals as well [37, 38]. In C. elegans, longevity triggered by increased dosage of JNK or reduced TGF-β receptor activity is mediated by DAF-16/FoxO, which thereby links the insulin/IGF-1, TGF-β and JNK pathways and autophagy in aging control. For example, BEC-1/Atg6/Beclin 1, which is an essential component of the autophagic machinery, but functions in diverse cellular processes, also mediates the effects of TGF-β signaling both on reproductive growth and cell size [7, 32]. In mammals, JNK1 is required for starvation-induced autophagy [39]. Although it is not yet demonstrated whether autophagic degradation is required for life span extension triggered by hyperactive JNK signaling or compromised TGF-β signaling, autophagy genes are likely to mediate their effects on aging (Fig. 1).
Insulin/IGF-1 signaling and the cellular energy sensor TOR kinase, another negative regulator of autophagy, constitute a common signaling axis to control aging in yeast, worms, flies and mice [40–43]. The activity of TOR depends on a cascade consisting of Akt/protein kinase B, the tuberous sclerosis complexes 1 and 2 (TSC1 and TSC2), and the Ras homologue enriched in brain (Rheb) [44]. In yeast, TORC1 (Tor kinase complex 1) cooperatively regulates autophagy with two other nutrient-sensing pathways including protein kinase A (PKA) and Sch9 [45]. Furthermore, the AMP-activated protein kinase (AMPK), which links the cellular AMP:ATP ratio and insulin/IGF-1 signaling to life span in nematodes [46], acts upstream of TOR to promote autophagy in human cell lines and flies [47, 48] (Fig. 1). Consistent with these findings, inactivation of autophagy genes inhibits life span extension in TOR-deficient worms [9, 27]. This implies that TOR controls life span through adjusting autophagy.
TOR also regulates cell growth and proliferation by altering mRNA translation [44]. TOR promotes protein synthesis through activation of ribosomal S6 kinase (S6K) and inactivation of 4E-binding protein, which is an inhibitor of the eukaryotic translation initiation factor 4E (eIF-4E) (Fig. 1). Overexpression of dominant-negative forms of S6K extends life span in Drosophila [41], and lowering protein synthesis through loss of a specific eIF4E isoform increases longevity and oxidative stress resistance in C. elegans [49]. Autophagic degradation is important for the removal of damaged proteins. Therefore, the regulation of autophagy may be tightly linked to protein turnover and involved in translation-dependent control of aging.
The Ras-PKA (cAMP-dependent protein kinase A) pathway also controls cell proliferation, stress response and aging. In yeast, mutations that attenuate Ras–PKA signaling or that increase the activity of the mitogen-activated protein (MAP) kinase ERK2 serve to increase both replicative and chronological life span [50, 51]. In good accordance with these results, in human cell lines Ras can activate autophagy through activation of the MAP kinase signaling pathway [52], while in yeast three autophagy-related proteins have been identified as substrates of PKA inhibition: Atg1, Atg13 and Atg18 [51]. Thus, Ras-PKA signaling may directly regulate autophagic degradation. Moreover, constitutive activation of PKA suppresses autophagy even under starvation conditions. The observation that mice in which the gene encoding type 5 adenylyl cyclase (which converts ATP to cAMP) has been knocked out live 30% longer than their wild-type littermates [53] suggests that Ras–PKA signaling also regulates life span in mammals. Given the fact that attenuation of Ras–PKA signaling is associated with longevity and influences autophagic activity, it can be speculated that the life span effect of Ras–PKA is mediated through autophagy genes in various organisms (Fig. 1).
Calcineurin (CaN) is a serine/threonine phosphatase that is activated by Ca2+/calmodulin and interacts with multiple signaling systems in diverse cellular processes. One such process is aging, in which CaN exerts its function downstream of FoxO [54]. According to a recent report, longevity response in CaN-defective mutant nematode strains is also autophagy gene-dependent [55] (Fig. 1).
Sirtuin-type chromatin remodeling factors are implicated in aging regulation. In nematodes, for example, DAF-16/FoxO binds SIR-2.1 sirtuin to promote the transcription of target genes involved in longevity [56] (Fig. 1). Consistently, nematodes with increased dosage of SIR-2.1 are long-lived [57]. Sir2 also promotes longevity in yeast and Drosophila [58]. The mammalian Sirt2 deacetylates FoxO [59], and hence may act as a determinant of aging. Furthermore, in the mouse, Sirt1 is activated by the chemical resveratrol, which has an anti-aging effect in a range of organisms [60]. Recent data demonstrate that Sirt1 is able to induce autophagy through deacetylating chromatin proteins at several autophagy-related loci [61]. The transcriptional activity of certain autophagy genes is thus under the direct control of Sirt1. Hence, autophagy may also be required for sirtuin-dependent life span extension in both invertebrates and vertebrates (Fig. 1) [62].
Another conserved regulatory protein that is involved in aging control is the tumor suppressor p53, which can influence life span in mammalian systems and nematodes. Inhibiting the cytoplasmic form of p53 induces autophagy in human, mouse and nematode cells [63], and this intervention increases organismal survival at least in C. elegans [64]. Consistently, the life span-extending effect of p53 deficiency requires autophagy genes in this organism [64]. However, p53 can also act a transcription factor when it functions together with p19/Arf. Interestingly, simultaneously elevated levels of p53 and Arf confer strong cancer resistance and delay aging in mice [6]. Hence, p53 influences life span in a complex way, apparently, independent of its actual cellular form, interacting with the autophagic machinery (see Box 2) (Fig. 1).
Box 2. p53 and aging.
The relationship between p53 and aging appears to be complex. In C. elegans, loss-of-function mutations in the insulin/IGF-1 signaling pathway which increase life span also inhibit tumor cell growth through activating a p53-mediated cell death pathway [5]. Consistently, enhanced activity of the tumor suppressor p53, together with its positive regulator Arf, increases tumor resistance and delays aging in mice [6]. These results suggest the co-evolution of tumor susceptibility and longevity: animal species that acquire tumor earlier in their lifetime normally have shorter mean life spans than those doing so later. Furthermore, loss of cytoplasmic p53 can induce autophagy in human, mouse and nematode cells under conditions of hypoxia and nutrient depletion [63], and this effect positively influences longevity in nematodes [64]. As the longevity response in p53/CEP-1-deficient worms depends on autophagy genes [64], p53 may regulate aging via autophagy. Indeed, in human cell lines, p53 is able to modulate autophagy in a DRAM (damage-regulated autophagy modulator)-dependent manner to execute a full cell death response [65]. In another C. elegans paradigm, however, mutational inactivation of p53/cep-1 decreases the survival of L1-stage larvae exposed to prolonged starvation [66]. Thus, the role of p53 in life span control may also be condition-dependent, and the underlying mechanism remains to be elucidated.
In addition to FoxO, several other conserved transcription factors control aging through the modulation of mechanisms for cellular maintenance. For example, the nematode heat shock factor HSF-1 shares several target genes with DAF-16/FoxO, many of which are implicated in longevity [67]. In addition, human HSF1 is directly activated by the deacetylase and longevity factor SIRT1 [68]. As autophagy has a crucial role in the cellular response to heat stress in various organisms, and is regulated by FoxO and SIRT1 in mammals, it may also mediate the effect of the HSF-1 family proteins on aging in divergent animal species. The NRF1- (nuclear respiratory factor 1) type transcription factors participate in the cellular defense against oxidative stress in organisms from worms to mammals [69]. SNF-1, the nematode ortholog of NRF1, is required for increased longevity triggered by dietary restriction [70]. Hypoxic/ischemic injury is a pathological condition in which autophagy plays an important role. This implies that autophagy genes may mediate longevity caused by SNF-1 activity. In C. elegans, the longevity response to caloric restriction also requires the FoxA-like transcription factor PHA-4 (Fig. 1), which functions partially redundantly with DAF-16/FoxO in life span determination [71]. PHA-4 is crucial for increased autophagy and longevity in mutant worms with inherent dietary restriction that is due to abnormal pharyngeal pumping [27].
Mitochondria produce ROS as a byproduct of oxidative respiration. A decreased rate of respiration generates fewer ROS and, owing to a general decline in ATP levels, may reduce the rate of protein turnover. In agreement with these data, when the activity of the mitochondrial electron transport chain is lowered, life span is extended in yeast and worms [72]. Depletion of autophagy proteins blocks life span extension in nematodes with reduced respiration [9], indicating that mitochondrial function affects aging via, at least in part, the autophagic pathway. However, the mechanism by which autophagy is linked to the mitochondrial respiratory chain in aging control remains unknown. In human cell lines, starvation stimulates formation of ROS, which may directly oxidize and inactivate the cysteine protease Atg4 required for autophagosome formation [3]. Thus, respiration may influence autophagy through the production of ROS. Alternatively, certain mitochondrial proteins, such as Bcl-2, have the ability to bind specific autophagy proteins, thereby modulating autophagic activity [73, [74]. p53 can also mediate mitochondria dysfunction-triggered autophagy activation.
In summary, autophagy appears to influence the aging process in organisms from yeast to mammals by acting downstream of multiple longevity pathways (Fig. 1). In other words, autophagy appears to mediate the effects that longevity pathways and regulatory proteins have on life span. According to our present knowledge, there is no alternative for another cellular mechanism that could effectively eliminate damaged, superfluous macromolecules and organelles (note that proteins, after they get ubiquitinated, can also be degraded by the proteasomal system).
Concluding remarks
The lifelong accumulation of cellular damage drives the aging process, and leads to age-related disease and, ultimately, death [1]. Thus, aging is considered as a collection of damage-induced, mostly independent degenerative processes, such as arthritis, cancer, dementia, neuronal demise (including Alzheimer, Parkinson and Huntington diseases), stroke, hearth failure and sarcopenia [2]. If a cure, for example, for cancer that killed abnormally proliferating cells were found, it would only increase the chance of an individual to acquire another type of such pathologies, e.g., Alzheimer disease. We propose that enhancing or prolonging the activity of mechanisms eliminating cellular damage decreases not only the rate at which damage accumulates, but protects, at least for a certain time, the individual from the incidence of degenerative processes. In this sense, understanding both the exact role of cellular degradation processes in aging and their mechanisms may provide us the potential to modulate our aging process.
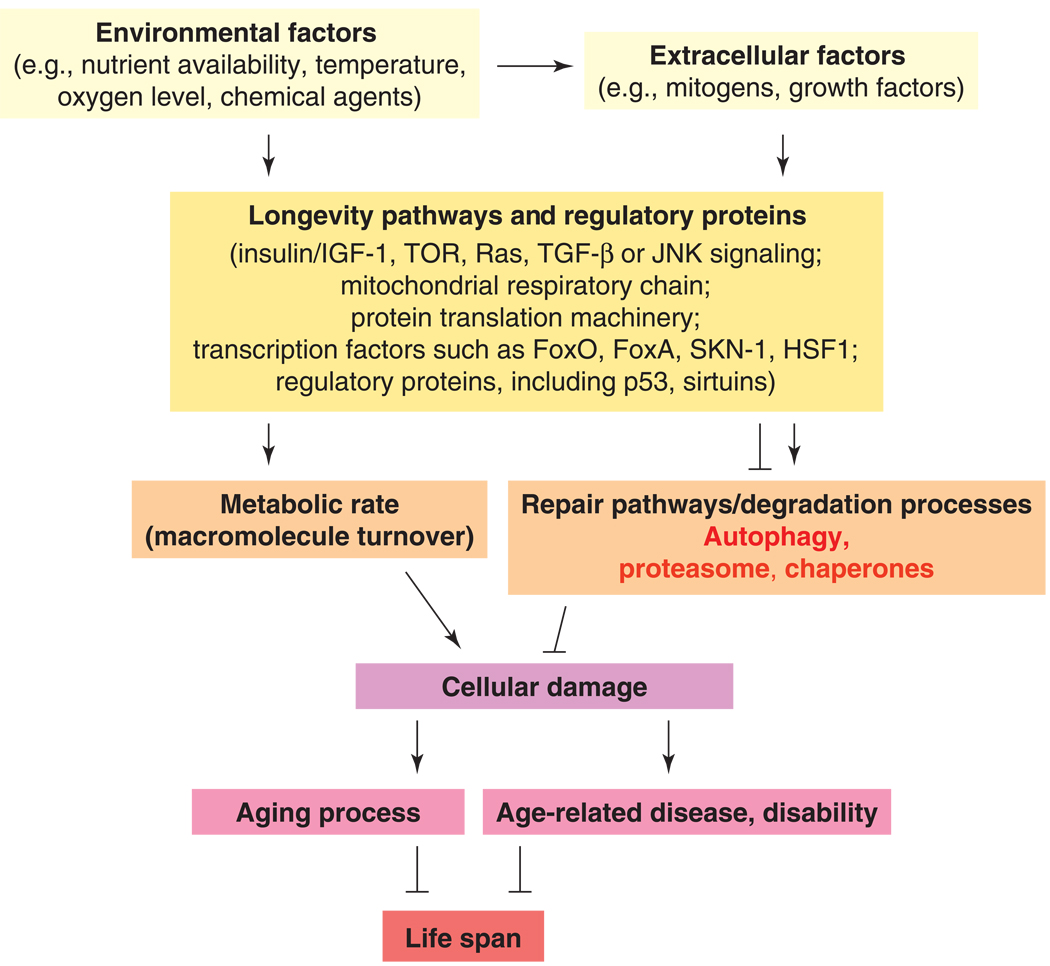
The overall rate at which molecular damage accumulates is influenced by longevity pathways and regulatory proteins that modulate metabolic rate (molecule turnover) and processes for cellular maintenance (Fig. 2). Autophagy, the ubiquitin-proteasome system and chaperones operate as the major homeostatic factors to eliminate damage from the cytosol. Whereas the proteasome system and chaperones only target specific proteins, autophagy is required for the bulk degradation of any type of damaged macromolecules or organelles. The activity of longevity pathways is modulated by various environmental and intrinsic cues. Thus, our model for the signaling network of aging readily accommodates genetic and environmental influences on longevity (Fig. 1 and Fig. 2). For example, nutrient supply stimulates, among others, insulin/IGF-1 signaling, which in turn increases growth rate and downregulates degradation: the generation of cellular damage increases, whereas its removal decreases. As a result, the rate at which damage accumulates is enhanced, resulting in acceleration in the aging process and shortening in life span. The improving level of nutrition in the developed countries, however, is coupled with increasing human longevity. It is likely, therefore, that improvements in medicine are counteracting a nutrient-associated tendency to succumb to diseases that generally occur at advanced ages.
Figure 2. The simplified mechanisms of aging control both by environmental and genetic factors.
The lifelong accumulation of random, unrepaired cellular damage drives the aging process in divergent eukaryotic organisms. The overall rate at which damage accumulates is influenced by metabolic rate and processes for maintenance and repair. Autophagy has a major role in eliminating cellular damage, but the proteasome system and molecular chaperones are also important. The activity of degradative processes is modulated by longevity pathways and regulatory proteins (transcription and chromatin factors). Environmental cues and intrinsic factors influence the activity of longevity pathways. Arrows indicate activations; bars represent inhibitory interactions. Both environmental and intrinsic factors may also directly affect the accumulation of cellular damage (not shown). Similarly, compromised degradation processes may lead to disease independent of cellular damage (not indicated).
In addition to aging, autophagy influences cell growth, division and survival by interacting with signaling systems that control these processes (Fig. 1). Thus, the linkage of longevity, proliferation and cell loss to the same cellular pathway – autophagy – has general relevance to the observation that most animal species become more susceptible to cancer and various degenerative diseases as they age. Is there a hope to delay the incidence of a wide range of such aging-related pathologies in humans? Autophagy, with a major role in the elimination of cellular damage and in the renewal–rejuvenation of cellular components, appears to function as a cellular mechanism underlying longevity in divergent eukaryotic organisms. In addition, it is implicated in various human degenerative diseases characterized by massive cell loss or uncontrolled proliferation. Hence, autophagy serves as a potential target for decelerating the aging process. Prolonging autophagic activity by pharmacological means in elderly people may extend the limit of their healthy life period, and thereby be an important area for further investigations. Recent data obtained from flies and mice indicate that positive changes in longevity and organ function are effected by modifying the activity of single (key) autophagy genes [24, 31]. This implies that applying autophagy modulators for adjusting our aging rate and preserving a healthy life is likely to be just a question of time.
Future directions
Further studies are needed to test the relevance of our model shown in Fig. 1 on human longevity and fill in further gaps in our knowledge of the fundamental link between autophagy and aging proposed here. First, where it is still unknown, the nature of regulatory interactions between longevity pathways and the autophagic machinery should be explored in invertebrate models of aging. As an example, it should be assessed whether the life span-extending effect of the Sirt1/2-HSF1 axis depends on autophagy genes. Second, one should determine key autophagy-related genes in mammals whose prolonged activation can result in life span extension. This would require the generation of genetically manipulated mammalian models with increased, but otherwise normally regulated, levels of autophagy. Third, it would be informative to examine the requirement of the autophagic machinery for the expression of increased longevity in long-lived mutant mouse strains or in mice treated with the longevity drug rapamycin or resveratrol. Fourth, novel pharmacological interventions should be developed with the ability to increase autophagic activity in mammals. Preferably, drug candidates that directly interfere with a component of the biochemical pathway of autophagy are needed instead of those acting upstream of the pathway. We mention rapamycin as an example. Although it can function as a potent autophagy inducer, this potential longevity drug works upstream of the pathway, and thereby may exert undesired side effects, such as perturbation of cell growth and protein turnover which is evident in TOR—the kinase target of rapamycin—deficient mutant animals.
Figure Box 1.
Acknowledgements
This work was supported by grants from the Ministry of Health (ETT 167/2006), the Hungarian Scientific Research Funds (OTKA K68372, K75843, NK78012 and PD75477) to T.V, M.S. and K.T-V., the National Office for Research and Technology (TECH_08_A1/2-2008-0106), and the National Institutes of Health (GM53396) to D.J.K. T.V. and K.T-V. are grantees of the János Bolyai scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflicts of interest that relate to the publication of this paper.
References
- 1.Kirkwood TBL. A systematic look at an old problem. Nature. 2008;451:644–647. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- 2.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 3.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 5.Pinkston JM, et al. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 6.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–380. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 7.Meléndez A, et al. Autophagy genes are essential for dauer development and life span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tóth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in. Caenorhabditis elegans Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 10.Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 11.Kurapati R, et al. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J. Gerontol. A. 2000;55:B552–B559. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama K, et al. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- 13.Ghazi A, et al. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonoki A, et al. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Vicente M, et al. Protein degradation and aging. Exp.Gerontol. 2005;40:622–633. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima M, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vittorini S, et al. The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J. Gerontol. A Biol. Sci. Med Sci. 1999;54:B318–B323. doi: 10.1093/gerona/54.8.b318. [DOI] [PubMed] [Google Scholar]
- 19.Cuervo AM, Dice JF. Age-related decline in chaperone mediated autophagy. J. Biol. Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 20.Donati A, et al. Effect of aging and anti-aging caloric restriction on the endocrine regulation of rat liver autophagy. J. Gerontol. A Biol. Sci. Med. Sci. 2008;63:550–555. doi: 10.1093/gerona/63.6.550. [DOI] [PubMed] [Google Scholar]
- 21.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 22.Juhász G, et al. Atg7-dependent autophagy promotes neuronal health, stress tolerance and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang C, et al. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidative stress in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 25.Hars ES, et al. Autophagy regulates aging. C elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 26.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension. C elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 27.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction. C elegans. PLoS Genet. 2008;4:24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvers AL, et al. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009 doi: 10.1111/j.1474-9726.2009.00469.x. in press PMID: 19302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada S, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannigan AM, Gorski SM. Macroautophagy: the key ingredient to a healthy diet? Autophagy. 2009;5:140–151. doi: 10.4161/auto.5.2.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aladzsity I, et al. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177:655–660. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, et al. FoxO3 co-ordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Wang MC, et al. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 35.Oh SW, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor DAF-16. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw WM, et al. The C. elegans TGF-β dauer pathway regulates longevity via insulin signalling. Curr. Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, et al. Activation of c-Jun-N-terminal kinase and decline of mitochondrial pyruvate dehydrogenase activity during brain aging. FEBS Lett. 2009;583:1132–1140. doi: 10.1016/j.febslet.2009.02.043. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonafè M, Olivieri F. Genetic polymorphism in long-lived people: cues for the presence of an insulin/IGF-pathway-dependent network affecting human longevity. Mol. Cell Endocrinol. 2009;299:118–123. doi: 10.1016/j.mce.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 39.Wei Y, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vellai T, et al. Influence of TOR kinase on lifespan C. elegansNature 2003. p426, 620 [DOI] [PubMed] [Google Scholar]
- 41.Kapahi P, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 43.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 45.Yorimitsu T, et al. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Apfeld J, et al. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan. C. elegans. GenesDev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang J, et al. The energy-sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 48.Lippai M, et al. SNF4Aγ, the Drosophila AMPK γ subunit is required for regulation of developmental and stress-induced autophagy. Autophagy. 2008;4:476–486. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- 49.Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Fabrizio P, et al. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- 51.Budovskaya YV, et al. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogier-Denis E, et al. Erk1/2-dependent phosphorylation of Gα-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J. Biol. Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- 53.Yan L, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 54.Dong MQ, et al. Quantitative mass spectrometry identifies insulin signaling targets. C elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 55.Dwivedi M, et al. C. elegans. Autophagy. 2009. Autophagy genes mediate the effect of calcineurin on life span in. In press, PMID: 19279398. [DOI] [PubMed] [Google Scholar]
- 56.Berdichevsky A, et al. Celegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 57.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 58.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 59.Wang F, et al. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 60.Borra MT, et al. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 61.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salminen A, Kaarniranta K. SIRT1: Regulation of longevity via autophagy. Cell Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 63.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tavernarakis N, et al. The effects of p53 on whole organism aging are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 65.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 66.Derry BW, et al. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 67.Hsu A-L, et al. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 68.Westerheide SD, et al. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An JH, Blackwell TK. SKN-1 links C. elegans mesodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 71.Panowski SH, et al. PHA-4/Foxa mediates diet-restriction induced longevity of. C elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 72.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–23401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 73.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Takács-Vellai K, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death. C elegans. Curr. Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]