Abstract
p73 is a member of the p53 family often overexpressed in human cancer. Its regulation, particularly following DNA damage, is different from that of p53. Following DNA damage, we found induction of p73 at both the protein and mRNA levels. Furthermore, by using different p73 promoter fragments, we found a role for E2F1 in mediating transcription of p73. However, this observation alone does not account for the observed DNA damage-induced activation of p73 in the cells used in these experiments. By analyzing the p73 promoter sequence, we revealed a new mechanism of p73 induction associated with the removal of transcriptional repression from the p73 promoter. We found, in fact, that treatment of cells with DNA damaging agents induced nuclear export of the transcription factor C-EBPα and blockage of this export abolished drug-induced p73 activation. We also show that C-EBPα has a direct repressive activity on transfactor E2F1, and for this repression the binding of C-EBPα to its consensus sequence in the DNA is required. These data suggest that in normal conditions a repressor complex involving C-EBPα, E2F1 and perhaps other proteins is present on the p73 promoter. This repressor complex is destroyed following damage by removal of C-EBPα from nuclei.
INTRODUCTION
The cellular response to DNA damage involves the activation of proteins which in turn activate downstream cascades leading to DNA repair or apoptosis depending on the type of damage and the cellular context.
The product of the tumor suppressor gene p53 is one of the early activated proteins following DNA damage (1,2). Its activation is mainly post-transcriptional and involves a block of ubiquitination-dependent degradation (3). The activation of p53 is considered particularly important in determining whether a cell will arrest the cell cycle to try to allow repair of the induced lesion or activate the apoptotic pathway if the damage is unrepairable.
The p73 gene shares relatively high sequence homology with p53, particularly in its DNA binding domain (4) and, in fact, is able, although with some differences in specificity, to activate the same downstream genes activated by p53 (4,5). There are, however, some important differences between p53 and p73, one of which is different activation following DNA damage. Whilst p53 is activated by all known forms of DNA damage, p73 is activated by treatment with cytotoxic agents, including doxorubicin (DX) and cis-dichloro-diamino platinum (DDP), but not, for example, by UV light (6–9). Furthermore, the mechanisms responsible for the reported induction of p73 are different to those able to activate p53.
Considering that in more than half of human tumors the p53 gene is inactivated (10), and hence its response to DNA damage impaired, it is important to verify whether p73, which is very rarely found to be mutated in human tumors (11–13), can be activated after damage and at least partially substitute for p53 in the cellular response to damage.
In addition, p73 is frequently overexpressed in cancer cells, including neuroblastoma cells (14–17). Changes in the methylation status of the p73 promoter have been reported in some tumors but not in other (7,18). Other mechanisms, involving the rate of transcription, could be responsible for the increased levels of p73 found in human tumors.
In the present study, we have analyzed the ability of DNA damage to induce activation of p73 and present evidence for a new mechanism that, at least in the model system used in these experiments, is associated with the removal of C-EBPα repression of transcription factor E2F1 on the p73 promoter.
MATERIALS AND METHODS
Cells and treatments
The neuroblastoma cell line SH-SY5Y was grown in Dulbecco’s modified Eagle’s medium, the ovarian carcinoma cell line SKOV3 was grown in RPMI 1640 medium and the human colocarcinoma cell line HCT116 was grown in Iscove’s modified Dulbecco’s medium. All media were supplemented with 10% fetal bovine serum (FBS). DX was supplied by Pharmacia (Nerviano, Italy), DDP was purchased from Sigma (Milan, Italy). The drugs were dissolved in medium just before use. Different concentrations of the drugs were used for the experiments. Leptomycin B (LMB) was obtained from Sigma and used at a concentration of 20 ng/ml.
Real time PCR
An aliquot of 200 ng of total RNA was retrotranscribed in 20 µl with a TaqMan Reverse Transcription Kit (Applied Biosystems, UK) and 2 µl was amplified by real time PCR. Real time PCR was used for relative quantification of p73; actin was used as the internal control.
P73 was amplified using 5′-GGATTCCAGCATGGACGTC TT-3′ as forward primer and 5′-GCGCGGCTGCTCATCT-3′ as reverse primer (both 200 nM). Actin was detected using 5′-TCACCCACACTGTGCCCATCTAC-3′ as forward primer and 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ as reverse primer (both 200 nM). Reactions were performed in a total volume of 25 µl with SYBR Green PCR Master Mix (Applied Biosystems).
Western blotting analysis
Cell extracts, obtained from untreated and drug-treated cells, were prepared by lysing cells in 50 mM Tris–HCl pH 7.4, 250 mM NaCl, 0.1% Nonidet NP-40, 5 mM EDTA and 50 mM NaF, in the presence of aprotinin, leupeptin and phenylmethylsulfonyl fluoride (PMSF) as protease inhibitors, for 30 min on ice. Insoluble material was pelleted at 13 000 g for 10 min at 4°C and the protein concentration was determined using a Bio-Rad assay kit (Bio-Rad, Rome, Italy). An aliquot of 40 µg of total cellular proteins was separated by SDS–PAGE and electrotransferred to nitrocellulose. Immuno blotting was carried out with anti-p73 monoclonal antibodies (Oncogene Research, San Diego, CA), anti-p53 monoclonal antibody (DO-1; Santa Cruz Biotechnology, Heidelberg, Germany) and anti-actin, anti-C-EBPα and anti-p21 polyclonal antibodies (Santa Cruz Biotechnology). Antibody binding was revealed by peroxidase secondary antibodies and visualized using enhanced chemiluminescence (ECL; Amersham, Milan, Italy).
Protein expression in nuclei and the soluble fraction was determined as previously described (19). To test the appropriate separation of nuclei and cytoplasm, we measured the levels of the nuclear protein topoisomerase I (monoclonal antibodies kindly supplied by Dr Ivana Scovassi, CNR, Pavia, Italy) in the two fractions by western blotting analysis.
p73 promoter constructs and activity
The different fragments of the human p73 promoter (2.7 kb, 1.2 kb, 880 bp and 220 bp) linked to the luciferase gene were as previously described (20). The 1.8 kb promoter fragment was obtained by exonuclease treatment. Briefly, the 2.7 kb fragment was linearized with XhoI, treated for different times with exonuclease III, the ends blunted with S1 nuclease and Klenow fragment of DNA polymerase and the plasmid closed with DNA ligase. Insert sizes of the deletion mutants were assessed by restriction analysis and gel electrophoresis prior to direct sequencing. The 269 bp promoter fragment containing the putative p53 binding site was obtained from the 2.7 kb fragment by NotI + PstI digestion and religation in the pGVB2 plasmid (20) and was named Δ2444.
The different constructs were transfected into the three cell lines using the calcium phosphate precipitation method. Cells were transfected with 2 µg of p73 promoter constructs and 0.05 µg of pRL-SV40, used for internal normalization. At the end of transfection cells were treated with DX and reporter gene activities were evaluated after 24 h using the Dual Luciferase System (Promega, Milan, Italy). Results are expressed as a percentage of the reported control luciferase activity normalized by the renilla activity value. The means ± SD of three independent experiments are shown.
In co-transfection experiments the different p73 constructs were co-transfected with 1–10 µg of a plasmid encoding for C-EBPα (kindly provided by Dr M. Pitarque, University of Modena, Italy), wild-type (wt) human E2F1, a mutant E2F1 (E132) not able to bind DNA or a transactivation-deficient mutant (1–374) (kindly provided by Dr Helin, IEO, Milan, Italy). An aliquot of 0.05 µg of pRL-SV40 was again used for internal normalization and luciferase activity measured as reported above.
Mutations in the DNA binding sites for WT1 and C-EBPα have been produced by site-directed mutagenesis (Stratagene, La Jolla, CA). The presence of the desired mutations was confirmed by sequencing.
Titration experiments were performed by co-transfecting increasing concentrations of the C-EBPα expression vector together with fixed amounts of p73 promoter fragments and 0.05 µg of pRL-SV40 as internal control. Luciferase activity was assessed 24 h after transfection.
Oligonucleotides
Phosphorothioated double-stranded oligonucleotides containing the consensus C-EBPα binding site (5′-TGCAGATTG CGCAATCTGCA) and a mutant oligonucleotide (5′-TGCA GAGACTAGTCTCTGCA) were obtained from Sigma. SH-SY5Y cells were treated for 24 h with 20 µM oligo nucleotides and p73 mRNA levels were determined by real time PCR.
Immunoprecipitation
Cells (5 × 105) were transfected with 10 µg of human wt E2F1 and 10 µg C-EBPα using GENE PORTER (GTS, San Diego, CA) according to the manufacturer’s instructions or mock transfected by treatment with the same reagents minus plasmid DNA. Cells were harvested 24 h following transfection and lysed in 100 µl of the lysis buffer used for western blotting. Lysates were centrifuged for 5 min at 13 000 g at 4°C. Supernatants were incubated for 12 h with 5 µg of anti-C-EBPα antibody and then for 2 h with A/G agarose beads (Santa Cruz Biotechnology). Lysates with bound complexes were washed five times with RIPA buffer. Bound complexes were released by heating at 95°C for 5 min, resolved on 12% SDS–polyacrylamide gels and analyzed by western analysis with anti-E2F1 antibodies (Santa Cruz Biotechnology).
RESULTS
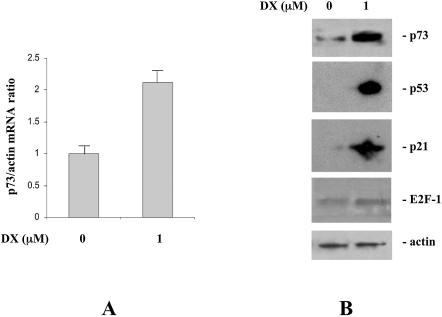
Treatment of SH-SY5Y neuroblastoma cells with the DNA damaging agent DX resulted in p73 mRNA induction (Fig. 1A). The same treatment conditions (1 µM for 24 h) were able to induce an accumulation of p73 at the protein level (Fig. 1B). The ability of DX to induce p73 was also tested in HCT116 cells, in which a 2.5-fold induction of p73 mRNA was found. Similar induction was obtained when another DNA damaging drug, DDP, was used in SH-SY5Y cells (see Supplementary Material, Annex 1A and 1B).
Figure 1.
(A) p73 mRNA levels analyzed in SH-SY5Y cells by real time PCR 24 h after treatment with DX (24 h). Data are expressed as fold induction over untreated cell mRNA levels. (B) Western blot analysis of p73, p53, p21 and E2F1 expression in SH-SY5Y cells untreated or 24 h after treatment with 1 µM DX.
In Figure 1B we compare the induction of p73 with that of other damage-inducible proteins such as p21, p53 and E2F1 in SH-SY5Y cells after treatment with 1 µM DX. The concentration of DX used, which was able to induce both p53 and p21, did not significantly modify the levels of E2F1.
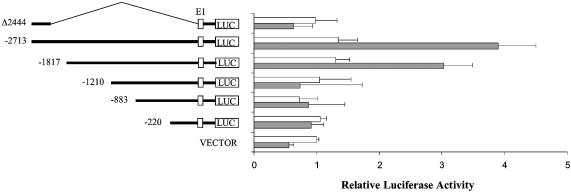
Trying to elucidate the mechanism(s) by which DX was able to induce activation of p73, we analyzed the transcription of different p73 promoter fragments in SH-SY5Y cells. By using the longest p73 promoter fragment available, spanning ∼2.7 kb 5′ to the initiation start site, we found in transient transfection experiments that DX induced the transcription of this fragment in SH-SY5Y cells (Fig. 2). The entire 2.7 kb long fragment contains three E2F1 binding sites and a putative p53-responsive site.
Figure 2.
Effect of DX on human p73 promoter activity. Constructs harboring various 5′ deletions of the human p73 promoter subcloned in the pGVB2 plasmid (20) were transfected into SH-SY5Y cells. Transfected cells were untreated or treated with 1 µM DX for 24 h before analysis of luciferase activity. For details see Materials and Methods.
Different deletion fragments originating from the 2.7 kb long fragment were also analyzed (Fig. 2). The 1.8 kb fragment, which lacks the putative p53 binding site and one of the three E2F1 binding sites, still maintained DX-dependent activation, being able to induce a more than 2-fold increase in luciferase activity over the basal level, similar to that obtained using the longer 2.7 kb fragment. The further deletion fragment, spanning 1.2 kb from the transcription start site (still containing two of the three E2F1 binding sites), did not show any change in activity in cells treated with DX. Similarly, the 880 and 220 bp long fragments showed similar activity in untreated and DX-treated cells. When the same experiments were performed in SH-SY5Y cells using DDP instead of DX, we found that DDP was able to induce transcription of the 1.8 kb fragment but not of the 1.2 kb fragment (Supplementary Material, Annex 1C).
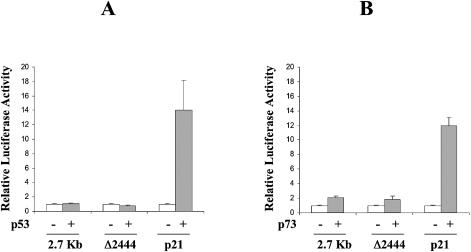
The construct containing the putative p53-responsive element with no E2F1 sites (fragment Δ2444) had low basal luciferase activity, not different from the PGL2 vector, which was not induced by DX (Fig. 2). Furthermore, this fragment was tested in co-transfection experiments with human p53 or p73 expression plasmids (Fig. 3). Under conditions in which a classical p53-responsive promoter, such as the p21-luc promoter, showed a clear p53 and p73 inducibility, the Δ2444 fragment as well as the entire 2.7 kb fragment did not.
Figure 3.
Activation of full-length p73 promoter (2.7 kb), a fragment of it containing the putative p53 binding site (Δ2444) and of p21 promoter by transfection of p53 or p73 expression vectors. Values are the means of three independent experiments each with three duplicates.
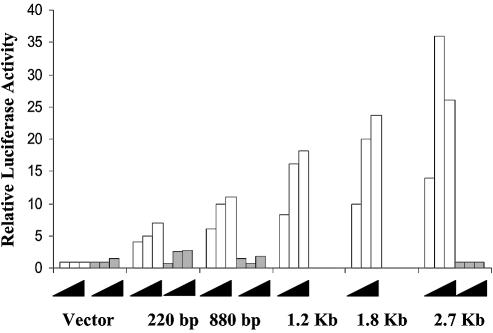
The same DNA constructs were tested for their ability to respond to E2F1. The different promoter fragments were co-transfected together with different amounts of a wt E2F1 expressing plasmid or with the same plasmid harboring a mutation (E132) abolishing the binding ability of E2F1. The results reported in Figure 4 clearly show that the longest p73 fragment shows a strong induction of its activity by co-transfection with E2F1 wt but not the mutant plasmid. Removal of one of the three E2F1 binding sites (1.8 and 1.2 kb) results in a slight decrease in luciferase activity, which declines further in the construct having only one site (880 kb). Moreover, by transfecting a truncated transactivaton-deficient version of E2F1 (1–374), still able to bind E2F1 sites, we could abolish DX-induced activation of the p73 promoter (data not shown), emphasizing, in agreement with previously reported data (21,22), that E2F1 plays a major role in DX-induced activation of p73. Considering the data obtained using the different promoter fragments together, they show that the 1.8 and 1.2 kb fragments both respond to E2F1 transfection but only the 1.8 kb fragment is sensitive to DX treatment. This suggests that there could be a transcriptional repressor sensitive to DX treatment in the 600 bp fragment contained in the 1.8 kb but not the 1.2 kb fragment.
Figure 4.
Activation of different human p73 promoter fragments by co-transfection with increasing amounts of E2F1 wt (white bars) or mutated E2F1 (E132, grey bars).
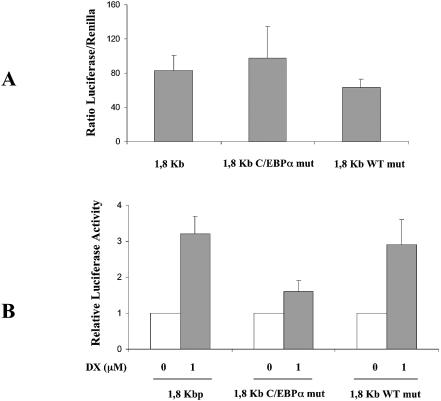
To test this hypothesis, we analyzed the 600 bp fragment sequence for the presence of DNA binding sites for other transcription factors and we concentrated our attention on two sites, the WT1 binding site located at position –1678/–1665 and the C-EBPα binding site located at position –1380/–1372. We first mutated the binding sites and used these constructs in transient transfection experiments. Figure 5A shows that mutation in the WT1 site slightly reduced the basal luciferase activity level compared to the wt 1.8 kb fragment while mutation in the C-EBPα binding site slightly enhanced this activity. Clear differences were observed when the response to DX treatment was analyzed. Figure 5B, in fact, shows that the 1.8 kb fragment with mutation in the WT1 binding site still responds to DX treatment, inducing transcriptional activation of luciferase comparable to that of the 1.8 kb fragment. When the fragment with a mutation in the C-EBPα binding site was used, DX was no longer able to induce activation of the promoter fragment, suggesting that an intact C-EBPα binding site is necessary for DX-dependent induction of this p73 promoter fragment. These results, however, do not exclude the possibility that C-EBPα could act as an activator of p73 promoter fragment activty and that its removal is associated with a decreased inducibility following DX treatment.
Figure 5.
(A) Basal luciferase activity normalized by renilla activity in SH-SY5Y cells transfected with 1.8 kb, 1.8 kb C-EBPα mut and 1.8 kb WT1 mut. (B) Induction of human wild-type 1.8 kb p73 promoter fragment and of the same fragment with a mutation in the C-EBPα binding site (C-EBPα mut) or in the WT1 binding site (WT1 mut) by 24 h treatment with DX.
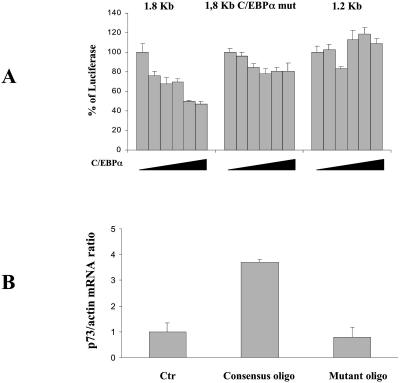
To further analyze whether the C-EBPα activity could be related to an inhibitory effect on E2F1, we co-transfected the 1.8 kb fragment with an E2F1 expression vector and increasing concentrations of C-EBPα expression vector in HCT116 cells. We used these cells because the transfection efficiency was higher than in SH-SY5Y cells and because DX in these cells was able to induce the p73 promoter at levels similar to those observed in neuroblastoma cells (data not shown). As seen in Figure 6A, a concentration-dependent reduction in E2F1-induced activation of the promoter fragment was found when the wt C-EBPα expression plasmid was used. This effect was lost when the C-EBPα mutated 1.8 kb fragment or the 1.2 kb fragment (lacking the C-EBPα binding site) was used. Furthermore, transfection of increasing amounts of C-EBPα in SH-SY5Y cells was able to block the ability of DX to induce p73 mRNA levels (see Supplementary Material, Annex 2).
Figure 6.
(A) Effect of increasing amount of C-EBPα expression vector on the activity of different p73 promoter fragments. Aliquots of 5 µg of the indicated p73 promoter fragments were co-transfected into SH-SY5Y cells together with 5 µg of E2F1 expression plasmid and with increasing amounts (1–10 µg) of C-EBPα expression plasmid. Luciferase activity was detected 48 h after transfection. (B) Effect of an oligonucleotide containing the binding site for C-EBPα on endogenous p73 mRNA levels. SH-SY5Y cells were treated with 20 µM oligonucleotide containing the consensus sequence and the levels of p73 measured by real time PCR 24 h after. As a control the same oligonucleotide containing a mutation in the C-EBPα binding site was used.
To test whether the block of C-EBPα activity was associated with an increased basal level of p73, we used oligonucleotides containing the binding site for C-EBPα which had previously been shown to be able to block C-EBPα activity (23). Treatment of cells with this oligonucleotide resulted in an increased level of p73 mRNA (Fig. 6B). As a control the same oligonucleotide with a mutation in the C-EBPα binding site did not show this effect (Fig. 6B).
Having shown that C-EBPα was able to repress E2F1-induced activation of the p73 promoter, we checked whether DX-induced p73 transcriptional activation was linked to C-EBPα.
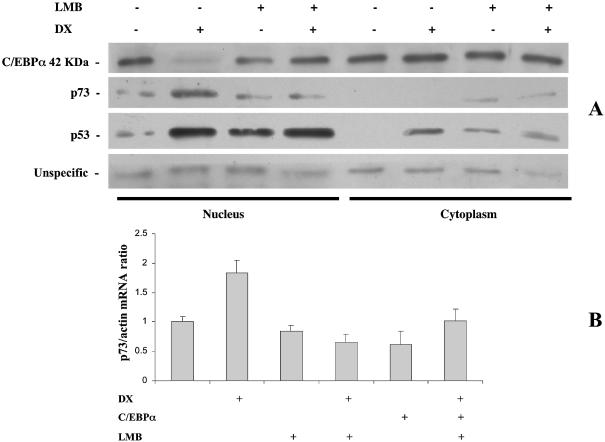
SH-SY5Y cells were treated with DX and the levels of C-EBPα analyzed in nuclei and cytoplasm. The appropriate separation of nuclei and cytoplasm was checked using anti-topoisomerase I antibodies, which were almost exclusively present in the nuclear fraction (see Supplementary Material, Annex 3). DX treatment was able to drastically reduce the nuclear levels of C-EBPα (Fig. 7A), while no significant differences could be observed in the cytoplasm.
Figure 7.
(A) Levels of C-EBPα, p53 and p73 in nuclei and cytoplasm of SH-SY5Y cells after treatment with 1 µM DX in the presence or absence of 20 ng/ml LMB. (B) p73 mRNA levels analyzed by real time PCR in SH-SY5Y cells treated with 1 µM DX in the absence or presence of 20 ng/ml LMB.
Pretreatment of cells with the nuclear export inhibitor LMB abolished the nuclear disappearance of C-EBPα. Furthermore, pretreatment of cells with LMB was able to block DX induction of p73 (Fig. 7A). This was also confirmed in real time PCR experiments, in which LMB treatment was able to prevent DX-induced activation of p73 (Fig. 7B). In both cases LMB alone did not modify p73 levels, while it was able to induce accumulation of p53 in the nuclei.
DISCUSSION
We have reported in this manuscript that treatment with DNA damaging agents such as DX and DDP is able to induce increased levels of p73 in cancer cells growing in vitro. This increase is observed at both the protein and RNA levels and we present evidence that these two drugs induce direct transcriptional activation of p73.
Even if a potential binding site for p53 is present in the p73 promoter fragment (24), we present data obtained using p73 promoter fragments lacking the putative p53 binding site that clearly indicate that DX-induced transcriptional activation of p73 is p53-independent. The situation is therefore different from that of the truncated DN form of p73, which uses a different promoter regulated by p53 (25,26).
The cellular response to DNA damage involves the activation of different proteins which in turn activate a cascade of signaling events. This brings about cell cycle arrest, to allow repair of the lesion, or apoptosis, depending on the kind of damage and cellular context.
The product of the tumor suppressor gene p53 is one of the key proteins reported to be a strong determinant of cancer cell response to anticancer agent treatment (27–30). Again, depending on the cellular context and on the type of drug used, p53 has been reported either to increase or to decrease the activity of those agents (31–34). p53 is normally expressed at a very low concentration, but is rapidly activated following stress induction, thus acting as a transcriptional factor inducing the expression of genes such as p21, bax, GADD45 and many others (2,35–38).
p73 behaves differently from p53, showing only a quantitatively moderate activation following damage. In particular, it has been reported that DDP, a drug also used in the present study, was able to induce p73 in a c-abl-mediated fashion through a mechanism involving stabilization of the protein (8,9,39).
As for the transcription of p73, the transcription factor E2F1 is one of the key regulators and, in fact, three different binding sites are present in the 5′ region of the p73 gene here analyzed (20–22,40). We confirmed in a different cell context that E2F1 expression is associated with increased p73 promoter activity and that the use of a transactivation-deficient form of E2F1 reduces the DX-associated activation of p73. Our results also indicate that DX-induced expression of p73 is not attributable to induced expression of E2F1, suggesting that other mechanisms leading to E2F1 activation are present. Furthermore, the use of promoter fragments with a different number of E2F1 binding sites suggested that other factors could play an important role, particularly as negative regulators of E2F1.
The minimal promoter fragment which seems to be responsible for the DX-induced transcriptional activation of p73 is the region located between –1800 and –1200 bp with respect to the transcription start site. This region contains putative binding sites for transcriptional factors such as WT1 and C-EBPα. We present evidence that C-EBPα negatively regulates the activity of E2F1, acting as a repressor. DNA damage induction is able to relieve the C-EBPα-mediated repression, thus allowing the transfactor E2F1 to activate p73 transcription. C-EBPα has been reported to act as a transcriptional repressor for other transcription factors, including E2F1 (41–45). Furthermore, it has already been reported that C-EBPα potentially binds to E2F1 (46) and we have data showing that in SH-SY5Y cells C-EBPα and E2F1 co-immunoprecipitate and that DX treatment is able to reverse this effect (see Supplementary Material, Annex 4). Our data suggest that C-EBPα repression of E2F1-mediated activation of p73 requires the binding of C-EBPα to its consensus sequence contained in the p73 promoter, suggesting the formation of a complex at the DNA site between C-EBPα, E2F1 and, perhaps, other proteins. This is confirmed by the evidence that DX treatment is able to induce nuclear export of C-EBPα, which would allow E2F1 to fully operate on its binding site. This is further corroborated by use of the crm1 inhibitor LMB, which by blocking the export of C-EBPα is able to block DX-induced activation of p73.
LMB has been shown to interfere with the nuclear export of both p53 and p73 (47,48). These results, although providing strong evidence that nuclear import–export is an important regulator of both p53 and p73 stability, have been produced in the absence of DNA damage. The lack of p73 induction following damage in cells pretreated with LMB therefore does not contradict previous reports.
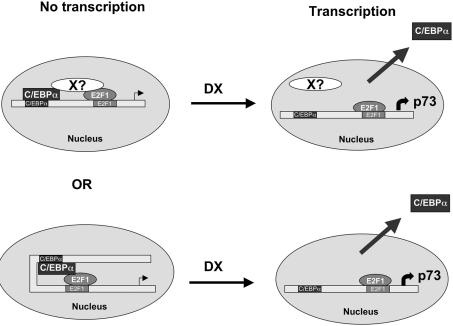
The mechanism by which DX induces the nuclear export of C-EBPα is at present unknown. We can envisage (Fig. 8) two possible mechanisms: one in which C-EBPα and E2F1, bound to their respective binding sites, directly contact each other; alternatively, an additional (or more than one) protein binds to both C-EBPα and E2F1 and acts as a repressor. In both cases DX would destroy the complex allowing full activation of E2F1. This represents a new mechanism of DNA damage-induced activation of p73, which fits with a model in which p73 has to be quickly activated following damage. In fact, removal of the repression of binding of already formed E2F1 induced by C-EBPα would not need the transcription of new E2F1 to induce p73. Additionally, these findings raise the possibility of using alternative ways to induce or repress transcription of p73.
Figure 8.
Proposed mechanisms of DX activity on C-EBPα-induced repression of E2F1.
Moreover, apart from the evidence that C-EBPα plays a role in the p73 response following damage, it would be interesting to test whether a relationship could be found between the expression of p73, often increased in human tumors (7,49), and the presence of C-EBPα. This would then provide a basis for new therapeutic strategies aimed at modifying/modulating p73 levels. This hypothesis is currently under investigation in the laboratory.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was partially supported by a grant from the Ministero della Salute. M.M. is a fellow of the Monzino Foundation. F.V. is a visiting scientist from The Institute of Cytology, Russian Academy of Science, St Petersburg, Russia.
REFERENCES
- 1.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 3.Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Regulation of p53 stability by Mdm2. Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 4.Kaghad M., Bonnet,H., Yang,A., Creancier,L., Biscan,J.C., Valent,A., Minty,A., Chalon,P., Lelias,J.M., Dumont,X. et al. (1997) Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell, 90, 809–819. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J., Jiang,J., Zhou,W. and Chen,X. (1998) The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res., 58, 5061–5065. [PubMed] [Google Scholar]
- 6.Costanzo A., Merlo,P., Pediconi,N., Fulco,M., Sartorelli,V., Cole,P.A., Fontemaggi,G., Fanciulli,M., Schiltz,L., Blandino,G. et al. (2002) DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell, 9, 175–186. [DOI] [PubMed] [Google Scholar]
- 7.Moll U.M., Erster,S. and Zaika,A. (2001) p53, p63 and p73—solos, alliances and feuds among family members. Biochim. Biophys. Acta, 1552, 47–59. [DOI] [PubMed] [Google Scholar]
- 8.Agami R., Blandino,G., Oren,M. and Shaul,Y. (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis [see comments]. Nature, 399, 809–813. [DOI] [PubMed] [Google Scholar]
- 9.Gong J.G., Costanzo,A., Yang,H.Q., Melino,G., Kaelin,W.G.,Jr, Levrero,M. and Wang,J.Y. (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage [see comments]. Nature, 399, 806–809. [DOI] [PubMed] [Google Scholar]
- 10.Hollstein M., Soussi,T., Thomas,G., von Brevern,M.C. and Bartsch,H. (1997) P53 gene alterations in human tumors: perspectives for cancer control. Recent Results Cancer Res., 143, 369–389. [DOI] [PubMed] [Google Scholar]
- 11.Nomoto S., Haruki,N., Kondo,M., Konishi,H. and Takahashi,T. (1998) Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res., 58, 1380–1383. [PubMed] [Google Scholar]
- 12.Takahashi H., Ichimiya,S., Nimura,Y., Watanabe,M., Furusato,M., Wakui,S., Yatani,R., Aizawa,S. and Nakagawara,A. (1998) Mutation, allelotyping and transcription analyses of the p73 gene in prostatic carcinoma. Cancer Res., 58, 2076–2077. [PubMed] [Google Scholar]
- 13.Han S., Semba,S., Abe,T., Makino,N., Furukawa,T., Fukushige,S., Takahashi,H., Sakurada,A., Sato,M., Shiiba,K. et al. (1999) Infrequent somatic mutations of the p73 gene in various human cancers. Eur. J. Surg. Oncol., 25, 194–198. [DOI] [PubMed] [Google Scholar]
- 14.Casciano I., Ponzoni,M., Cunsolo,C., Tonini,G. and Romani,M. (1999) Different p73 splicing variants are expressed in distinct tumour areas of a multifocal neuroblastoma. Cell Death Differ., 6, 391–393. [DOI] [PubMed] [Google Scholar]
- 15.Yokomizo A., Mai,M., Tindall,D.J., Cheng,L., Bostwick,D.G., Naito,S., Smith,D.I. and Liu,W. (1999) Overexpression of the wild type p73 gene in human bladder cancer. Oncogene, 18, 1629–1633. [DOI] [PubMed] [Google Scholar]
- 16.Tokuchi Y., Hashimoto,T., Kobayashi,Y., Hayashi,M., Nishida,K., Hayashi,S., Imai,K., Nakachi,K., Ishikawa,Y., Nakagawa,K. et al. (1999) The expression of p73 is increased in lung cancer, independent of p53 gene alteration. Br. J. Cancer, 80, 1623–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovalev S., Marchenko,N., Swendeman,S., LaQuaglia,M. and Moll,U.M. (1998) Expression level, allelic origin and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ., 9, 897–903. [PubMed] [Google Scholar]
- 18.Corn P.G., Kuerbitz,S.J., van Noesel,M.M., Esteller,M., Compitello,N., Baylin,S.B. and Herman,J.G. (1999) Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt’s lymphoma is associated with 5′ CpG island methylation. Cancer Res., 59, 3352–3356. [PubMed] [Google Scholar]
- 19.Vikhanskaya F., Clerico,L., Valenti,M., Stanzione,M.S., Broggini,M., Parodi,S. and Russo,P. (1997) Mechanism of resistance to cisplatin in a human ovarian-carcinoma cell line selected for resistance to doxorubicin: possible role of p53. Int. J. Cancer, 72, 155–159. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y., Inoue,T., Kamiyama,J., Tamura,Y., Ohtani-Fujita,N., Igata,E. and Sakai,T. (1999) Molecular cloning and functional characterization of the upstream promoter region of the human p73 gene. DNA Res., 6, 347–351. [DOI] [PubMed] [Google Scholar]
- 21.Irwin M., Marin,M.C., Phillips,A.C., Seelan,R.S., Smith,D.I., Liu,W., Flores,E.R., Tsai,K.Y., Jacks,T., Vousden,K.H. et al. (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature, 407, 645–648. [DOI] [PubMed] [Google Scholar]
- 22.Stiewe T. and Putzer,B.M. (2000) Role of the p53-homologue p73 in E2F1-induced apoptosis. Nature Genet., 26, 464–469. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg E., Li,F., Reisher,S.R., Wang,M., Gonzales,L.W., Ewing,J.R., Malek,S., Ballard,P.L., Notarfrancesco,K., Shuman,H. et al. (2002) Members of the C/EBP transcription factor family stimulate expression of the human and rat surfactant protein A (SP-A) genes. Biochim. Biophys. Acta, 1575, 82–90. [DOI] [PubMed] [Google Scholar]
- 24.Chen X., Zheng,Y., Zhu,J., Jiang,J. and Wang,J. (2001) p73 is transcriptionally regulated by DNA damage, p53 and p73. Oncogene, 20, 769–774. [DOI] [PubMed] [Google Scholar]
- 25.Vossio S., Palescandolo,E., Pediconi,N., Moretti,F., Balsano,C., Levrero,M. and Costanzo,A. (2002) DN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncogene, 21, 3796–3803. [DOI] [PubMed] [Google Scholar]
- 26.Kartasheva N.N., Contente,A., Lenz-Stoppler,C., Roth,J. and Dobbelstein,M. (2002) p53 induces the expression of its antagonist pt′DN, establishing an autoregulatory feedback loop. Oncogene, 21, 4715–4725. [DOI] [PubMed] [Google Scholar]
- 27.Lowe S.W. (1995) Cancer therapy and p53. Curr. Opin. Oncol., 7, 547–553. [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb T.M. and Oren,M. (1996) p53 in growth control and neoplasia. Biochim. Biophys. Acta, 1287, 77–102. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal M.L., Taylor,W.R., Chernov,M.V., Chernova,O.B. and Stark,G.R. (1998) The p53 network. J. Biol. Chem., 273, 1–4. [DOI] [PubMed] [Google Scholar]
- 30.Ko L.J., Prives,C., Maki,C.G., Huibregtse,J.M. and Howley,P.M. (1996) p53: puzzle and paradigm. Genes Dev., 56, 2649–2654. [DOI] [PubMed] [Google Scholar]
- 31.Fan S., Smith,M.L., Rivet,D.J., Duba,D., Zhan,Q., Kohn,K.W., Fornace,A.J.J. and O’Connor,P.M. (1995) Disruption of p53 function sensitizes breast cancer MCF-7 cells to cisplatin and pentoxifylline. Cancer Res., 55, 1649–1654. [PubMed] [Google Scholar]
- 32.Brown J.M. and Wouters,B.G. (1999) Apoptosis, p53 and tumor cell sensitivity to anticancer agents. Cancer Res., 59, 1391–1399. [PubMed] [Google Scholar]
- 33.Bunz F., Hwang,P.M., Torrance,C., Waldman,T., Zhang,Y., Dillehay,L., Williams,J., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (1999) Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest., 104, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vikhanskaya F., Vignati,S., Beccaglia,P., Ottoboni,C., Russo,P., D’Incalci,M. and Broggini,M. (1998) Inactivation of p53 in a human ovarian cancer cell line increases the sensitivity to taxol by inducing G2 arrest and apoptosis. Exp. Cell Res., 241, 96–101. [DOI] [PubMed] [Google Scholar]
- 35.Shieh S.Y., Ikeda,M., Taya,Y. and Prives,C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell, 91, 325–334. [DOI] [PubMed] [Google Scholar]
- 36.el Deiry W.S., Tokino,T., Velculescu,V.E., Levy,D.B., Parsons,R., Trent,J.M., Lin,D., Mercer,W.E., Kinzler,K.W. and Vogelstein,B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 37.Kastan M.B., Zhan,Q., el Deiry,W.S., Carrier,F., Jacks,T., Walsh,W.V., Plunkett,B.S., Vogelstein,B. and Fornace,A.J.J. (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 38.Bates S. and Vousden,K.H. (1996) p53 in signaling checkpoint arrest or apoptosis. Curr. Opin. Genet. Dev., 6, 12–18. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Z.M., Shioya,H., Ishiko,T., Sun,X., Gu,J., Huang,Y.Y., Lu,H., Kharbanda,S., Weichselbaum,R. and Kufe,D. (1999) p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature, 399, 814–817. [DOI] [PubMed] [Google Scholar]
- 40.Seelan R.S., Irwin,M., van der Stoop,P., Qian,C., Kaelin,W.G.,Jr and Liu,W. (2002) The human p73 promoter: characterization and identification of functional E2F binding sites. Neoplasia, 4, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haaxma C.A., Kim,P.K., Andrejko,K.M., Raj,N.R. and Deutschman,C.S. (2003) Transcription factors C/EBP-alpha and HNF-1alpha are associated with decreased expression of liver-specific genes in sepsis. Shock, 19, 45–49. [DOI] [PubMed] [Google Scholar]
- 42.Timchenko L.T., Iakova,P., Welm,A.L., Cai,Z.J. and Timchenko,N.A. (2002) Calreticulin interacts with C/EBPalpha and C/EBPbeta mRNAs and represses translation of C/EBP proteins. Mol. Cell. Biol., 22, 7242–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramji D.P. and Foka,P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J., 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slomiany B.A., D’Arigo,K.L., Kelly,M.M. and Kurtz,D.T. (2000) C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol., 20, 5986–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffensen K.R., Schuster,G.U., Parini,P., Holter,E., Sadek,C.M., Cassel,T., Eskild,W. and Gustafsson,J.A. (2002) Different regulation of the LXRalpha promoter activity by isoforms of CCAAT/enhancer-binding proteins. Biochem. Biophys. Res. Commun., 293, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 46.Johansen L.M., Iwama,A., Lodie,T.A., Sasaki,K., Felsher,D.W., Golub,T.R. and Tenen,D.G. (2001) c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol. Cell. Biol., 21, 3789–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lain S., Midgley,C., Sparks,A., Lane,E.B. and Lane,D.P. (1999) An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp. Cell Res., 248, 457–472. [DOI] [PubMed] [Google Scholar]
- 48.Inoue T., Stuart,J., Leno,R. and Maki,C.G. (2002) Nuclear import and export signals in control of the p53-related protein p73. J. Biol. Chem., 277, 15053–15060. [DOI] [PubMed] [Google Scholar]
- 49.Sayan A.E., Sayan,B.S., Findikli,N. and Ozturk,M. (2001) Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene, 20, 5111–5117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.