Abstract
Detailed comparisons of 16 editosome proteins from Trypanosoma brucei, Trypanosoma cruzi and Leishmania major identified protein motifs associated with catalysis and protein or nucleic acid interactions that suggest their functions in RNA editing. Five related proteins with RNase III-like motifs also contain a U1-like zinc finger and either dsRBM or Pumilio motifs. These proteins may provide the endoribonuclease function in editing. Two other related proteins, at least one of which is associated with U-specific 3′ exonuclease activity, contain two putative nuclease motifs. Thus, editosomes contain a plethora of nucleases or proteins presumably derived from nucleases. Five additional related proteins, three of which have zinc fingers, each contain a motif associated with an OB fold; the TUTases have C-terminal folds reminiscent of RNA binding motifs, thus indicating the presence of numerous nucleic acid and/or protein binding domains, as do the two RNA ligases and a RNA helicase, which provide for additional catalytic steps in editing. These data indicate that trypanosomatid RNA editing is orchestrated by a variety of domains for catalysis, molecular interaction and structure. These domains are generally conserved within other protein families, but some are found in novel combinations in the editosome proteins.
INTRODUCTION
Most mitochondrial mRNAs in trypanosomes undergo RNA editing to produce mature and functional mRNAs through a series of coordinated steps, which are catalyzed by a multi-protein complex (the editosome) that inserts and deletes uridylates (Us) as specified by guide RNAs (gRNAs) (1–3). Each gRNA has a 5′ region that is complementary to sequence that is adjacent to the block of sequence that is edited, as specified by the informational region of the gRNA. The gRNA also has a 3′ U-tail that is added post-transcriptionally, which may stabilize the gRNA/pre-mRNA interaction. During editing, the mRNA undergoes endonucleolytic cleavage at the editing site, 3′ terminal uridylyltransferase (TUTase) adds Us in insertion editing or 3′ exouridylylase (exoUase) removes Us in deletion editing, both at the 3′ end of the 5′ cleavage fragment, depending on the interaction with the gRNA; the processed 5′ fragments are then ligated. Each gRNA specifies the editing of several sites and multiple gRNAs are used to edit most mRNAs. Hence, the gRNAs must be displaced from each editing site and, ultimately, the mRNA, perhaps by RNA helicase (4,5).
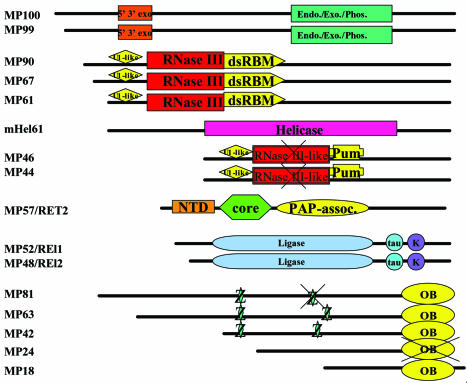
The number of proteins in the fully functional editosome in Trypanosoma brucei is not yet known, but 16 proteins that are stable components of the editosome have been identified (6,7). Some editosome proteins have motifs suggestive of nucleic acid interaction. Five of these, TbMP81, TbMP63, TbMP42, TbMP24 and TbMP18, are related to varying degrees and three of these have zinc fingers (8). TbMP18 has a single-stranded DNA binding motif, TbMP24 an S1 motif and TbMP46 a Pumilio RNA binding motif. Five other proteins, TbMP61, TbMP67, TbMP90, TbMP46 and TbMP44, also have varying degrees of similarity and contain RNase III or RNase III-like motifs, while TbMP100 and TbMP99 are related and share an AP endonuclease motif (6). Two closely related editosome proteins (TbREL1 and TbREL2) have been shown to be RNA editing ligases (9–12), another is an RNA helicase (5) and a 57 kDa editosome 3′ TUTase (TbRET2) has also been identified (13,14). Another 3′ TUTase, TbRET1, is essential for editing, but it does not appear to be stably associated with the editosome and thus may add Us to gRNA (13,14). A complex that was isolated from mitochondrial extracts of Leishmania tarentolae contains 13 proteins, six of which are homologous to TbMP81, TbMP63, TbREL1, TbREL2, TbMP42, TbMP24 and TbMP18 (15).
The catalytic characteristics of the two editing ligases, and their functions and patterns of physical association with other editosome proteins, indicate that insertion and deletion editing activities are segregated. There appear to be two distinct catalytic subcomplexes; one that contains TbREL1, TbMP63 and TbMP99 that can catalyze pre-cleaved (independent of endonuclease) in vitro deletion editing, while the other contains TbREL2, TbMP81 and TbRET2 and can catalyze pre-cleaved in vitro insertion editing (16).
While RNA editing occurs in all trypanosomatids, the mRNAs that are edited and the extent to which they are edited differs among the species. For example, two domains of ND7 mRNA are extensively edited in T.brucei and Trypanosoma cruzi while only the 5′ ends of these domains are edited in Leishmania (17). It has been proposed that the more extensive editing is ancestral to the more limited editing in monogenetic trypanosomatids and Leishmania (18,19). In addition, editing of the mRNA is itself regulated during the life cycle of these organisms, during which there are changes in the modes of energy generation (20–22). The mRNAs for cytochromes are preferentially edited in T.brucei procyclic insect forms in which significant energy is generated by cytochrome- mediated oxidative phosphorylation. In contrast, these mRNAs are not edited in mammalian bloodstream forms (BFs), in which energy is generated by glycolysis (23). Similarly, mRNAs for components of NADH dehydrogenase are preferentially edited in BFs. There may be differences in regulation of editing in T.cruzi and Leishmania since oxidative phosphorylation in these species appears to be essential for parasite survival throughout the life cycle, suggesting that cytochrome mRNAs may be edited in the mammalian stages as well. Editing is essential in the BFs of T.brucei (9), which is surprising, since some mutant BFs can survive the loss of mitochondrial DNA and, consequentially, mitochondrial gene expression (24,25). Hence, BF T.brucei are capable of genetic or physiological compensation for the loss of editing or the essential proteins may have additional roles.
This study reports the identification of T.cruzi and L.major homologs of 16 T.brucei genes for editosome proteins and the sequence similarity among them. It utilized the substantial data from the T.brucei, T.cruzi and L.major genome sequencing projects (http://www.geneDB.org/ and http://www.tigr.org/tdb/mdb/tcdb/) that have completed sequencing several chromosomes and provided (to date unfinished and unannotated) sequences of essentially the entire genomes of these three kinetoplastids. Related genes in other organisms, as well as functional motifs and structural similarities, were identified using a variety of algorithms, including BlastP and a more sensitive Hidden Markov Model (HMM). The conserved features as well as their novel arrangements suggest specific functions for the editosome proteins.
MATERIALS AND METHODS
The candidate editosome proteins were designated TbMP#, TcMP# and LmMP#, where # indicates the T.brucei predicted precursor protein mass in kDa. Whenever possible, the orthologs were obtained in silico from the L.major (http://www.sanger.ac.uk) and T.cruzi (http://www.tigr.org) genome sequence databases. Where this was not possible due to incomplete coverage within these databases, the genes were cloned by RT–PCR, making use of conserved splice leader sequences or polyadenylation sites as previously described (6). To identify the potential trypanosomatid orthologs of T.brucei two criteria were considered: (i) in a database similarity search of all of the proteome of T.cruzi or L.major the best hit with the highest expect value (E) and the most similar protein in length and sequence was identified; (ii) the T.brucei protein used in the first criterion was the best hit when either T.cruzi or L.major potential orthologs were used to query the proteome of T.brucei. The degree of coverage of coding regions within these genomes is sufficient to lead us to believe that we have obtained true orthologous genes, rather than homologs arising from duplication events.
We used the same general computational analyses for each protein, as detailed in previous publications (26–29). In brief, we identified sequence homologs by performing database searches using the NCBI Web interface to the program PSI-BLAST run with default parameters (http://www.ncbi.nlm.nih.gov/blast/) (30). This included searches against the PSSMs in the Conserved Domain Database (CDD). Next, we used pairwise PSI-BLAST alignments and the CDD alignments as the starting point to estimate a HMM for a protein domain/sequence family of interest. The SAM HMM algorithm was then used (31) to build models for the homologs identified in the initial sequence searching step (http://www.soe.ucsc.edu/research/compbio/sam.html) (28,29). We then employed the ensuing HMM to generate a multiple sequence alignment of the proteins used to estimate the model. Using Alscript version 2.0, these alignments were formatted for publication so that residues with similar properties are shaded in the same hue. These similarly shaded groups consist of [S, T, N and Q], [H, R and K], [D, E and C], [V, I, L, P, G and A] and [F, Y and W].
For a number of these protein domains, secondary structures were predicted using a variety of algorithms: PROFsec (http://cubic.bioc.columbia.edu/predictprotein/; 30); SAM-T02 (http://www.soe.ucsc.edu/research/compbio/HMM-apps/T02-query.html; 31); PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/; 32). Multiple EM for Motif Elicitation (MEME) was used to locate conserved ungapped short motifs within a group of related, unaligned sequences (32).
RESULTS AND DISCUSSION
Editing proteins in trypanosomatid species
The relationships among 16 homologous protein components of the T.brucei, T.cruzi and L.major editosomes were determined using Clustal-W (33) as summarized in Table 1 and diagrammed in Figure 1. The T.brucei and T.cruzi homologs were more similar to each other than to the L.major homologs, consistent with previous estimates of the phylogenetic relationships among these organisms (34). The L.major homologs are generally tens of amino acids larger than the corresponding T.brucei or T.cruzi proteins except for the absence of an ∼250 amino acid C-terminal region in LmMP99 (see below). For example, L.major MP90, MP81 and MP46 are, respectively, 204/243, 227/224 and 154/168 amino acids larger than their homologs in the other two species. The additional sequence is distributed throughout these proteins without any significant similarity to known motifs in the databases, which is consistent with a general observation in Leishmania proteins (P.Myler, personal communication).
Table 1. Candidate editosome proteins.
| Trypanosoma brucei proteina | Trypanosoma cruzi protein | Leishmania major protein | |||||
|---|---|---|---|---|---|---|---|
| Amino acids | I (%)d | S (%)e | Amino acidsb | I (%)d | S (%)e | Amino acidsc | |
| MP100 | 902 | 67 | 76 | 924 | 39 | 45 | 992 |
| MP99 | 907 | 65 | 75 | 899 | 40 | 55 | 656 |
| MP90 | 818 | 58 | 81 | 779 | 24 | 51 | 1022 |
| MP81 | 762 | 49 | 76 | 765 | 30 | 54 | 989 |
| MP67 | 596 | 48 | 77 | 587 | 24 | 51 | 631 |
| MP63 | 588 | 60 | 85 | 590 | 38 | 70 | 623 |
| MP61 | 538 | 70 | 87 | 532 | 39 | 70 | 587 |
| mHEL61p | 546 | 61 | 80 | 498 | 49 | 77 | 544 |
| MP57 (RET2) | 487 | 87 | 96 | 486 | 66 | 87 | 510 |
| MP52 (REL1) | 469 | 81 | 92 | 462 | 65 | 85 | 490 |
| MP48 (REL2) | 416 | 66 | 80 | 356 | 65 | 90 | 418 |
| MP46 | 414 | 54 | 80 | 400 | 25 | 52 | 568 |
| MP44 | 429 | 67 | 87 | 387 | 42 | 64 | 429 |
| MP42 | 448 | 59 | 85 | 395 | 45 | 73 | 448 |
| MP24 | 246 | 40 | 66 | 215 | 16 | 45 | 246 |
| MP18 | 229 | 81 | 92 | 161 | 44 | 64 | 229 |
aPre-processed mitochondrial protein followed by size in kilodalton.
b,cNumber of amino acids used in the alignment in T.cruzi or L.major, respectively.
dPercent identity.
ePercent similarity.
Figure 1.
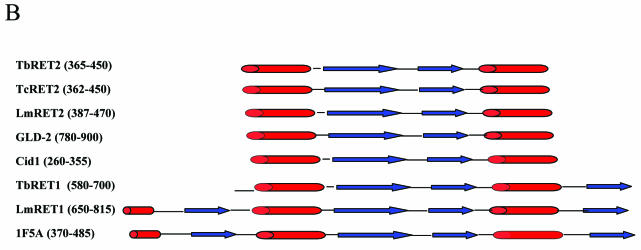
Domain architecture of the editosome proteins. Domain abbreviations: 5’ 3’ exo, 5′→3′ exoribonucleases; Endo./Exo./Phos., endonuclease/ exonuclease/phosphatase; RNase, ribonuclease; Z, C2H2 zinc finger; U1-like, U1-like zinc finger; dsRBM, double-stranded RNA binding motif; Pum, Pumilio domain; NTD, nucleotidyltransferase domain; core, poly(A) polymerase core; PAP-assoc., poly(A) polymerase-associated domain; tau, microtubule-associated τ; K, kinesin light chain; OB, OB fold. Crossed symbols indicate potentially disrupted domains.
Proteins with RNase III motifs
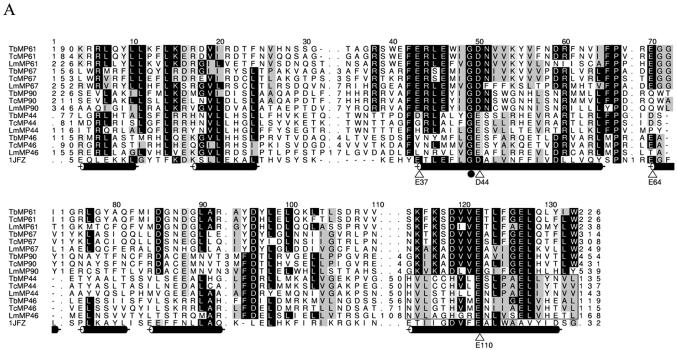
The canonical RNase III motif is present in MP61, MP67, MP90, MP44 and MP46 to varying degrees (Fig. 2A). All contain the G43 and E110 (numbered as per Aquifex aeolicus 1JFZ), which are invariantly conserved in RNase IIIs and have been shown to be important for activity in Escherichia coli and yeast (35). In addition, four interacting residues (E37 with E64 and D44 with E110) are conserved in MP61, MP67 and MP90, except for a substitution of E64 for Q in the MP90 homologs. However, the Q64 substitution may conserve the interactions in the catalytic site since it may still form the critical hydrogen bond with the carboxylate group of E37, as implied by the conservation of RNase III activity in an E.coli E37Q mutant (35). MP44 and MP46 have fewer matches to the canonical RNase III motif, do not conserve E37 or D44 nor invariably conserve E64, implying divergence in, or loss of, the catalytic site. Instead, many of the orthologs contain hydrophobic amino acids at E37 and E64, except for TbMP44, which has D substitutions that might still interact through hydrogen bonds. A similar substitution (E38V) rendered E.coli RNase III inactive (35). Thus, any cleavage by these proteins might entail intramolecular interactions of the semi-conserved E44 and E110.
Figure 2.
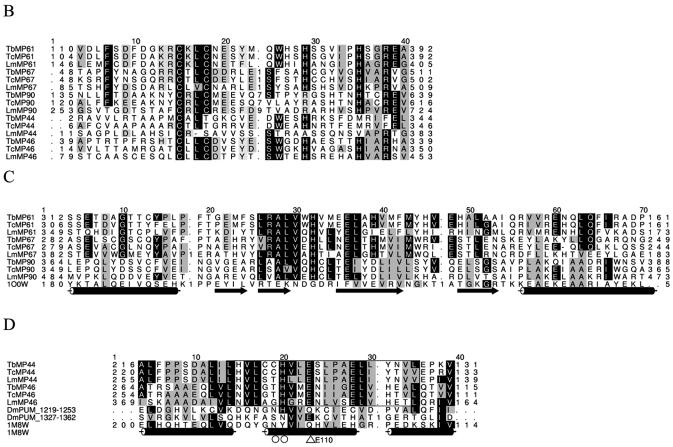
HMM-generated multiple sequence alignment of the RNase III and RNA binding domains in the RNase III family members of the editosome. (A) Alignment of the RNase III motifs. Cylinders denote the α-helices in the structure of the endonuclease domain from A.aeolicus (1JFZ) (35). Triangles indicate residues involved in the proposed RNA cleavage. The boxed region indicates the Pumilio RNA binding unit. (B) Alignment of the N-terminal region of RNase III family members containing the U1-like zinc finger domain. (C) Alignment of putative dsRBM of MP61, MP67, MP90 and RNase III from Thermotoga maritima (10OW) with α-helices denoted as cylinders and β-strands as arrows. (D) Alignment of homologous Pumilio regions of MP44, MP46 and D.melanogaster Pumilio protein (DmPUM, Swiss-Prot accession no. P25822). Cylinders denote the α-helices in the structure of 1M8W, the Pumilio-homology domain from human Pumilio1 (39). Important residues involved in RNA binding are indicated by circles.
Each of the five RNase III-like proteins in each species conserved a U1-like zinc finger domain (SM00451) (Fig. 2B). A U1-like zinc finger domain is located at the N-terminal end of human snRNP protein C, and its binding to U1 snRNP is dependent on the presence of both U1 snRNA and one or more of the U1 snRNP proteins (36). This implies that these RNase III-like proteins may interact with one or more of the editosome proteins as well as substrate RNAs. The loss of editing and disruption of the editosome and loss of its proteins upon inactivation of TbMP44 expression is consistent with this notion (37). The MP61, MP67 and MP90 homologs also contain the double-stranded RNA binding motif (dsRBM) commonly found in RNase III proteins, with the conservation mainly localized more C-terminal (Fig. 2C). Proteins with the similar type B dsRBM conserve C-terminal sequence and although they bind dsRNA less well, they are still required for dsRNA binding (38). In contrast, dsRBMs were not detected in MP44 and MP46 proteins. However, searches of the Pfam collection identified a region in these two proteins that has similarity to the Pumilio RNA binding domain (PF00806) and overlaps the C-terminus region of the RNase III motif (Figs 1 and 2A and D). This Pumilio-like region contains a glutamic acid that corresponds to E110 in A.aeolicus. An E1346K mutation of the equivalent amino acid leads to both the disruption of RNA binding and unfolding of the protein in Drosophila melanogaster Pumilio protein (DmPUM) (39) and to substrate uncoupling in RNase III proteins (35). MP44 and MP46 are the first examples of proteins with a suggested overlap between the RNase III motif and a subunit of the Pumilio RNA binding domain. The Pumilio RNA binding domain is found in members of the Puf family of proteins, which regulate gene expression by binding to untranslated regions of their mRNA targets. Fly, human, frog and mouse Pumilio proteins bind to two conserved regions, termed box A (GUUGU) and box B (AUUGUA), with the UGU triplets in each box being important for recognition and the amino acids indicated by the circles and triangles in Figure 2D contacting RNA bases (39). Conservation at analogous positions in MP44 and MP46 suggests that they may recognize similar RNA sequences.
Taken together these data suggest that MP61, MP67, MP90, MP44 and MP46 represent a novel subfamily of RNase III proteins with functions associated with RNA editing. The endonuclease that cleaves substrate mRNA during editing has not been identified but one or more of these five proteins are obvious candidates for this function. Endonuclease activity has not been demonstrated for any of these proteins and thus it is uncertain if some or all of these proteins contain such activity. Of the five proteins, the greater conservation of the RNase III catalytic motif in MP61, MP67 and, to a lesser degree, in MP90, along with the presence of a dsRBM, indentifies these three proteins as those most likely to possess endonuclease activity. The divergence of the RNase III motif in MP44 and MP46, and lack of the dsRBM but presence of Pumilio motifs, suggests either a modification or loss of endonuclease activity. Loss of RNase III activity may not mean loss of ancestral RNase III functions such as RNA and protein recognition and binding. Alternatively, all five may have endonuclease activity, perhaps with different specificities due to a requirement for the cleavage of the thousands of different insertion and deletion editing sites that are encountered by the editosome.
Available evidence suggests that only the substrate mRNA of the mRNA/gRNA duplex is cleaved and at a single site (typically immediately 5′ to the anchor RNA duplex) during each round of editing. In contrast, RNase IIIs, which are divided into three classes based upon the presence of one or two RNase III domains and of accessory domains, cleave both RNA strands (40). Prokaryotic RNase IIIs have a single endonuclease domain linked to a dsRBM (class I) and are functionally and structurally well characterized. They act as homodimers that introduce four strand cleavages producing a 9 bp dsRNA with 2 base 3′ overhangs (35) but cleave a single strand of T phage substrates in an internal loop (41). Eukaryotic RNase IIIs produce larger products as a consequence of protein structure, subunit interaction and substrate specificity (42). The prokaryotic RNase III homodimers have four catalytic sites, two resulting from intermolecular interaction between E37 and E64 from each monomer (A.aeolicus positions) and the other two resulting from interaction within each monomer between D44 and E110 (see Fig. 2A) (35). The interactions between these negatively charged amino acids is stabilized by metal ion binding. Hence, although MP44 and MP46 retain, or have conservative replacements of, the amino acids that interact in at least one of the catalytic sites, it appears unlikely that they cleave both mRNA and gRNA, especially at two sites each. One possibility is that the lack of E37 and E64 in MP44 and MP46 has eliminated their ability to form multiple catalytic sites, resulting in selective cleavage of only one strand, i.e. in the mRNA immediately 5′ to the anchor duplex. MP61, MP67 and MP90 may function in the cleavage of other editing sites, perhaps not adjacent to the anchor or during re-editing. Other possibilities include cleavage of gRNA, perhaps during editing or after its use, or cleavage of polycistronic pre-mRNAs (43). In addition, the specificity of the editing endonuclease activity appears to be complex since nucleotide substitutions flanking the cleavage site (editing site) can block or alter the site of cleavage and this may be reflected in the abundance of partially edited RNAs with incompletely edited ‘junction’ regions (44–46).
TUTases
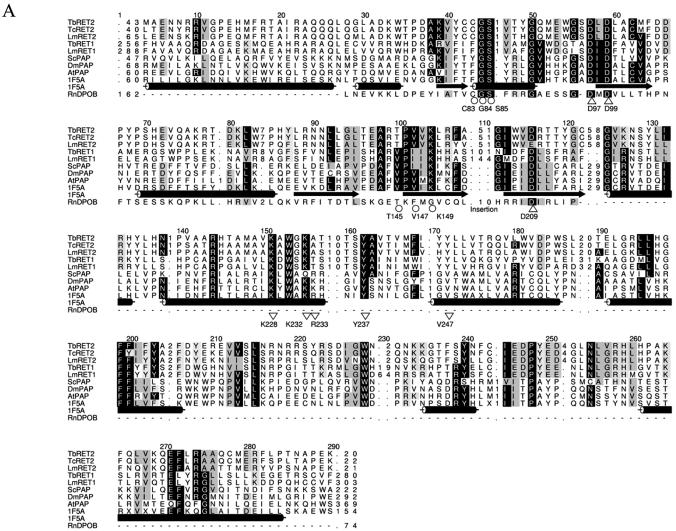
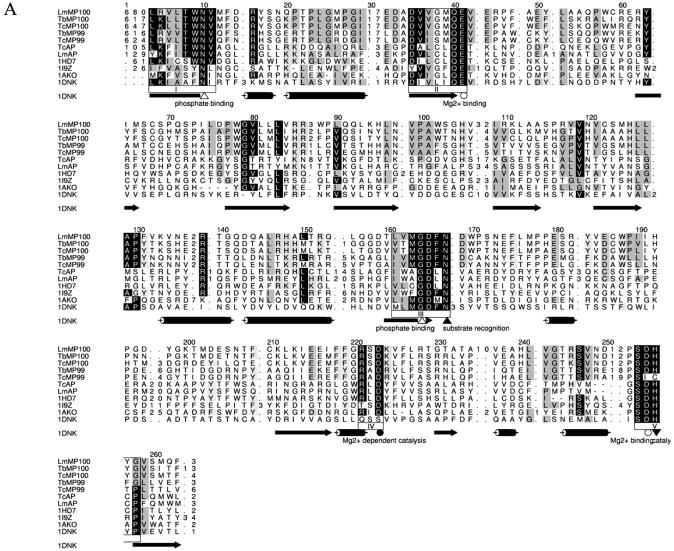
Searching of the Pfam database indicated that the editosome TUTases (i.e. RET2s) belong to the poly(A) polymerase (PAP) superfamily of nucleotidyltransferases. Classical PAPs have separate catalytic, central and RNA recognition motif (RRM)-like domains, from N- to C-terminal (47). Multiple sequence alignment of the RET2s and eukaryotic PAPs showed conservation of a nucleotidyltransferase signature motif (G[G/S]X9–13DX[D/E]), corresponding to G84, S85, D97 and D99 as well as the downstream D209 (except for a conservative substitution of glutamate in LmRET2), that is essential for catalytic activity (Fig. 3A). The conserved nucleotidyltransferase motif is most similar to that in DNA polymerase β family members, which are involved in eukaryotic base excision repair. The critical residues in DNA polymerase β that interact with nucleotides are also conserved in the RET2s at C83 and T145, consistent with TUTase function, since DNA polymerase β acts in both a template-dependent and in a stepwise manner rather than processively (48).
Figure 3.
Editosome TUTases. (A) HMM-generated multiple sequence alignment of the catalytic, central and PAP-associated domains of the RET2 TUTases, TbRET1 (GenBank accession no. AAK38334.1), LmRET1 (GenBank accession no. AY029069.1) and related nucleotidyltransferases from S.cerevisiae (ScPAP; Swiss-Prot accession no. P2968), D.melanogaster (DmPAP; DDBJ/EMBL/GenBank accession no. AAF36978) and Arabidopsis thaliana (AtPAP; DDBJ/EMBL/GenBank accession no. CAB80002). The sequence of rat DNA polymerase β (RnDPOB; Swiss-Prot accession no. P06766) is also included in the alignment. Circles and triangles highlight important conserved residues. The secondary structure elements of PAP of bovine origin (1F5A; Swiss-Prot accession no. P25500) are indicated as cylinders for α-helices and arrows for β-strands. (B) Secondary structure analysis of the C-terminal PAP-associated domains. Secondary structures for the RETs were predicted with PROFsec (see Materials and Methods for details) and aligned with cytoplasmic poly(A) polymerase from Caenorhabditis elegans (GLD-2; GenBank accession no. AAM94369), cytoplasmic poly(A) polymerase from Schizosaccharomyces pombe (Cid1; NP_594901) and PAP of bovine origin (1F5A; Swiss-Prot accession no. P25500). α-helices are represented by cylinders and β-strands are represented by arrows.
Several amino acids (D97, D99, D209, T145, V147 and K149) that can provide for nucleotide selection by interacting with the nucleotide in the catalytic domain or by metal coordination of the nucleotide are conserved among the RET2s. However, unlike most characterized proteins with a nucleotidyltransferase signature, the RET2s have a large (∼50 amino acid) insertion between aspartate D99 and D209, as previously reported, although a different aspartate was identified (15). The conservation of amino acids that are critical for PAP function along with a sizeable insertion suggests that the TUTases may have retained the primer-dependent nucleotide transfer mechanism but evolved specificity for uridine products. This suggests a mechanism consistent with the experimentally demonstrated stereochemical inversion of the UMP α-phosphate (49) upon addition of U to the 3′ end of RNA (probably by an in-line attack of the 3′-OH on the incoming UTP without a covalent enzyme–nucleotide intermediate) and release of pyrophosphate.
The RET2s also conserve elements of the central (core) domain of PAP/25A (Fig. 3A). These include K228 and K232, which have been proposed to interact with the RNA phosphate backbone, and Y237 and K228, which interact with ATP in mammalian PAP (50). Some amino acids that interact with the RNA (R233) or ATP (V145 and V247) in bovine PAP are not conserved in RET2s, perhaps reflecting the UTP and RNA substrate specificities of the editing TUTases.
Unlike the canonical PAPs, the RET2s do not contain a known RRM. However, Pfam analysis revealed a PAP-associated domain of unknown function in the C-terminal region (TbRET2 residues 365–450). Our recent in vitro data show that TbRET2 has TUTase activity which is enhanced by a binding partner, TbMP81 (13), indicating that TbRET2 can interact with RNA in the absence of the RRM motif and this may be enhanced when associated with its binding partner. Similarly, several cytoplasmic PAPs (GLD-2 and Cid1) which lack conventional RRMs, but contain the C-terminal PAP-associated domain, can interact with RNA and this interaction is enhanced in the presence of a binding partner protein (51,52). The C-termini of the RET2s have predicted HββH secondary structures, as do GLD-2 and Cid1 (Fig. 3B). The two central β-strands are topologically similar to those in the mid region of mammalian PAP RBD, which provide an interaction surface for RNA (53). Hence, we propose that the PAP-associated domain along with the conserved residues in the central domain constitute a novel RNA binding motif within the RET2s.
The RET1 U-specific nucleotidyltransferases (i.e. TUTases) such as LmRET1 and TbRET1 are not components of the editosome, but also conserve the G[G/S]X9–13DX[D/E] nucleotidyltransferase signature and the D473 aspartate (numbered with respect to TbRET1) within the catalytic region (Fig. 3A) (see above and 15). They also contain insertions of 150 and 193 amino acids, respectively, between aspartates D312 and D473, similar to the large insertions in the RET2s. However, the RET1s are more similar to PAPs than the RET2s and do not conserve similarity to DNA pol β regions, consistent with their template-independent polymerase activity. The predicted HββHβ and HβHββHβ secondary structures of the PAP/25A-associated domains of TbRET1 and LmRET1, respectively, are topologically more similar to that of the bovine PAP RRM than that in the RET2s (Fig. 3B). This suggests that RET1s (TbMP108 and LmMP120) may differ from RET2s in substrate binding specificity. Interestingly, although suggested to form a homo-tetramer in vivo, no other interacting partners have been reported for either RET1s, although in vivo interactors may exist. In addition, the conserved C2H2 type zinc fingers in TbRET1 and LmRET1 (amino acids 193–217 and 225–249, respectively) may enhance the RNA binding of these two proteins, perhaps eliminating the requirement for an interacting partner with RNA binding properties. These data are consistent with the suggested functional differences between RET2 and RET1 TUTases, namely addition of Us to mRNA during insertion editing (dsRNA substrates) versus addition of Us to the 3′ ends of gRNAs (ssRNA substrates).
MP99/MP100
TbMP100 and TbMP99 proteins have 28% overall identity and 46% similarity and sequences are highly conserved between the homologous MP99/MP100 proteins in T.brucei, L.major and T.cruzi except that LmMP99 has ∼250 fewer C-terminal amino acids. Using HMM, the C-terminal region of the MP99/MP100 homologs (absent in LmMP99) were found to be similar to a domain that is conserved in a large endonuclease/exonuclease/phosphatase (EEP) protein family (Pfam03372.5) (Fig. 4A). This family includes magnesium-dependent endonucleases such as DNase I, apurinic/apyrimidinic (AP) DNA-repair endonucleases such as exonuclease III (ExoIII) and the major human AP endonuclease (HAP1), and several phosphatases of lipid second messengers such as synaptojanin and inositol polyphosphate 5-phosphatases. DNase I and related proteins are Mg2+-dependent endonucleases, which have sequence-dependent non-specific nicking activity (54,55). AP endonucleases repair AP sites arising from hydrolytic breakage of the N-glycosylic bond, which can occur spontaneously in vivo, as an intermediate in DNA base excision repair, or as a result of damage by active oxygen species, alkylating agents or ionizing radiation (56,57). ExoIII exhibits 3′-repair diesterase, 3′→5′ exonuclease, 3′-phosphomonoesterase and RNase activities in addition to the endonuclease function (58). The HAP1 AP endonuclease lacks the 3′→5′ exonuclease activity of the E.coli protein, but has phosphodiesterase, RNase H and 3′-phosphatase activities (59,60). Inositol polyphosphate 5′-phosphatase and related proteins are Mg2+-dependent phosphatases, which can remove the phosphate from the 5′-position of the inositol ring of lipid second messengers (61).
Figure 4.
(A) HMM-generated multiple sequence alignment of the C-terminal region of the MP99 and MP100 putative exonuclease component of the editosome together with a number of related proteins in other organisms for which structures have been solved experimentally: Homo sapiens AP endonuclease (1HD7), S.pombe phosphatidylinositol phosphate phosphatases (1I9Z), E.coli exonuclease III (1AKO) and Bos taurus deoxyribonuclease I (1DNK). Cylinders denote the α-helices in the structure of 1DNK, arrows denote β-barrels. Highly conserved residues important for phosphate binding are represented as open, upwards pointing arrowheads. Those which coordinate Mg2+ are shown as open circles. Upwards pointing closed arrowheads designate residues involved in substrate recognition. Residues required for Mg2+-dependent catalysis are depicted as closed circles. Residues involved in catalysis are depicted by downwards pointing arrowheads. (B) HMM-generated multiple sequence alignment of the N-terminal region of the MP99 and MP100 proteins together with members of a 5′→3′ exonuclease domain family of proteins. Shown are mouse 5′→3′ exoribonuclease (Xrn2), the multifunctional exonuclease from S.cerevisiae (KEM1), a putative nuclease from the invertebrate iridescent virus 6 dsDNA virus (EXO2) and a Plasmodium yoelii protein with a putative 5′→3′ exonuclease domain (PY00898).
Several EEP family proteins have been characterized extensively, both structurally and functionally (58). These nucleases and metallo-dependent phosphatases share a similar 2-fold symmetric, four-layered α/β sandwich structure with the active site located on one side of the β-sheet, which assembles several conserved acidic amino acids, although their β-sheet topology and the environments around the active sites differ (62–64). Three high-resolution X-ray crystal structures of HAP1 in complex with abasic DNA have been resolved, allowing a detailed reaction mechanism for the AP endonucleases to be proposed (63). The target phosphate is oriented by a H/D pair and a conserved N. Nucleophilic attack is initiated via a conserved D, which activates a water molecule, forming the attacking nucleophile. Cleavage is achieved via inversion of the phosphate configuration. A conserved E then coordinates a Mg2+ ion that stabilizes the transition state and leaving group. Despite having relatively low sequence identity (especially at the N-terminus) and different cleavage preferences, these amino acids are well conserved amongst proteins belonging to the EEP family (see for example 63,65–67) (Fig. 4A). Although the overall degree of sequence similarity between the MP99/MP100 proteins and EEP family members is relatively low, these highly conserved amino acids are conserved (Fig. 4A). Several other amino acids (many being aromatic) are also completely conserved among the MP99 and MP100 proteins.
Secondary structure analyses of these proteins produced predictions reminiscent of the four-layered α/β sandwich structure expected for this family of proteins although some discrepancies were identified, perhaps due to the estimated 20–25% inaccuracy of the prediction methods used (data not shown). Surprisingly, given the loss of this region in the LmMP99 protein, the remaining MP99 proteins and the MP100 proteins show approximately the same degree of conservation of these structures. Three amino acids, F266, W280 and L282, in HAP1 form a hydrophobic pocket proposed to be involved in AP substrate specificity, since substitution of these amino acids results in a decrease in abasic incision potency and an increase in 3′→5′ DNA exonuclease activity (68). The aromatic amino acid is absent at the first position and the hydrophobicity of the latter amino acid is significantly diminished in the MP99 and MP100 proteins, suggesting that these proteins have no particular preference for AP sites. Searches with the MP99 and MP100 proteins identified two trypanosomatid AP endonucleases, LmAP and TcAP, that complement exonuclease III and dUTPase- deficient E.coli in expression screens and act on AP DNA (69). Perhaps these proteins, which are distinct from MP99 and MP100, retain this specificity. The substantial conservation in MP99 and MP100 of the amino acids that are absolutely conserved residues in the active site of EEP family members suggests that this region in MP99 and MP100 has Mg2+-stabilized, non-AP-specific nuclease activity. The absence of this domain in L.major MP99 indicates that the function of this protein diverges from the other MP99/MP100 proteins.
The N-terminal regions of the MP99 and MP100 orthologs are similar to a putative 5′→3′ exonuclease domain in the N-terminal regions of a large family of proteins (examples of which are shown in Fig. 4B). This domain is phylogenetically widespread and has diverse functions, including key roles in some processing pathways. For example, mouse Xrn2 5′→3′ exoribonuclease appears to trim the 5′ ends of rRNAs and snoRNAs during their maturation (70); KEM1 is a Mg2+-dependent multifunctional nuclease with 5′→3′ exonuclease activity and suspected roles in RNA turnover, DNA recombination and DNA replication (71,72). Despite weak overall homology, several amino acids, including G and charged amino acids D and E that can form hydrogen bonds, are highly conserved amongst these proteins. Although the conserved amino acids have not yet been experimentally shown to be critical for the 5′→3′ exonuclease activity, their conservation suggests this possibility. Whether MP99 and MP100 have a 5′→3′ exonuclease function in the editing complex is unknown. The current model for RNA editing does not require such an activity. However, if this region is indeed a second nuclease domain, this constitutes a novel domain arrangement.
In summary, the endo and exonuclease motifs identified in MP99 and MP100 suggest nuclease functions. Recent data suggest that TbMP99 is a U-specific editing exonuclease (16). The significance of the absence of the C-terminal EEP domain in L.major and presence of the related MP100 protein is unclear. MP99 and MP100 may be redundant, perhaps with LmMP100 functionally compensating for the absent domain in LmMP99. The functional significance of the EEP domain is also uncertain. The RNase III family of proteins seem more likely candidates for endonuclease function, but this function for them along with MP99 and MP100 cannot be excluded. Perhaps multiple endonucleases are required for the variety of substrate characteristics presented by the numerous editing sites. Alternatively, endonuclease functions may be associated with steps other than cleaving mRNA editing sites prior to U addition or removal, such as gRNA and/or polycistronic pre-mRNA processing or degradation of unedited RNAs.
REL1 (MP52) and REL2 (MP48)
The REL1 and REL2 ligases rejoin the mRNA fragments after completion of the preceding catalytic steps in deletion and insertion editing, respectively (10,16). In addition, the former, but not the latter, is essential for editing (9,10,73). Further more, TbREL1 and TbREL2 specifically interact with TbMP63 and TbMP81, respectively, but the domains responsible for the interactions have not yet been identified. Whether these differences reflect distinct catalytic properties of the ligases and/or their physical location in specific catalytic subunits of the editosome is unknown.
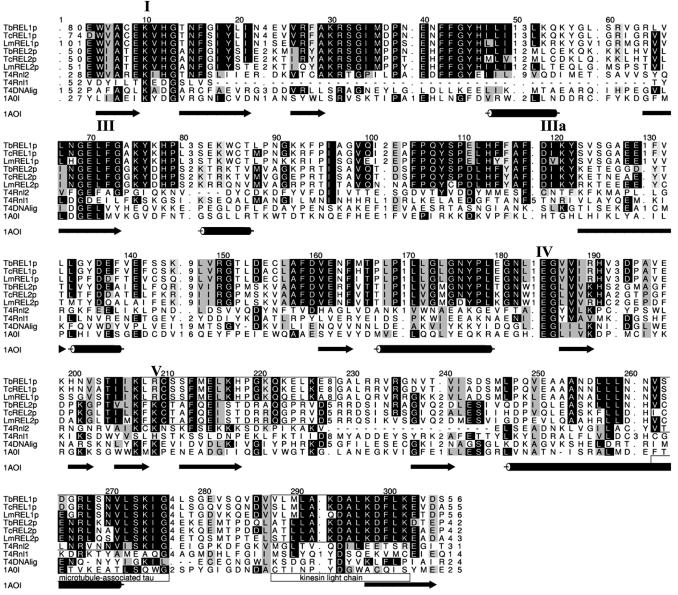
Multiple alignment (Fig. 5) of the six trypanosomatid RELs with two T4 RNA ligases and the T4 and T7 DNA ligases (the T7 DNA ligase crystal structure has been determined; 74) shows a very high conservation among the RELs and their closest similarity to T4Rnl2 (75). The nucleotidyltransferease motifs I, III, IIIa, IV and V (76), which are conserved among all DNA ligases and RNA capping enzymes, were the most conserved regions among these ligases using the MEME software (window size 6–15), consistent with their catalytic activities. Overall sequence conservation with these ligases is low but several key amino acids in the ligase motifs are mostly conserved among the proteins. The notable exception is T4Rnl2, which shares additional MEME motifs with similarity to the RELs, e.g. between ligase motifs I and III (columns 30–45) and in the C-terminal regions of the proteins (columns 262–277), representing previously unrecognized regions of conservation. In the absence of known biological function or substrate specificity of T4Rnl2 (77) it is difficult to speculate what commonality between T4Rnl2 and the RELs might be reflected in this conservation. It appears unlikely to be required for RNA ligation per se since it is not shared with T4Rnl1.
Figure 5.
HMM-generated multiple sequence alignment of REL1 and REL2 ligases with primary sequences of Enterobacteria phage T4 RNA ligase 2 (T4Rnl2, NP_049790), T4 RNA ligase 1 (T4Rnl1, NP_049839) and T4 DNA ligase (T4DNAlig, LQBP34). Arrows and cylinders represent β-strands and α-helices, respectively, in the secondary structure of T7 DNA ligase (1A0I). The five conserved ligase motifs, I, III, IIIa, IV and V, are indicated. The two boxes overlapping motif V represent regions that are divergent between REL1 and REL2 ligases. Also indicated by boxes are regions with similarity to motifs found in microtubule-associated tau proteins (IPB0011084D) and kinesin light chain repeats (IPB002151C), which were identified by MEME.
Structural predictions have suggested that the REL C-terminal regions differ substantially from the C-terminal region of DNA ligases, e.g. from T7 and Chlorella virus (16). Three-dimensional structures of the DNA ligases reveal two distinct domains, an adenylation domain with motifs I–IV, linked by motif V to an OB fold domain, with motif VI (78,79). The OB fold domain is proposed to play a role in substrate binding and specificity, in addition to other functions (80). Schnaufer et al. (16) argue against the presence of such an OB fold domain in these proteins based on the structural predictions for the RELs, and suggest that their C-terminal regions might facilitate interaction with their binding partners, which could provide OB fold domains in trans. Similar structural predictions for T4Rnl2 suggest structural similarity to the RELs that includes the C-terminal region (A. Schnaufer, unpublished results). Our identification of a conserved motif in this region supports the notion of a common function for the C-terminal region of RELs and T4Rnl2. LAMA analysis of the MEME motif comprising columns 262–277 indicates similarity to a motif found in microtubule-associated tau proteins (IPB0011084D). This similarity is in a domain that facilitates protein–protein interaction, consistent with the role proposed above for the C-terminal region of the RELs.
We were unable to detect ligase motif VI in either the RELs or the T4 RNA ligases. This motif is located in the OB fold domain in DNA ligases and RNA capping enzymes, contains the consensus sequence RX1–3D[K/R/N] (81), and is important for the formation of the ligase–adenylate intermediate in Chlorella virus (82). It is not known whether motif VI is generally dispensable for RNA ligases or whether a cryptic motif VI is present in the RNA ligases or their binding partners. Although these enzymes are active on their own (9,83), ligase activity of at least TbREL2 is enhanced by its binding partner TbMP81 (16); it is not known which of the three catalysis steps is accelerated.
In addition to the ligase motifs, RELs share several regions of high sequence conservation, such as a MEME motif comprising columns 288–303 in their C-terminal regions. This motif may be specific to the RELs since it is not shared with T4Rnl2. It has considerable similarity to one of the kinesin light chain repeats (IPB002151C), which has been implicated in protein–protein interactions. Interaction of TbREL1 with TbMP63 and TbREL2 with TbMP81 was highly specific in co-immunoprecipitation studies but somewhat promiscuous in yeast two-hybrid experiments (16), suggesting semi- conservation between REL1s and REL2s of some interacting regions and divergence of others. The boxed regions in Figure 5 (columns 207–227) have been identified by MEME as highly specific for REL1s versus REL2s and therefore might represent such divergent regions responsible for specific interactions. In addition, these regions overlap ligase motif V, shown by mutational studies in E.coli and T7 DNA ligases to be involved in DNA binding, formation of ligase–adenylate and nick sealing (84,85). These differences in domain V between REL1 and REL2 may affect their specificities (see above) and enzymatic characteristics, namely that TbREL1 has a higher affinity for ATP than TbREL2 (11) and some tolerance for gaps and overhangs, at least when within their respective subcomplexes (16). These characteristics may be intrinsic to the ligases or be conferred by the binding partners. While an attractive hypothesis, the proposed function of the C-terminal regions of the RELs in protein–protein interaction has not been confirmed experimentally.
Related proteins MP81, MP63, MP42, MP24 and MP18
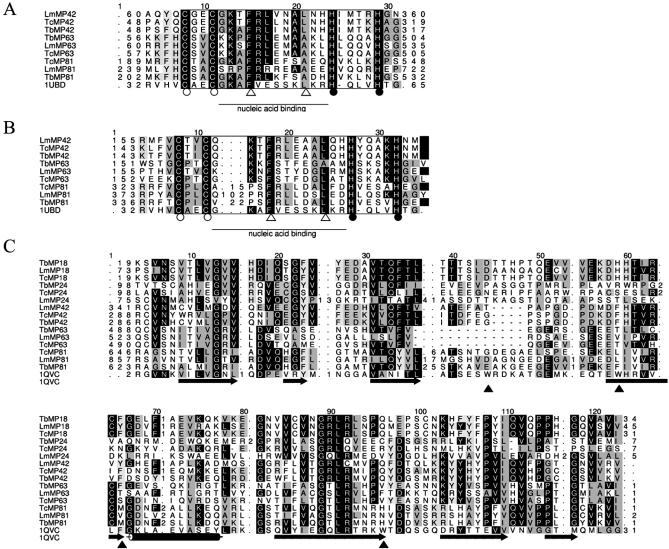
The MP81, MP63, MP42, MP24 and MP18 orthologs were identified by homology searches, except the LmMP24 gene, which was identified due to conservation of gene order in T.brucei; it is located immediately upstream of the LmMP18 gene, suggesting that MP18 and MP24 arose through gene duplication. This annotation was confirmed through multiple sequence alignment (Fig. 6C). These five proteins have varying degrees of amino acid sequence identity (6). The MP81, MP63 and MP42 orthologs conserve two C2H2 zinc finger motifs (Fig. 6A and B). C2H2 zinc fingers occur in a variety of proteins, including nucleic acid and chromatin binding transcription factors and translation initiation factors (86–88). Zinc-centered domains have also been implicated in protein interaction (89). The general motif X-C-X1–5-C-X3-*-X5-*-X2-H-X3–6-[H] typically has polar and basic amino acids between the second C and first H and F and L at the positions indicated by asterisks, which stabilizes the fold in green plants, animals and prokaryotes. An example, the human Ying-Yang 1 protein (1UBD), is shown in Figure 6A. This fold consists of an α-helix and two β-sheets with the two C and two H bound to a Zn atom in a tetrahedral array, yielding a finger-like projection that can interact with nucleotides in the major groove of a nucleic acid (87). The MP81, MP63 and MP42 orthologs conserve F and L or A at the asterisks (open triangles in Fig. 6A and B). The C-terminal C2H2 motifs in the MP81s have additional amino acids, suggesting that they may not retain typical function. The remaining C2H2 zinc finger motifs in MP81, MP63 and MP42 may bind the pre-mRNAs and gRNAs during editing and/or bind proteins to the editing complex (16).
Figure 6.
Related proteins MP18, MP24, MP42, MP63 and MP81. (A and B) HMM-generated multiple sequence alignment of the two C2H2 zinc finger binding domains conserved in the MP81, MP63 and MP42 homologs. Part of the sequence of the human Ying-Yang 1 zinc finger-containing protein (1UBD) is also shown in these figures. The Zn-coordinating C and H amino acids are indicated with open circles and closed circles, respectively. Open arrowheads indicate conserved residues important for correct folding of the zinc finger. An underscore indicates the nucleic acid binding region. (C) HMM-generated multiple sequence alignment of the SSB region identified within the C-terminal regions of MP18, MP24, MP42, MP63 and MP81, together with the SSB region from the classical single-stranded DNA binding protein monomer in E.coli (1QVC). Cylinders denote the α-helices in the structure of 1QVC, arrows denote β-barrels. Closed arrowheads indicate the aromatic residues believed to form a non-sequence-specific binding platform, where general stacking interactions between bases and aromatic side chains are capable of stabilizing RNA–protein complexes.
The five protein orthologs share conserved C-terminal regions (Fig. 6C) that show similarity to the single-strand binding (SSB) protein family domain (90). Proteins with SSB domains, some of which have been extensively characterized (91), have various functions, including preventing DNA secondary structure formation and primase priming, enhancing DNA synthesis rate and error repair by DNA polymerase III holoenzyme, enhancing DNA duplex unwinding by DnaB helicase, and in DNA repair and recombination (92–94). Sequence conservation among SSB domain-containing proteins is generally low, even when they perform similar functions, but they conserve the OB fold (95,96), a structural domain that has seven (or five)-stranded β-sheets that are coiled to form a closed β-barrel and is capped by an α-helix that is typically located between the fourth and fifth strands (95,97,98). Comparison of several secondary structure predictions of the five related proteins with the crystal structure of the 1QVC E.coli SSB protein shows that some have the same general predicted structural organization as in the OB fold domain (16; data not shown). All the algorithms predict the same pattern of β-sheets and α-helices as in the E.coil SSB protein OB fold for MP18s, less so for MP42s, MP63s and MP81s (although the inconsistencies may be due to imprecision in the algorithms) and least so for MP24s, which have inconsistencies for all of the species examined.
The ssDNA binding by OB fold proteins appears to entail a non-sequence-specific stacking interaction between aromatic amino acids on the β-sheets with bases, and cationic interactions between other amino acids with the phosphate–ribose backbone and nitrogenous bases (99). In the E.coli SSB protein, W40, W54, F60 and W88 help form a hydrophobic cluster and mutation of some of these amino acids reduces ssDNA binding (95,96,100). These amino acids are not conserved in the five editosome proteins but in many instances aromatic amino acids occur at nearby positions, suggesting that a hydrophobic cluster may be present. Similarly, positively charged K, H and R amino acids are generally present, notably in the MP18s and MP42s, perhaps corresponding to amino acids near the aromatic amino acids in the OB fold domain that in E.coli appear to form a positively charged cleft that interacts with ssDNA (101–104). In addition, amino acids such as G, E and P that are conserved among SSB proteins and commonly found in turn sequences (perhaps with protein flexibility function) are also conserved. Hence, despite the limited amino acid sequence conservation, the apparent preservation of structural organization and key types of amino acids suggests that at least MP18, MP42, MP63 and MP81 have active OB fold domains.
The characteristics of the zinc finger and OB fold motifs, which imply both nucleic acid and protein binding by MP63 and MP81, along with their presence in catalytically active editing subcomplexes (16), suggests that the OB fold provides substrate recognition and binding for the catalysts as well as for coordination of the order of the catalytic steps. The differences between MP63 and MP81 are likely to account for the differential partner protein and substrate recognition within the subcomplexes.
mHel61p
The L.major and T.cruzi homologs have 49.6 and 61.1% amino acid sequence identity, respectively, to T.brucei mHel61p, a catalytically active RNA helicase (5,105) (Fig. 7). They contain several motifs that are conserved among helicases, including Prp5p and the ATPase domain of translation initiation factor 4a of Saccharomyces cerevisiae. All contain the DEAD sequence that identifies them as DEAD-box RNA helicases within superfamily II of RNA and DNA helicases. These helicases have diverse roles in replication, recombination and repair, transcription, translation, ribosomal structure and biogenesis (106). ATP-dependent DEAD-box helicases are prominent components of the pre-mRNA splicing machinery with roles in the dynamics of spliceosome components and RNA–RNA interactions (107,108) while other family members may have roles in mediating RNA–protein interactions in the spliceosome (109).
Figure 7.
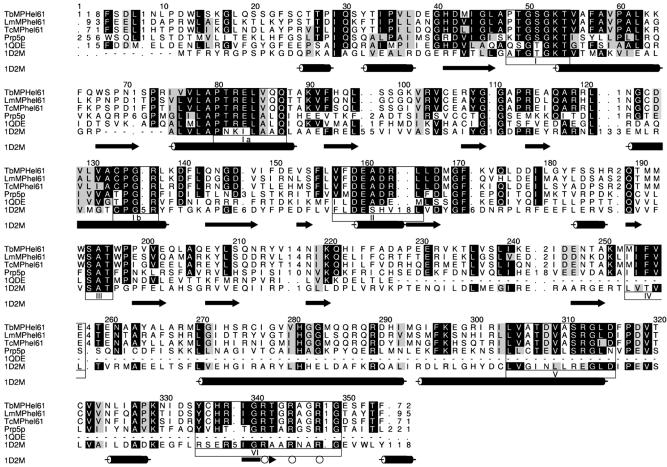
HMM-generated multiple sequence alignment of the editing complex helicases, together with the sequence of the DEAD-box helicase from S.cerevisiae (Prp5p), the sequence of the ATPase domain of the prototype DEAD-box translation initiation factor 4a from S.cerevisiae (1QDE) and the sequence from the T.thermophilus UvrB protein (1D2M). Cylinders and arrows denote the α-helices and β-strands, respectively, in the structure of 1D2M. The most highly conserved motifs are boxed and numbered I–VI. Open circles indicate residues that are believed to be involved in non-specific DNA binding.
Motif I (A-T/S G T/SGKT) corresponds to an ATP binding motif of a pocket that binds the phosphates of ATP in most nucleotide binding proteins and is commonly found in nucleic acid unwinding DEAD-box helicases (110). Motif II (VlDEaD_m) corresponds to a motif that interacts with the β and γ phosphates via an Mg2+ and coordinates ATP hydrolysis by addition of a water molecule (111–117). The V and DEAD amino acids are completely conserved among the mHel61p proteins. Motifs Ia (PTRELa-Q) and, less so, Ib (TPGRl) are conserved among DEAD-box proteins. Analysis of crystal structures suggests that these motifs bind the substrate through the sugar–phosphate backbone (115–118). The Ia motif is completely conserved and the core of the Ib motif and amino acids with structural similarity (e.g. size) with those in the motif are conserved in the mHel61p proteins. Motifs IV (liF-T/S) and V (LvaTdvaaRGlD) are also conserved in the mHel61p proteins. They also appear to function in substrate binding; motif V with the sugar–phosphate backbone and also, possibly, with ATP. Motif III (SAT) binds the γ phosphate (115,116) and may link ATP hydrolysis with the unwinding activity (119). Motif VI (Y-HRiGRT/ SgR-G) is required for RNA binding and ATP hydrolysis in mammalian eIF4A (119), but is not required for RNA binding in the DExH protein NPH-II (120), and has a role in protein–RNA interactions (113,121). Initial non-specific contacts entail electrostatic interactions between the three conserved R residues and the nucleic acid γ phosphate backbone (122), and mutation of these residues affects substrate binding, which, in turn, leads to reduced ATPase activity (115,116, 123). Motifs VI and III in combination appear to link ATP binding and hydrolysis with the conformational changes required for the activity of these helicases.
The mHel61p helicases have greatest overall similarity to the S.cerevisiae spliceosome Prp5p helicase (124) and high regional similarity to the H2 domain of UvrB (1D2M), a nucleotide excision repair enzyme in Thermus thermophilus Hb8 (122,125–127). Prp5p may associate with the spliceosome during 17S U2 assembly and dissociate after catalyzing a conformational change (124,128). Similarly, the association of TbmHel61p with the editosome may be loose or transitional (6). Hence, the helicase may catalyze conformational changes, perhaps associated with RNA binding, or full or partial displacement of gRNAs during editing, perhaps by unwinding gRNA/pre-mRNA duplexes, a common activity of helicases (5). Although inactivation of TbmHel61p gene expression in T.brucei reduces editing efficiency, editing is not eliminated in null mutants, indicating that the helicase is not essential and suggesting that other helicases may compensate for its loss; DEAD-box protein candidates have been identified (7).
Concluding remarks
Analyses of editosome proteins identified several domains and novel arrangements thereof, as summarized in Figure 1. A number of these proteins appear to have arisen from ancestral genes by gene duplication prior to the divergence of Leishmania and Trypanosoma and to have diverged from more ancient ancestral proteins by processes including mutation, domain shuffling and sequence additions and deletions, the latter of which may be compensated for by interactions with partner proteins in the complex. Sequence divergence appears to have retained structure in some cases, presumably to retain but adapt functions. The endonuclease, nucleotidyltransferase, exonuclease, ligase and helicase catalytic components of the editosome share features with proteins involved in other cellular processes, notably DNA repair. Speculation that enzymes for DNA repair originally arose as components in a RNA-based ancestral cell (129,130) implies that the editing and DNA repair enzymes may have arisen from a common ancestral RNA repair system.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH grants 1R21AI053784-01 to R.S. and AI14102 to K.S.
REFERENCES
- 1.Blum B., Bakalara,N. and Simpson,L. (1990) A model for RNA editing in kinetoplastid mitochondria: ‘guide’ RNA molecules transcribed from maxicircle DNA provide the edited information. Cell, 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 2.Stuart K.D., Panigrahi,A.K., Schnaufer,A., Drozdz,M., Clayton,C. and Salavati,R. (2002) Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond. B Biol. Sci., 357, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfonso J.D., Thiemann,O. and Simpson,L. (1997) The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res., 25, 3751–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung S.S. and Koslowsky,D.J. (1999) Mapping contacts between gRNA and mRNA in the trypanosome RNA editing. Nucleic Acids Res., 27, 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missel A., Souza,A.E., Norskau,G. and Göringer,H.U. (1997) Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol., 17, 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panigrahi A.K., Schnaufer,A., Ernst,N., Wang,B., Carmean,N., Salavati,R. and Stuart,K. (2003) Identification of novel components of Trypanosoma brucei editosomes. RNA, 9, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart K. and Panigrahi,A.K. (2002) RNA editing: complexity and complications. Mol. Microbiol., 45, 591–596. [DOI] [PubMed] [Google Scholar]
- 8.Panigrahi A.K., Schnaufer,A., Carmean,N., Igo,R.P.,Jr, Gygi,S., Ernst,N., Palazzo,S.S., Weston,D., Aebersold,R., Salavati,R. et al. (2001) Four related proteins of the T. brucei RNA editing complex. Mol. Cell. Biol., 21, 6833–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnaufer A., Panigrahi,A.K., Panicucci,B., Igo,R.P.,Jr, Wirtz,E., Salavati,R. and Stuart,K.D. (2001) An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science, 291, 2159–2162. [DOI] [PubMed] [Google Scholar]
- 10.Huang C.E., Cruz-Reyes,J., Zhelonkina,A.G., O’Hearn,S., Wirtz,E. and Sollner-Webb,B. (2001) Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J., 20, 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Reyes J., Zhelonkina,A.G., Huang,C.E. and Sollner-Webb,B. (2002) Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol., 22, 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McManus M.T., Shimamura,M., Grams,J. and Hajduk,S.L. (2001) Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA, 7, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst N.L., Panicucci,B., Igo,R.P.,Jr, Panigrahi,A.K., Salavati,R. and Stuart,K. (2003) TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell, 11, 1525–1536. [DOI] [PubMed] [Google Scholar]
- 14.Aphasizhev R., Aphasizheva,I. and Simpson,L. (2003) A tale of two TUTases. Proc. Natl Acad. Sci. USA, 100, 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aphasizhev R., Aphasizheva,I., Nelson,R.E., Gao,G., Simpson,A.M., Kang,X., Falick,A.M., Sbicego,S. and Simpson,L. (2003) Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J., 22, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnaufer A., Ernst,N., O’Rear,J., Salavati,R. and Stuart,K. (2003) Separate insertion and deletion sub-complexes of the Trypanosoma brucei RNA editing complex. Mol. Cell, 12, 307–319. [DOI] [PubMed] [Google Scholar]
- 17.Simpson L., Wang,S.H., Thiemann,O.H., Alfonzo,J.D., Maslov,D.A. and Avila,H.A. (1998) U-insertion/deletion edited sequence database. Nucleic Acids Res., 26, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arts G.J. and Benne,R. (1996) Mechanism and evolution of RNA editing in kinetoplastida. Biochim. Biophys. Acta, 1307, 39–54. [DOI] [PubMed] [Google Scholar]
- 19.Maslov D.A., Avila,H.A., Lake,J.A. and Simpson,L. (1994) Evolution of RNA editing in kinetoplastid protozoa. Nature, 368, 345–348. [DOI] [PubMed] [Google Scholar]
- 20.Feagin J.E., Jasmer,D.P. and Stuart,K. (1987) Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell, 49, 337–345. [DOI] [PubMed] [Google Scholar]
- 21.Feagin J.E. and Stuart,K. (1988) Developmental aspects of uridine addition within mitochondrial transcripts of Trypanosoma brucei. Mol. Cell. Biol., 8, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koslowsky D.J., Bhat,G.J., Perrollaz,A.L., Feagin,J.E. and Stuart,K. (1990) The MURF3 gene of T. brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell, 62, 901–911. [DOI] [PubMed] [Google Scholar]
- 23.Priest J.W. and Hajduk,S.L. (1994) Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr., 26, 179–192. [DOI] [PubMed] [Google Scholar]
- 24.Stuart K. and Feagin,J.E. (1992) Mitochondrial DNA of kinetoplastids. Int. Rev. Cytol., 141, 65–88. [DOI] [PubMed] [Google Scholar]
- 25.Schnaufer A., Domingo,G.J. and Stuart,K.D. (2002) Natural and induced dyskinetoplastid trypanosomatids: how to live without mitochondrial DNA. Int. J. Parasitol., 32, 1071–1084. [DOI] [PubMed] [Google Scholar]
- 26.Grate L.R., Bhattacharyya,C., Jordan,M.I. and Mian,I.S. (2003) Integrated analysis of transcript profiling and protein sequence data. Mech. Ageing Dev., 124, 109–114. [DOI] [PubMed] [Google Scholar]
- 27.Mian I.S. (2002) Comparative sequence analysis of ribonucleases HII, III, II, PH and D. Nucleic Acids Res., 25, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh S.M., Steinberg-Neifach,O., Mian,I.S. and Lue,N.F. (2002) Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell, 1, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozutsumi Y., Segal,M., Normington,K., Gethering,M.-J. and Sambrook,J. (1988) The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature, 332, 462–464. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogh A., Brown,M., Mian,I.S., Sjolander,K. and Haussler,D. (1994) Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol., 235, 1501–1531. [DOI] [PubMed] [Google Scholar]
- 32.Bailey T.L. and Gribskov,M. (1998) Methods and statistics for combining motif match scores. J. Comput. Biol., 5, 211–221. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickerman K. (1994) The evolutionary expansion of the trypanosomatid flagellates. Int. J. Parasitol., 24, 1317–1331. [DOI] [PubMed] [Google Scholar]
- 35.Blaszczyk J., Tropea,J.E., Bubunenko,M., Routzahn,K.M., Waugh,D.S., Court,D.L. and Ji,X. (2001) Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 9, 1225–1236. [DOI] [PubMed] [Google Scholar]
- 36.Gunnewiek J.M., van Aarssen,Y., Wassenaar,R., Legrain,P., van Venrooij,W.J. and Nelissen,R.L. (1995) Homodimerization of the human U1 snRNP-specific protein C. Nucleic Acids Res., 23, 4864–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B., Ernst,N., Palazzo,S.S., Panigrahi,A.K., Salavati,R. and Stuart,K. (2003) TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot. Cell, 2, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green S.R., Manche,L. and Mathews,M.B. (1995) Two functionally distinct RNA-binding motifs in the regulatory domain of the protein kinase DAI. Mol. Cell. Biol., 15, 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., McLachlan,J., Zamore,P.D. and Hall,T.M. (2002) Modular recognition of RNA by a human pumilio-homology domain. Cell, 110, 501–512. [DOI] [PubMed] [Google Scholar]
- 40.Filippov V., Solovyev,V., Filippova,M. and Gill,S.S. (2000) A novel type of RNase III family proteins in eukaryotes. Gene, 245, 213–221. [DOI] [PubMed] [Google Scholar]
- 41.Krinke L. and Wulff,D.L. (1990) The cleavage specificity of RNase III. Nucleic Acids Res., 18, 4809–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson A.W. (1999) Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev., 23, 371–390. [DOI] [PubMed] [Google Scholar]
- 43.Grams J., McManus,M.T. and Hajduk,S.L. (2000) Processing of polycistronic guide RNAs associated with RNA editing complexes in Trypanosoma brucei. EMBO J., 19, 5525–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piller K.J., Rusché,L.N., Cruz-Reyes,J. and Sollner-Webb,B. (1997) Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA, 3, 279–290. [PMC free article] [PubMed] [Google Scholar]
- 45.Lawson S., Igo,R.P.,Jr, Salavati,R. and Stuart,K.D. (2000) The specificity of nucleotide removal during RNA editing in Trypanosoma brucei. RNA, 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salavati R., Panigrahi,A.K., Morach,B.A., Palazzo,S.S., Igo,R.P. and Stuart,K. (2002) Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol., 120, 23–31. [DOI] [PubMed] [Google Scholar]
- 47.Martin G., Keller,W. and Doublie,S. (2000) Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J., 19, 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 49.Frech G.C. and Simpson,L. (1996) Uridine insertion into preedited mRNA by a mitochondrial extract from Leishmania tarentolae: stereochemical evidence for the enzyme cascade model. Mol. Cell. Biol., 16, 4584–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin G., Keller,W. and Doublie,S. (2000) Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J., 19, 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Eckmann,C.R., Kadyk,L.C., Wickens,M. and Kimble,J. (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature, 419, 312–316. [DOI] [PubMed] [Google Scholar]
- 52.Read R.L., Martinho,R.G., Wang,S.W., Carr,A.M. and Norbury,C.J. (2002) Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl Acad. Sci. USA, 99, 12079–12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- 54.Drew H.R. and Travers,A.A. (1984) DNA structural variations in the E. coli tyrT promoter. Cell, 37, 491–502. [DOI] [PubMed] [Google Scholar]
- 55.Suck D. (1994) DNA recognition by DNase I. J. Mol. Recognit., 7, 65–70. [DOI] [PubMed] [Google Scholar]
- 56.Lindahl T. (1990) Repair of intrinsic DNA lesions. Mutat. Res., 238, 305–311. [DOI] [PubMed] [Google Scholar]
- 57.Doetsch P.W. (1995) What’s old is new: an alternative DNA excision repair pathway. Trends Biochem. Sci., 20, 384–386. [DOI] [PubMed] [Google Scholar]
- 58.Mol C.D., Kuo,C.F., Thayer,M.M., Cunningham,R.P. and Tainer,J.A. (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature, 374, 381–386. [DOI] [PubMed] [Google Scholar]
- 59.Demple B., Herman,T. and Chen,D.S. (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA, 88, 11450–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barzilay G. and Hickson,I.D. (1995) Structure and function of apurinic/apyrimidinic endonucleases. Bioessays, 17, 713–719. [DOI] [PubMed] [Google Scholar]
- 61.Mitchell C.A., Brown,S., Campbell,J.K., Munday,A.D. and Speed,C.J. (1996) Regulation of second messengers by the inositol polyphosphate 5-phosphatases. Biochem. Soc. Trans, 24, 994–1000. [DOI] [PubMed] [Google Scholar]
- 62.Suck D., Lahm,A. and Oefner,C. (1988) Structure refined to 2Å of a nicked DNA octanucleotide complex with DNase I. Nature, 332, 464–468. [DOI] [PubMed] [Google Scholar]
- 63.Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 64.Beernink P.T., Segelke,B.W., Hadi,M.Z., Erzberger,J.P., Wilson,D.M.,III and Rupp,B. (2001) Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J. Mol. Biol., 307, 1023–1034. [DOI] [PubMed] [Google Scholar]
- 65.Gorman M.A., Morera,S., Rothwell,D.G., de La Fortelle,E., Mol,C.D., Tainer,J.A., Hickson,I.D. and Freemont,P.S. (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J., 16, 6548–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erzberger J.P., Barsky,D., Scharer,O.D., Colvin,M.E. and Wilson,D.M.,III (1998) Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res., 26, 2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dlakic M. (2000) Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci., 25, 272–273. [DOI] [PubMed] [Google Scholar]
- 68.Hadi M.Z., Ginalski,K., Nguyen,L.H. and Wilson,D.M.,III (2002) Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol., 316, 853–866. [DOI] [PubMed] [Google Scholar]
- 69.Perez J., Gallego,C., Bernier-Villamor,V., Camacho,A., Gonzalez-Pacanowska,D. and Ruiz-Perez,L.M. (1999) Apurinic/apyrimidinic endonuclease genes from the trypanosomatidae Leishmania major and Trypanosoma cruzi confer resistance to oxidizing agents in DNA repair-deficient Escherichia coli. Nucleic Acids Res., 27, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kastenmayer J.P. and Green,P.J. (2000) Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA, 97, 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson A.W. and Kolodner,R.D. (1994) The activity of the Saccharomyces cerevisiae strand exchange protein 1 intrinsic exonuclease during joint molecule formation. J. Biol. Chem., 269, 3664–3672. [PubMed] [Google Scholar]
- 72.Bashkirov V.I., Solinger,J.A. and Heyer,W.D. (1995) Identification of functional domains in the Sep1 protein (= Kem1, Xrn1), which is required for transition through meiotic prophase in Saccharomyces cerevisiae. Chromosoma, 104, 215–222. [DOI] [PubMed] [Google Scholar]
- 73.Drozdz M., Palazzo,S.S., Salavati,R., O’Rear,J., Clayton,C. and Stuart,K. (2002) TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J., 21, 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subramanya H.S., Doherty,A.J., Ashford,S.R. and Wigley,D.B. (1996) Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]
- 75.Ho C.K. and Shuman,S. (2002) Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA, 99, 12709–12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shuman S. and Ru,X.M. (1995) Mutational analysis of vaccinia DNA ligase defines residues essential for covalent catalysis. Virology, 211, 73–83. [DOI] [PubMed] [Google Scholar]
- 77.Yin S., Ho,C.K. and Shuman,S. (2003) Structure–function analysis of T4 RNA ligase 2. J. Biol. Chem., 278, 17601–17608. [DOI] [PubMed] [Google Scholar]
- 78.Subramanya H.S., Doherty,A.J., Ashford,S.R. and Wigley,D.B. (1996) Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]
- 79.Odell M., Sriskanda,V., Shuman,S. and Nikolov,D.B. (2000) Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell, 6, 1183–1193. [DOI] [PubMed] [Google Scholar]
- 80.Doherty A.J. and Wigley,D.B. (1999) Functional domains of an ATP-dependent DNA ligase. J. Mol. Biol., 285, 63–71. [DOI] [PubMed] [Google Scholar]
- 81.Sriskanda V. and Shuman,S. (1998) Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res., 26, 4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sriskanda V. and Shuman,S. (1998) Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res., 26, 3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palazzo S.S., Panigrahi,A.K., Igo,R.P.,Jr, Salavati,R. and Stuart,K. (2003) Kinetoplastid RNA editing ligases: complex association, characterization, and substrate requirements. Mol. Biochem. Parasitol., 127, 161–167. [DOI] [PubMed] [Google Scholar]
- 84.Sriskanda V., Schwer,B., Ho,C.K. and Shuman,S. (1999) Mutational analysis of Escherichia coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4. Nucleic Acids Res., 27, 3953–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doherty A.J. and Dafforn,T.R. (2000) Nick recognition by DNA ligases. J. Mol. Biol., 296, 43–56. [DOI] [PubMed] [Google Scholar]
- 86.Chou A.Y., Archdeacon,J. and Kado,C.I. (1998) Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl Acad. Sci. USA, 95, 5293–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leon O. and Roth,M. (2000) Zinc fingers: DNA binding and protein–protein interactions. Biol. Res., 33, 21–30. [DOI] [PubMed] [Google Scholar]
- 88.Laity J.H., Lee,B.M. and Wright,P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol., 11, 39–46. [DOI] [PubMed] [Google Scholar]
- 89.Alberts I.L., Nadassy,K. and Wodak,S.J. (1998) Analysis of zinc binding sites in protein crystal structures. Protein Sci., 7, 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sancar A., Williams,K.R., Chase,J.W. and Rupp,W.D. (1981) Sequences of the ssb gene and protein. Proc. Natl Acad. Sci. USA, 78, 4274–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lohman T.M. and Ferrari,M.E. (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem., 63, 527–570. [DOI] [PubMed] [Google Scholar]
- 92.Lohman T.M. and Overman,L.B. (1985) Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem., 260, 3594–3603. [PubMed] [Google Scholar]
- 93.Meyer R.R. and Laine,P.S. (1990) The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev., 54, 342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alani E., Thresher,R., Griffith,J.D. and Kolodner,R.D. (1992) Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol., 227, 54–71. [DOI] [PubMed] [Google Scholar]
- 95.Raghunathan S., Kozlov,A.G., Lohman,T.M. and Waksman,G. (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature Struct. Biol., 7, 648–652. [DOI] [PubMed] [Google Scholar]
- 96.Bochkareva E., Belegu,V., Korolev,S. and Bochkarev,A. (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J., 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Curth U., Urbanke,C., Greipel,J., Gerberding,H., Tiranti,V. and Zeviani,M. (1994) Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem., 221, 435–443. [DOI] [PubMed] [Google Scholar]
- 98.Brill S.J. and Bastin-Shanower,S. (1998) Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol., 18, 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murzin A.G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferrari M.E., Fang,J. and Lohman,T.M. (1997) A mutation in E. coli SSB protein (W54S) alters intra-tetramer negative cooperativity and inter-tetramer positive cooperativity for single-stranded DNA binding. Biophys. Chem., 64, 235–251. [DOI] [PubMed] [Google Scholar]
- 101.Bandyopadhyay P.K. and Wu,C.W. (1978) Fluorescence and chemical studies on the interaction of Escherichia coli DNA-binding protein with single-stranded DNA. Biochemistry., 17, 4078–4085. [DOI] [PubMed] [Google Scholar]
- 102.Hollis T., Stattel,J.M., Walther,D.S., Richardson,C.C. and Ellenberger,T. (2001) Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl Acad. Sci. USA, 98, 9557–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hyland E.M., Rezende,L.F. and Richardson,C.C. (2003) The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. J. Biol. Chem., 278, 7247–7256. [DOI] [PubMed] [Google Scholar]
- 104.Chen J., Smith,D.L. and Griep,M.A. (1998) The role of the 6 lysines and the terminal amine of Escherichia coli single-strand binding protein in its binding of single-stranded DNA. Protein Sci., 7, 1781–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Missel A. and Göringer,H.U. (1994) Trypanosoma brucei mitochondria contain RNA helicase activity. Nucleic Acids Res., 22, 4050–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 108.Ruby S.W., Chang,T.H. and Abelson,J. (1993) Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev., 7, 1909–1925. [DOI] [PubMed] [Google Scholar]
- 109.Kistler A.L. and Guthrie,C. (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev., 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- 111.Walker J.E., Auffret,A.D., Carne,A., Gurnett,A., Hanisch,P., Hill,D. and Saraste,M. (1982) Solid-phase sequence analysis of polypeptides eluted from polyacrylamide gels. An aid to interpretation of DNA sequences exemplified by the Escherichia coli unc operon and bacteriophage lambda. Eur. J. Biochem., 123, 253–260. [DOI] [PubMed] [Google Scholar]
- 112.Fry D.C., Kuby,S.A. and Mildvan,A.S. (1986) ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc. Natl Acad. Sci. USA, 83, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.J., Nishi,K., Schnier,J. and Slonimski,P.P. (1989) Birth of the D-E-A-D box. Nature, 337, 121–122. [DOI] [PubMed] [Google Scholar]
- 114.Smith C.A. and Rayment,I. (1996) Active site comparisons highlight structural similarities between myosin and other P-loop proteins. Biophys. J., 70, 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin C. and Kim,J.L. (1999) Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol., 73, 8798–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Caruthers J.M., Johnson,E.R. and McKay,D.B. (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA, 97, 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell, 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 118.Chang T.H., Latus,L.J., Liu,Z. and Abbott,J.M. (1997) Genetic interactions of conserved regions in the DEAD-box protein Prp28p. Nucleic Acids Res., 25, 5033–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pause A., Methot,N. and Sonenberg,N. (1993) The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol., 13, 6789–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gross C.H. and Shuman,S. (1996) The QRxGRxGRxxxG motif of the vaccinia virus DExH box RNA helicase NPH-II is required for ATP hydrolysis and RNA unwinding but not for RNA binding. J. Virol., 70, 1706–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmid S.R. and Linder,P. (1991) Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol., 11, 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin J.J., Phillips,A.M., Hearst,J.E. and Sancar,A. (1992) Active site of (A)BC excinuclease. II. Binding, bending, and catalysis mutants of UvrB reveal a direct role in 3′ and an indirect role in 5′ incision. J. Biol. Chem., 267, 17693–17700. [PubMed] [Google Scholar]
- 123.Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. [DOI] [PubMed] [Google Scholar]
- 124.Wells S.E. and Ares,M.,Jr (1994) Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 6337–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsu D.S., Kim,S.T., Sun,Q. and Sancar,A. (1995) Structure and function of the UvrB protein. J. Biol. Chem., 270, 8319–8327. [DOI] [PubMed] [Google Scholar]
- 126.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 127.Grossman L. and Thiagalingam,S. (1993) Nucleotide excision repair, a tracking mechanism in search of damage. J. Biol. Chem., 268, 16871–16874. [PubMed] [Google Scholar]
- 128.Will C.L., Urlaub,H., Achsel,T., Gentzel,M., Wilm,M. and Luhrmann,R. (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J., 21, 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mushegian A.R. and Koonin,E.V. (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA., 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]