Introductory Paragraph
Several pathogenic bacteria, including Listeria monocytogenes, use an F-actin motility process to spread between mammalian cells1. Actin ‘comet tails’ propel Listeria through the cytoplasm, resulting in bacteria-containing membrane protrusions that are internalized by neighboring cells. The mechanism by which Listeria overcomes cortical tension to generate protrusions is unknown. Here, we identify bacterial and host proteins that directly regulate protrusions. We show that efficient spreading between polarized epithelial cells requires the secreted Listeria virulence protein InlC. We next identify the mammalian adaptor protein Tuba as a ligand of InlC. InlC binds to a C-terminal SH3 domain in Tuba, which normally engages the human actin regulatory protein N-WASP2. InlC promotes protrusion formation by inhibiting Tuba and N-WASP, most likely by impairing binding of N-WASP to the Tuba SH3 domain. Tuba and N-WASP are known to control the structure of apical junctions in epithelial cells3. We demonstrate that, by inhibiting Tuba and N-WASP, InlC makes taut apical junctions become slack. Experiments with myosin II inhibitors indicate that InlC-mediated perturbation of junctions accounts for the role of this bacterial protein in protrusion formation. Collectively, our results suggest that InlC promotes bacterial dissemination by relieving cortical tension, thereby enhancing the ability of motile bacteria to deform the plasma membrane into protrusions.
Cell-cell spread plays a critical role in Listeria virulence by allowing dissemination in host tissues, and protection from humoral immune responses1,4. Spreading occurs after internalized bacteria escape from host membrane vacuoles and enter the cytoplasm. In the cytosol, the Listeria surface protein ActA induces formation of F-actin ‘comet tails’ that propel bacteria. Motile bacteria ultimately encounter the host plasma membrane, deforming it into protrusions. Finally, pathogen-containing protrusions are engulfed by adjacent mammalian cells5.
Listeria-induced F-actin tail assembly is well understood1. In contrast, little is known about the mechanism of protrusion formation. A prevailing view is that the force provided by actin-dependent motility might be the only factor governing protrusion generation6. However, studies leading to this view have generally employed simple mammalian cell lines that lack the capacity to polarize. Most tissues infected by Listeria contain polarized cells that form tight barriers. These tissues consist of enterocytes lining the intestine, hepatocytes, and cells of the choroid plexus or brain endothelium, which form the blood-brain barrier4. Polarized cells exhibit a thick radial network of cortical F-actin and myosin connected to the apical junctional complex- a structure consisting of tight junctions and adherens junctions7. This actomyosin network could potentially restrain Listeria spreading by generating cortical tension that counteracts the ‘pushing’ force produced by motile bacteria. Hence, protrusion formation in polarized tissues might require the intervention of unidentified Listeria proteins that function after ActA to control properties of the host plasma membrane.
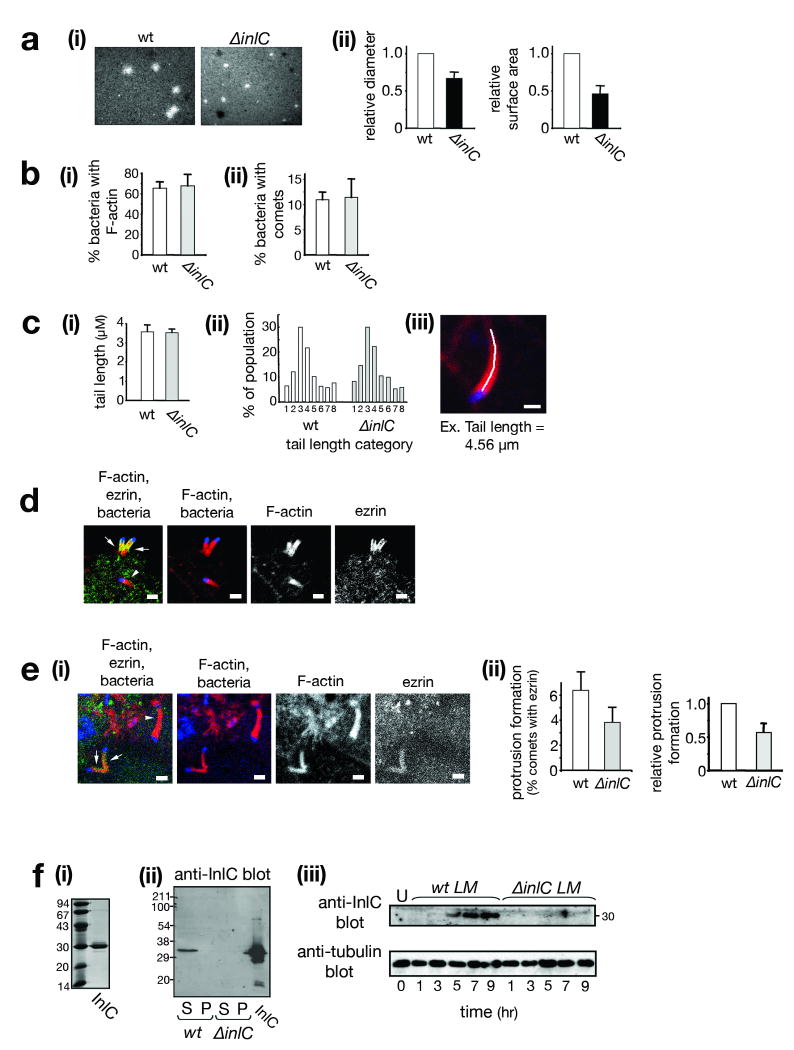
Listeria is a food-borne pathogen4, and infection of intestinal enterocytes is the first step in listeriosis8. Given the crucial role of enterocytes in disease, we investigated cell-cell spread in Caco-2 BBE1 cells, a human enterocyte cell line that polarizes in culture9. Using a plaque assay10, we found an important role for the Listeria virulence protein InlC11 in spreading. A Listeria strain deleted for inlC (ΔinlC) made plaques that were ∼65% the diameter of those made by the isogenic wild-type strain (Fig. 1a).
Figure 1. InlC is needed for efficient spreading and protrusion formation in a polarized cell line.
(a) InlC promotes spreading. Caco-2 BBE1 cells in transwells were infected with wild-type (wt) or ΔinlC Listeria strains. (i): Representative images of plaque assays. (ii): Average relative plaque diameters or surface areas ± s.d. (n=3). (b). InlC does not affect F-actin tail formation. (i). Average percentage (%) ± s.d. (n=3) of intracellular wild-type or ΔinlC bacteria with symmetric F-actin or comet tails. P = 0.45. (ii). Average percentage (%) ± s.d. (n=3) of intracellular bacteria with comet tails only. P = 0.46. (c) InlC does not influence tail length. (i). Average tail lengths (μm) ± s.d. (n=3). P = 0.84. (ii). Distributions of tail lengths: <1 μm (1), 1.00-1.99 μm (2), 2.00-2.99 μm (3), 3.00-3.99 μm (4), 4.00-4.99 μm (5), 5.00-5.99 μm (6), 6.00-6.99 μm (7), 7.00-14.00 μm (8). (iii). Tail length measurement. F-actin: red; bacterium: blue. Tail length is measured as a free-hand trace. Scale bar, 1 μm. (d) Ezrin localizes to protrusions. Confocal microscopy was performed on subconfluent Caco-2 BBE1 cells infected with wild-type Listeria. The left panel is a merged image showing F-actin (red), ezrin (green), and bacteria (blue). Arrows indicate ezrin-containing comet tails in protrusions projecting into space. The arrowhead indicates an ezrin-negative tail in the cell body. (e) InlC promotes protrusion formation. Scale bars, 2 μm. (i). Representative image of wild-type Listeria in cells in transwells. F-actin, ezrin, and bacteria are colored as in d. Arrows and arrowheads indicate F-actin tails containing or lacking ezrin, respectively. (ii). Quantification of ezrin recruitment. Protrusion formation is expressed as the average percentage ± s.d. (n=7) of total F-actin tails containing ezrin (left panel), or as relative values (right panel). P = 0.008. (f) InlC is expressed in infected human cells. (i) InlC used to generate antibodies. (ii) Anti-InlC Western blots of supernatants (S) or pellets (P) from broth cultures of wild-type (wt) or ΔinlC strains. (iii) Caco-2 BBE1 cells were left uninfected (U) or infected with Listeria strains for increasing times. Cell lysates were Western blotted with antibodies against InlC or tubulin.
The ∼ 35% reduction in spreading of the ΔinlC mutant is of sufficient magnitude to account for the known role of InlC in virulence in mice. Deletion of inlC causes an approximately 50-fold increase in LD5011. The Listeria plcB gene encodes a phospholipase that contributes to spreading by mediating escape from host vacuoles4. Null mutations in plcB or inlC result in quantitatively similar reductions in plaque size and increases in LD5011,12. In addition, certain hypomorphic mutations in actA cause spreading and virulence defects that mirror those resulting from deletion of inlC11,13.
InlC could promote spreading by affecting intracellular bacterial replication, by controlling comet tail assembly, or by directly regulating protrusion formation. As previously reported in a Caco-2 cell line11, wild-type and ΔinlC mutant strains of Listeria exhibited nearly identical intracellular growth rates from 1-5 h post-infection in Caco-2 BBE1 cells (data not shown). These results indicate that InlC is dispensable for cytosolic replication. In addition, we were unable to detect a role for InlC in comet tail formation. At 5 h post-infection, the wild-type and ΔinlC strains displayed similar proportions of intracellular bacteria decorated with F-actin (Fig. 1b), and produced comet tails that were essentially identical in length (Fig. 1c).
We next examined if InlC controls protrusion formation. Protrusion efficiency was measured by evaluating bacterial comet tails that contain the ERM family protein ezrin14. Ezrin is present in comet tails in protrusions but absent from those in the cell body15,16 (Fig. 1d), making the protein a useful marker for protrusions. Compared to the wild-type strain, the ΔinlC mutant had a 43% reduction in the proportion of comet tails containing ezrin (Fig. 1e). These findings indicate a role for InlC in protrusion formation.
We confirmed the above results by using a second assay that directly detects bacterial protrusions. Cells were transfected with a vector expressing actin fused to EGFP (EGFP-actin). The efficiency of transfection was ∼25%, resulting in situations in which EGFP-labeled Caco-2 BBE1 cells were adjacent to unlabeled cells. Protrusions projecting from cells expressing EGFP-actin into EGFP-actin –negative cells were readily observed (Fig. S1a). Comet tails in the cell body were also apparent. The efficiencies of protrusion formation by wild-type or ΔinlC bacteria were determined as the proportion of total comet tails in protrusions. The ΔinlC strain exhibited a 38 % reduction in protrusions compared to the wild-type strain (Fig. S1b). These results with the EGFP-actin –based assay are in good agreement with those from the ezrin -based assay (Fig. 1e). Collectively, the findings in Figures 1e and S1 indicate that InlC is needed for efficient protrusion formation. Since F-actin tails appear unaffected by InlC (Fig. 1b,c), the role of InlC in protrusion production is likely direct.
InlC is a secreted protein found entirely in culture supernatants11 (Fig. 1f). Consistent with its role in spreading, InlC mRNA and protein accumulate in Caco-2 BBE1 cells during Listeria infection11 (Fig. 1f). Interestingly, InlC has an amino-terminal Leucine-Rich Repeat (LRR) domain11. The LRR domain is a protein fold commonly involved in protein-protein interactions17. L. monocytogenes expresses several virulence proteins with LRR domains, at least two of which bind to mammalian receptors18. The expression pattern of InlC and presence of an LRR domain suggested that InlC might engage a cytoplasmic host ligand.
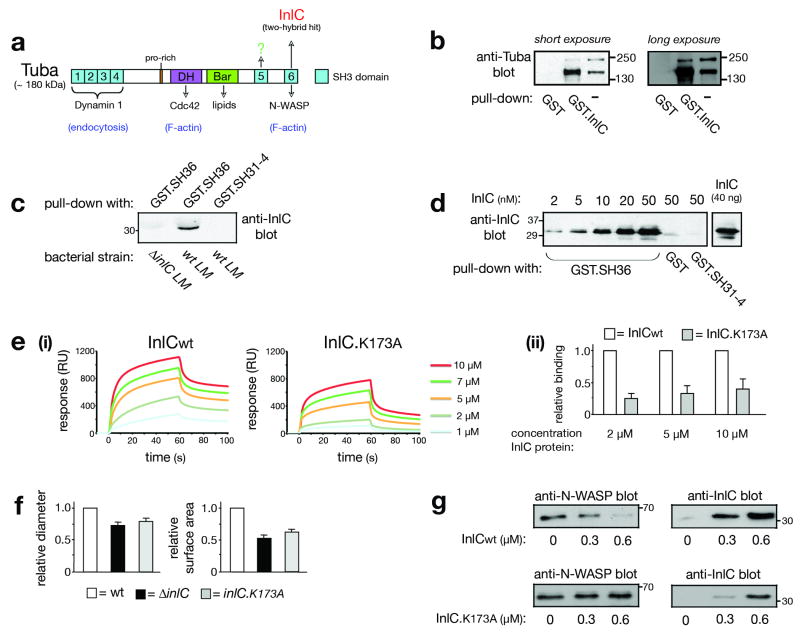
To identify such ligands, a yeast two-hybrid system was employed to screen a mouse embryonic cDNA library using InlC as bait. Multiple hits were obtained for a cDNA encoding an isolated SH3 domain that is normally part of a large adaptor protein called Tuba19 Fig. 2a). Importantly, Tuba is expressed in the intestine, liver, placenta, and brain19, tissues infected by Listeria4. Tuba contains a Bar domain with the potential to bind lipids, a Dbl homology (DH) domain that activates the GTPase Cdc42, and six SH3 domains. The first four SH3 domains (SH31-4) bind Dynamin I, a GTPase involved in endocytosis19. Ligands of the fifth SH3 domain (SH35) have yet to be reported. The last SH3 domain (SH36) of Tuba interacts with N-WASP, a regulator of actin polymerization2. In the two-hybrid screen, SH36 was identified as the InlC-interacting domain.
Figure 2. InlC interacts with the mammalian adaptor protein Tuba.
(a) Structure of Tuba. SH3, Dbl homology (DH), and Bin-Amphiphysin-Rvs (Bar) domains are indicated. The last SH3 domain (SH36) interacts with InlC or human N-WASP19. Other ligands of the first four SH3 (SH31-4), DH, or Bars domain are indicated. In addition to full-length Tuba (∼180 kDa), isoforms lacking SH31-4, but retaining the DH, Bar, SH35, and SH36 domains, exist19,20. (b) InlC associates with ∼ 180 and ∼150 kDa Tuba isoforms from Caco-2 BBE1 cells. A GST-InlC fusion protein was incubated with cell lysates and precipitated. Precipitates were immunoblotted with antibodies against the Tuba SH36 domain. (c) The Tuba SH36 domain associates with secreted InlC. Caco-2 BBE1 cells were infected with wild-type (wt) or ΔinlC strains of L. monocytogenes (LM) for 7 h. Cell lysates were used for precipitation with GST proteins containing SH36, or the SH31-4 region as a control. (d) InlC interacts directly with SH36. GST proteins containing SH36, SH31-4, or GST alone (10 nM) were incubated with the indicated concentrations of InlC and precipitated. InlC was detected by immunoblotting. The last lane is a loading control. (e) An RxxK sequence in InlC participates in binding to SH36 (i). Binding of wild-type InlC protein (InlCwt) or InlC.K173A, as measured by SPR. Representative data from a single experiment is shown. Data from 3-4 experiments were used to estimate equilibrium dissociation constants (KD) of 9.0 ± 3.5 and 58 ± 12.3 μM for wild-type InlC and InlC.K173A, respectively. P = 0.0007. (ii). Relative binding of wild-type (wt) InlC and InlC.K173A to SH36. The data is average ± s.e.m. (n=3). (f) An RxxK sequence in InlC is needed for efficient spreading. Average relative plaque diameters (i) or surface areas (ii) ± s.d. (n=3) of wild-type (wt), ΔinlC, or inlC.K173A strains of Listeria are presented. (g) InlC displaces N-WASP from SH3-6. 10 nM GST-SH36 was incubated with 10 nM of N-WASP in the presence of the indicated concentrations of wild-type InlC (InlCwt) or InlC.K173A for 2.5 h. GST-SH36 was precipitated, and N-WASP (left panels) or InlC (right panels) detected by immunoblotting.
As previously reported3, two isoforms of Tuba were detected in Caco-2 BBE1 cells (Fig. 2b). Based on size and reactivity with antibodies recognizing SH31-4 or SH36, the larger protein (∼180 kDa) is likely full-length Tuba (Fig. S2). The smaller protein (∼ 150 kDa) is likely an isoform that lacks the first four SH3 domains, but retains the rest of Tuba, including SH36 (Fig. S2). Splicing isoforms of Tuba lacking SH31-4 and retaining the DH, Bar, SH35, and SH36 domains have been reported19,20. We found that InlC bound both the 150 and 180 kDa Tuba isoforms (Fig. 2b). Interaction between Tuba and InlC expressed in infected cells was also detected by co-precipitation with the SH36 domain of Tuba (Fig. 2c). Importantly, experiments with purified proteins (Fig. S3a,b) indicated that InlC interacts directly with SH36, but not with any other SH3 domain in Tuba (Fig. 2d; data not shown). The affinity (KD) of InlC for SH36, as determined by Surface Plasmon Resonance (SPR) studies (Fig. 2e), was 9.0 ± 3.5 micromolar. This KD is within the range of affinities of previously characterized SH3 domain ligands (1-100 μM)21.
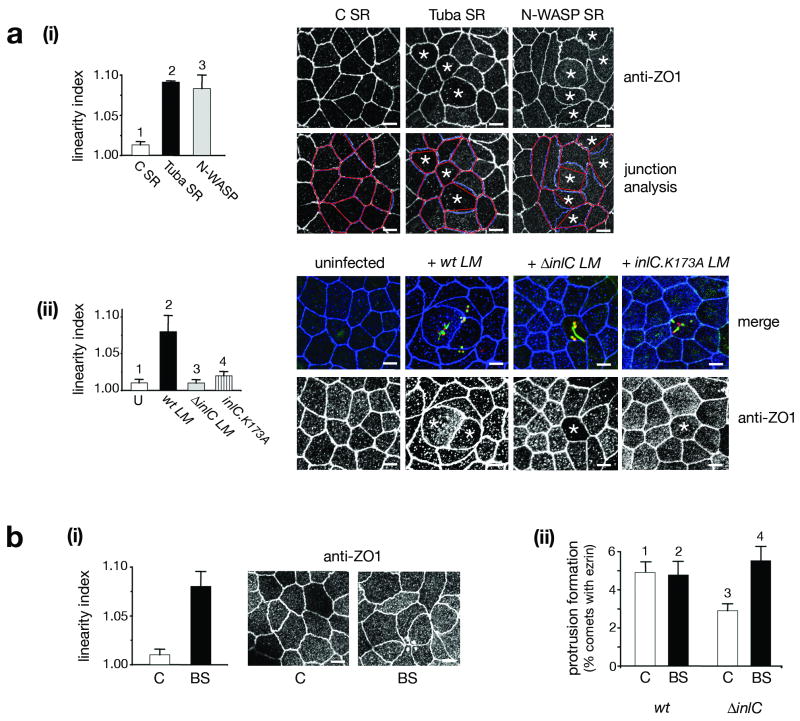
We next investigated the role of Tuba in Listeria protrusion formation. We envisioned two ways in which Tuba and InlC might act. First, Tuba might promote protrusion formation, with InlC positively regulating Tuba. Second, Tuba might antagonize protrusions, with InlC relieving Tuba-mediated inhibition. Interestingly, experiments involving RNA interference (RNAi) indicated an antagonistic role for Tuba. Specifically, knockdown of Tuba allowed the ΔinlC mutant strain to make protrusions as efficiently as wild-type Listeria (Fig. 3a,b). Importantly, neither the proportion of intracellular bacteria with comet tails nor the length of those tails was affected by Tuba depletion (Fig. 3c; data not shown). These findings indicate that, in the absence of InlC, Tuba antagonizes protrusion formation. InlC expressed by wild-type Listeria relieves Tuba-mediated inhibition.
Figure 3. Tuba and N-WASP control protrusion formation.
Caco-2 BBE1 cells were treated with control (C) siRNA or siRNA molecules targeting Tuba or N-WASP. About 72 h after transfection, cells were infected with wild-type (wt) or ΔinlC strains of Listeria for ∼ 5 h prior to Western blotting (a) or fixation and labeling for analysis of protrusions (b) or F-actin tails (c). (a) siRNA-mediated depletion of Tuba or N-WASP. Western blots were performed using lysates from cells treated with ‘siTuba-1’ or ‘siNWASP-1’ siRNA (Methods). Top panels display anti-Tuba or anti-N-WASP immunoblots, whereas the bottom panels show anti-tubulin blots of the stripped membranes. (b) Quantification of recruitment of ezrin to comets. Protrusion efficiency is expressed as the average percentage ± s.d. of F-actin tails that contain ezrin (Y-axis). The data are from 3-4 experiments, depending on the condition. Statistical analysis revealed significant differences (p<0.05) in the following column pairs: i+iv, iv+v, and iv+vi. *Note that the apparent difference in protrusions in cells treated with control or N-WASP siRNA (columns i and iii) is not statistically significant (p>0.05). (c) Comet tail lengths in Caco-2 BBE1 cells depleted for Tuba or N-WASP. Tail lengths (μm) are average ± s.d. values from three experiments. Statistical analysis revealed no significant difference between any of the columns (p = 0.53). The data in parts a, b, and c are from experiments using ‘siTuba-1’ or ‘siNWASP-1’ siRNA molecules (Methods). siRNA molecules that targeted different regions in Tuba or N-WASP mRNA (‘siTuba-2’ or ‘siNWASP-2; Methods) produced similar results (data not shown).
Additional experiments indicated that InlC promotes spreading by binding to SH36 in Tuba. A variety of peptide motifs engage SH3 domains21. The majority of SH3 domains studied to date interact with proline-rich sequences of the type PxxP (where × is any amino acid). Less frequently, other motifs such as PxxDY, RKxxYxxY, or RxxK bind SH3 domains. Although InlC lacks PxxP motifs, the bacterial protein does have an RxxK sequence (RNNK) at amino acids 170-173. We constructed a mutant InlC protein with a lysine-to-alanine substitution in this RNNK sequence (InlC.K173A). In other RxxK-containing proteins, the conserved lysine is needed for efficient interaction with SH3 domains21. We therefore reasoned that the K173A mutation in InlC might compromise binding. Indeed, Surface Plasmon Resonance studies indicated that the K173A mutation reduces the affinity of binding to SH36 (Fig. 2e). Circular Dichroism revealed that the binding defect was not due to structural effects on InlC (data not shown). Importantly, a Listeria strain expressing InlC.K173A was defective in cell-cell spread (Fig. 2f) and in formation of bacterial protrusions (Fig. S4a). Expression of the mutant InlC protein in infected Caco-2 BBE1 cells was similar to that of wild-type InlC (data not shown). Collectively, the results in Figures 2e,f and S4a indicate that interaction of InlC with SH36 in Tuba is essential for efficient spreading of Listeria.
The actin regulatory protein N-WASP interacts with Tuba, and mediates Tuba's biological activities3,19,20. N-WASP uses PxxP peptides in its central proline-rich domain to associate with SH36 in Tuba19. Since InlC and N-WASP both bind SH36, we wondered if InlC inhibits Tuba by affecting N-WASP. Two lines of evidence supported this hypothesis. First, depletion of N-WASP caused effects on Listeria protrusion formation that mirrored those resulting from Tuba depletion (Fig 3a,b). Second, InlC displaced N-WASP from the Tuba SH36 domain in vitro (Fig. 2g). The InlC.K173A mutant protein was less effective than wild-type InlC in displacing N-WASP. Collectively, the results in Figures 3 and 2f suggest that InlC promotes spreading by interfering with Tuba- N-WASP complexes.
Recent findings indicate that Tuba- N-WASP complexes control the structure of cell-cell junctions in enterocytes3. RNAi-induced depletion of Tuba or N-WASP results in curved apical junctions, as indicated by staining for the tight junction protein ZO-1. It was proposed that these curved junctions reflect a role for Tuba and N-WASP in generating cortical tension that imparts rigidity to the plasma membrane.
Since InlC impairs Tuba and N-WASP during spreading (Fig. 3b), we wondered whether InlC might also perturb apical junctions. We first confirmed that Tuba and N-WASP control apical junction morphology in Caco-2 BBE1 cells (Fig. 4ai). The effect of depletion of Tuba or N-WASP on junctional morphology was quantified by calculation of linear indices, a measure of the extent to which cell borders deviate from ideal linearity3. Interestingly, infection of Caco-2 BBE1 cells with wild-type Listeria resulted in a phenocopy of the curved junctional architecture normally observed upon depletion of Tuba or N-WASP (Fig. 4aii). In comparison to the situation with wild-type bacteria, bacteria lacking inlC (ΔinlC) or expressing InlC.K173A were defective in altering junction morphology. These findings indicate that InlC is necessary for Listeria-induced perturbations in apical junctions. Interestingly, transfection experiments indicated that InlC was also sufficient to alter cell junctions in the absence of bacteria (Fig. S5). Taken together, these results demonstrate that InlC perturbs the morphology of cell junctions by impairing Tuba and N-WASP. The “slack” junctions induced by wild-type Listeria likely reflect diminished cortical tension.
Figure 4. InlC, Tuba, and N-WASP control the morphology of apical junctions.
(a) Role of Tuba, N-WASP, and InlC in junction structure. (i). Effect of depletion of Tuba or N-WASP. Caco-2 BBE1 cells were treated with control siRNA (C SR) or Tuba-1 siRNA (Tuba SR) for ∼ 72 h prior to labeling for ZO1. Left Panel: Average linearity indices (LI) ± s.d. from three experiments. Differences in the column pairs 1+2 and 1+3 are statistically significant (p<0.001). Top right panels: ZO1 staining in cells transfected with control, Tuba, or N-WASP siRNA. Note cells in the Tuba- or N-WASP- depleted populations with curved junctions (marked with *). Bottom right panels: Examples of junction analysis. The linear index (LI) is the ratio of junction length (blue lines) to the distance between vertices (red lines). (ii). Infection with Listeria expressing InlC perturbs apical junction morphology. Caco-2 BBE1 cells were left uninfected or infected with wild-type (wt), ΔinlC, or inlC.K173A strains of L. monocytogenes (LM) for 5 h. Left panel: LI analysis of junctions from 3-6 experiments. Differences in the pairs 1+2, 2+3, and 2+4 are statistically significant (p<0.001). Top right panels: Representative images of uninfected or infected cells. Bacteria are in red, F-actin is in green, and ZO1 is in blue. Bottom right panels: ZO1 staining from the same fields of view. Asterisks indicate cells containing intracellular bacteria decorated with F-actin. Note curved junctions in cells infected with wild-type Listeria. Junctions in cells with ΔinlC or inlC.K173A bacteria have markedly less curvature. (b) Myosin II restrains Listeria protrusion formation. Caco-2 BBE1 cells were treated with DMSO or 50 μM blebbistatin for 1 h, prior to analysis of apical junctions (i) or protrusions (ii). (i). Effect of blebbistatin on junctions. Right panel: Typical images of apical junctions. Left panel: Average linear index values ± s.d. from three experiments. P = 0.0026. (ii) Effect of blebbistatin on Listeria protrusions. Average percentages of comet tails with ezrin ± s.d. from three experiments are presented. Differences in the pairs 1+3, 2+3, and 3+4 are statistically significant (p<0.05). All scale bars are 5 μm.
The effect of InlC on apical junctions could account for the role of this bacterial protein in protrusion formation. In this case, InlC might diminish cortical tension at junctions, making it easier for motile bacteria to deform the membrane into protrusions. Alternatively, InlC might control spreading by affecting unknown activities of Tuba and N-WASP that are unrelated to junctions. In order to distinguish between these two possibilities, we perturbed junctions through a mechanism independent of Tuba or N-WASP. Specifically, the myosin II inhibitor blebbistatin22 was used to alter cell junctions. As previously reported for other inhibitors of myosin II3, incubation of cells with blebbistatin resulted in curved apical junctions (Fig. 4bi). These findings likely reflect a role for actomyosin-mediated contractility in cortical tension. Importantly, blebbistatin treatment suppressed the defect in protrusion formation normally caused by deletion of inlC (Fig. 4bii) or by the inlC.K173A mutation (S4b). Similar results on apical junctions and Listeria spreading were caused by Y27632, another chemical that impairs myosin II –mediated contractility23 (data not shown). Taken together, the findings in Figures 3, 4, and S4b indicate that InlC promotes bacterial spreading most likely by affecting junctional architecture.
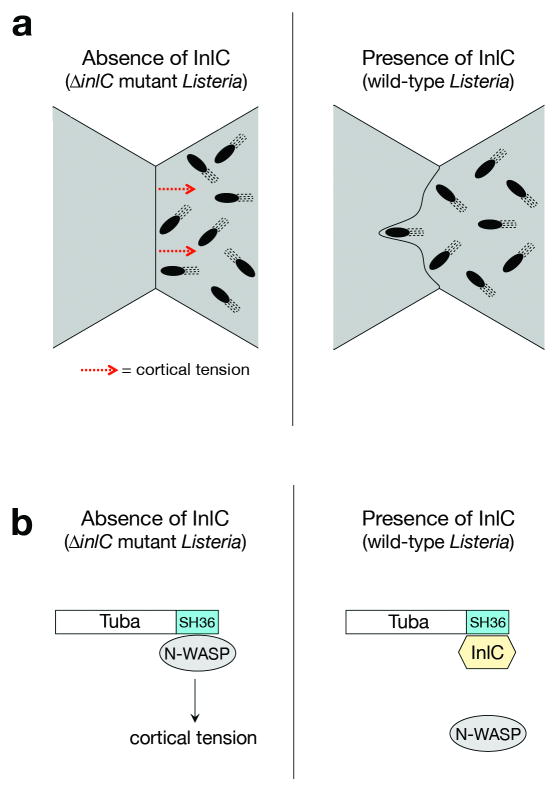
Our findings provide the first evidence for a bacterial protein-host ligand interaction that directly controls the accumulation of protrusions during spreading. An interesting issue, to be addressed in future work, is whether InlC affects the initiation or persistence of protrusions. Listeria enhances its spreading in polarized epithelial cells by employing a remarkable strategy that alters the structure of apical junctions (Fig. 5). A key question is how Tuba, N-WASP, and InlC control cell junctions. One possibility is that Tuba and N-WASP promote endocytosis at apical junctions, thereby limiting membrane surface area and causing junctions to become taut. By disrupting Tuba- N-WASP complexes, InlC would interfere with membrane internalization. A second possibility is that Tuba and N-WASP promote contractility by contributing to the actomyosin ring linked to apical junctions7,24. Activated N-WASP stimulates actin polymerization through the Arp2-3 complex2. It is possible that binding of the Tuba SH36 domain activates N-WASP, as previously reported for the SH3 domains of WISH, Grb2, and Nck2. In this scenario, InlC might antagonize the ability of Tuba to enhance N-WASP –mediated actin polymerization. Future studies should answer whether InlC promotes Listeria spreading by controlling endocytosis or actin polymerization. Regardless of the mechanism involved, our work establishes a new paradigm for how intracellular pathogens can promote their cell-cell spread by manipulating properties of host cell junctions.
Fig. 5. Model for InlC-mediated cell-cell spread of Listeria.
(a) InlC controls protrusion formation by altering the morphology of apical junctions in polarized epithelial cells. Left panel: Uninfected host cells or cells infected with the ΔinlC mutant of Listeria exhibit linear apical junctions. Junction linearity is likely due to cortical tension, which inhibits the ability of motile bacteria to deform the plasma membrane into protrusions. Right panel: cells infected with wild-type Listeria. InlC causes normally taut apical junctions to become slack, most likely by attenuating cortical tension. Diminished tension allows more efficient protrusion formation by motile bacteria. (b) Putative molecular mechanism of InlC-mediated protrusion formation. Left panel: In the absence of InlC, Tuba and N-WASP form a complex in the host cell that generates cortical tension at apical junctions. Right panel, InlC alleviates Tuba- N-WASP –mediated tension by disrupting Tuba- N-WASP complexes.
Methods
Strains and cell lines
The wild-type L. monocytogenes strain EGD and the isogenic inlC deletion mutant (ΔinlC) were previously described11. The inlC.K173A strain was constructed by allelic replacement. Briefly, a lysine-to-alanine substitution of codon 173 (K173A) in inlC was generated using a Quikchange kit (Stratagene). Primers used were 5′-CTATTCGTAATAATGCATTAAAAAGTATTGTG-3′ and 5′-CACAATACTTTTTAATGCATTATTACGAATAG-3′. The mutant allele was verified by DNA sequencing and subcloned into pLSV101. Homologous insertion into the genome of L. monocytogenes strain EGD was as described25. Clones containing the desired mutation were identified by sequencing of genomic DNA isolated by the polymerase chain reaction (PCR).
The human enterocyte cell line Caco-2 BBE1 (ATTC; CRL-2102) was grown in DMEM with 4.5 g/L glucose, 2 mM glutamine, 10% FBS, and 0.01 mg/ml human transferrin. For all experiments with the exception of Figure 1d, cells were grown in 6 well plates on transwell permeable supports (Costar, 0.4 μm pore size). Cell monolayers were verified to have a transepithelial resistance of at least 150 ohms. For Figure 1d, cells were grown on glass coverslips and infected while subconfluent. The passage number of Caco-2 BBE1 cells was always between 46 and 60.
Plasmids
Plasmids expressing full-length human Tuba or GST fusion proteins containing SH31-4, SH35, or SH36 domains in Tuba19 were provided by Dr. P. DeCamilli (HHMI). Plasmids expressing 6xHis tagged rat N-WASP deleted for its EVH1 domain27 or EGFP-tagged actin were from Drs. J. Taunton (UCSF) or G.P. Downey (University of Colorado), respectively. The pEBB-based plasmid expressing epitope-tagged β-galactosidase was previously described26. Plasmids constructed for this study are pET21a-based plasmids encoding wild-type InlC or InlC.K173A with a C-terminal 6xHis tag, a pGex- based plasmid encoding GST-InlC, a plasmid containing LexA fused to the C- terminus of InlC used in yeast 2-hybrid studies, plasmids with Ha-tagged wild-type ‘mini-Tuba’ and ‘mini-Tuba’ deleted for the SH36 domain, and a plasmid containing myc-tagged InlC in the mammalian expression vector pEBB26. Each of these plasmids was generated through PCR amplification of the insert followed by ligation into the appropriate vector backbone using standard molecular biology approaches. pET21a encoding InlC with a lysine-to-alanine substitution in codon 173 (InlC.K173A) was made using a QuikChange kit and the primers 5′-TCTTATCTATTCGTAATAATGCGTTAAAAAGTATTGTGATGC-3′ and 5′-GCATCACAATACTTTTTAACGCATTATTACGAATAGATAAGA-3′. The inserts in all plasmids were verified by DNA sequencing.
siRNAs, antibodies, and other reagents
siRNA molecules directed against Tuba were ‘siTuba-1’ (5′-AUAUGCAGAUGGUGAUUAAUU-3′) or ‘siTuba-2’ (5′-GAGCUUGAGGGAACAUACAAGAUUU -3′)3. siRNAs used to deplete N-WASP were ‘siNWASP-1’ (5′-UCAAAUUAGAGAGGGUGCUCAGCUAUU -3′), or ‘siNWASP-2’ (5′-UCAAAUUAGAGAGGGUGCUCAGCUAUU -3′). As a negative control, a ‘non-targeting’ siRNA that contains two or more mismatches with all sequences in the human genome (5′-UAGCGACUAAACACAUCAAUU-3′) was used. Polyclonal antibodies recognizing recombinant InlC, the last SH3 domain in Tuba (SH36), or the first four SH3 domains in Tuba (SH31-4) were raised in rabbits. Commercially available antibodies were mouse anti-ezrin (Zymed; 35-7300), mouse-anti-ZO1 (Zymed; 33-9100), mouse anti-tubulin (Sigma; T5168), rabbit anti-N-WASP (Cell Signaling; 4848), and rabbit anti-myc (Covance; PRB-150P).
Plaque assays
Plaque assays with Caco-2 BBE1 cells were performed as described10, except that cells were grown on transwells for ∼ 7 days prior to infection. After infection for 1 h, cells were overlayed with medium containing 0.7% agar and gentamicin (10 μg/ml), and incubated at 37°C in 5% C02 for another ∼ 3 days before detection of plaques. In each experiment, typically 25 plaques were measured for each bacterial strain.
RNA interference
Approximately 6 × 104 Caco-2 BBE1 cells were grown in transwells for ∼ 24 h prior to transfection with 100 nM siRNA using LF2000 (Invitrogen). About 72 h after transfection, cells were solubilized for analysis of target protein expression or infected with Listeria strains.
Immunofluorescence microscopy
Cells were grown in transwells for ∼ 96 h prior to infection. In the case of Figures S1 and S5, cells were transfected with plasmids expressing EGFP-actin, epitope-tagged InlC, or β-galactosidase approximately 24 h after seeding, and grown for another ∼72 h before infection for 5 h or processing for cell junction analysis. Cells were fixed in 3.7% paraformaldehyde. After excision of transwell filters, samples were permeabilized, labeled, and mounted as described26. Primary antibodies used were mouse anti-ezrin, mouse anti -ZO1, rabbit anti-myc, and rabbit anti-Listeria R1126. F-actin was detected with phallodin coupled to BodyP FL or Alexa 647. Samples were analyzed using a Zeiss LSM510 laser scanning confocal microscope equipped with argon (488 nm), helium-neon 1 (543 nm), and helium neon 2 (633 nm) lasers, using the 100× objective.
Analysis of F-actin comet tails
Zeiss LSM 510 software (version 3.2 SP2) was used to measure F-actin tail length. Comets were defined as asymmetrically distributed F-actin visibly extending from one bacterial pole28. In each experiment, at least 50 comet tails of wild-type or ΔinlC strains were measured. For analysis of F-actin recruitment (Fig. 1b), cells were infected for 5 h with Listeria strains pre-labeled with a Texas Red succinimidyl ester conjugate26, followed by fixation. Labeling of intracellular or extracellular bacteria was as described26. F-actin was labeled with phalloidin coupled to BodyP FL. F-actin recruitment efficiencies were expressed as the percentage of intracellular bacteria decorated with F-actin. Both bacteria displaying symmetric F-actin or asymmetric comet tails were scored. In each experiment, intracellular wild-type bacteria were analyzed until approximately 50 comet tails and 250 bacteria with symmetric F-actin were scored. An equivalent number of intracellular ΔinlC bacteria were analyzed.
InlC expression in infected cells
Caco-2 BBE1 cells in transwells were infected with wild-type or ΔinlC strains of Listeria for the indicated times. One h after the start of infection, medium containing gentamicin was added to eliminate extracellular bacteria. The time axis in Figure 1f indicates the total infection time, including the period preceding gentamicin treatment.
Apical junction studies
Zeiss LSM 510 software was used to measure linear indices in cells labeled for ZO1, as described3. For each condition, 50-110 cell junctions were analyzed. In experiments not involving bacterial infection, cells were randomly selected. In experiments involving infection, only cells displaying Listeria that recruited F-actin were analyzed. In experiments involving epitope-tagged InlC or β-galactosidase, only cells expressing the tagged transgenes were analyzed.
Analysis of protrusions
Two methods were used to assess protrusion formation. These methods involved either detection of protrusions by labeling for ezrin, or direct examination of protrusions in cells expressing EGFP-actin.
Ezrin -based approach
Caco-2 BBE1 cells were infected with wild-type or ΔinlC Listeria strains. At 5 h post-infection, cells were fixed, permeabilized, and labeled with mouse anti-ezrin and phalloidin-Alexa 647. Samples were analyzed using confocal microscopy. In each of the seven experiments used for Figure 1e, comets in cells infected with wild-type Listeria were analyzed until ∼25 ezrin-positive tails were scored. Similar numbers of total comets were analyzed in cells infected with the ΔinlC mutant. In each of the four experiments used for Figures 3b, 4b, and S4b, ∼ 20 ezrin-positive comets were scored in control siRNA or DMSO -treated cells infected with wild-type bacteria. Similar numbers of total comets were analyzed for all other conditions. In each of the three experiments used for Figure S4a, ∼ 30 ezrin-positive comet tails were scored in cells infected with wild-type Listeria. Similar numbers of total comet tails were analyzed for all other bacterial strains.
EGFP-actin –based approach
Cells were transfected with a plasmid expressing EGFP-actin. 72 h later, cells were infected with wild-type or ΔinlC strains. At 5 h post-infection, cells were fixed, permeabilized, and labeled with anti-L. monocytogenes antiserum R1126 and anti-rabbit-Cy5. Confocal microscopy was used to acquire images of 1μm serial sections encompassing the entire height of the cell monolayer. Comet tails that projected from EGFP-positive cells into flanking EGFP-negative cells were scored as protrusions. Tails located entirely within EGFP-positive cells were also counted. In each experiment, 200-300 total F-actin tails were analyzed for each bacterial strain.
Yeast two-hybrid screen
The DupLex-A yeast two-hybrid system (Origene) was used to screen a mouse embryonic cDNA library (Origene; DLM-110) using InlC with the LexA DNA binding domain fused to its C-terminus as bait. Approximately 1 × 107 independent cDNA clones were screened. The same InlC-interacting clone was isolated on three occasions.
Protein purification
Recombinant InlC or InlC.K173A mutant proteins containing C-terminal 6xHis tags were expressed in E. coli strain BL21 λDE3 and purified according to standard procedures. GST fusion proteins were expressed and purified essentially as described26. Purification of recombinant rat N-WASP protein deleted for its EVH1 domain was as described27.
Pull-down experiments
GST pull-downs were done essentially as described26.
Surface Plasmon Resonance
A Biacore X instrument was used to measure binding of InlC or InlC.K173A to the SH36 domain of Tuba. SH36 protein was linked to CM5 biosensor chips by amine coupling. A reference chip lacking protein was also prepared. Analyte injections were carried out at 25°C for 1 min (association phase), followed by a ∼1 min period of injection with buffer only (dissociation phase). Corrected binding data were generated by subtracting the RU values obtained with the reference chip from those measured with the SH36 chip. Data was analyzed using BiaEvaluation software (version 4.0.1). Association (ka) and dissociation (kd) rates were determined by fitting to a ‘two state binding with conformational change’ model. Equilibrium dissociation constants (KD) were calculated as kd/ka. Relative binding data in Figure 2eii were determined by normalizing the maximum RU value obtained with each concentration of the InlC.K173A mutant protein to that reached with the same concentration of wild-type InlC.
Statistical analysis
Statistical analysis was performed using InStat (version 2.2; GraphPad Software). When comparing data sets from two conditions, a Student's t-test was used. When comparing data from three or more conditions, ANOVA was performed. The Tukey-Kramer test was used as a post-test. A p value of 0.05 or lower was considered as significant.
Supplementary Material
Acknowledgments
We thank J. Brumell, H. Sarantis, A. Wilde, and R. Collins for reviewing the manuscript. We are grateful to N. Freitag, T. Otani, E. Leung, and K. Nemec for advice on assays or protein purification. Y. Shen is thanked for help with plasmid construction. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (MT-15497) and National Institutes of Health (1R21AI076881-01) awarded to K.I., and a CIHR grant (MOP-15499) awarded to S.D.G.
Footnotes
Author Contributions: K.I., T.R., and S.D.G. designed research; T.R. and B.G. performed the research; T.R. and K.I. analyzed data; M.H., S.M.A. and W.G. contributed novel reagents; K.I. and T.R. wrote the paper.
Competing Financial Interests: The authors declare that they have no competing financial interests.
References
- 1.Gouin E, Welch WD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Miki H, Takenawa T. Regulation of actin dynamics by WASP family proteins. J Biochem. 2003;134:309–313. doi: 10.1093/jb/mvg146. [DOI] [PubMed] [Google Scholar]
- 3.Otani T, Ichii T, Aono S, Takeichi M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J Cell Biol. 2006;175:135–46. doi: 10.1083/jcb.200605012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez-Boland JA, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins JR, et al. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J Cell Biol. 1999;146:1333–1349. doi: 10.1083/jcb.146.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monack DM, Theriot JA. Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cell Microbiol. 2001;3:633–647. doi: 10.1046/j.1462-5822.2001.00143.x. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi J, Takai Y. Molecular perspective on tight-junction assembly and epithelial polarity. Adv Drug Deliv Rev. 2005;57:815–855. doi: 10.1016/j.addr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Lecuit M, et al. A transgenic model for Listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 9.Peterson MD, Mooseker MS. Characterization of the enterocyte-like brush border cytoskeleton. J Cell Sci. 1992;102:581–600. doi: 10.1242/jcs.102.3.581. [DOI] [PubMed] [Google Scholar]
- 10.Sun AN, Camilli A, Portnoy DA. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun. 1990;58:3770–3778. doi: 10.1128/iai.58.11.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelbrecht, et al. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–37. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith GA, et al. The two distinct phospholipases C of Listeria monocytogenes have ovelapping roles in escape from a vacuole and cell-to-cell spread. Infec Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith GA, Theriot J, Portnoy D. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fievet B, Louvard D, Arpin M. ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta. 2007;1773:653–660. doi: 10.1016/j.bbamcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Pust S, Morrison H, Wehland J, Sechi AS, Herrlich P. Listeria monocytogenes exploits ERM protein functions to efficiently spread from cell to cell. EMBO J. 2005;24:1287–1300. doi: 10.1038/sj.emboj.7600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sechi AS, Wehland J, Small JV. The isolated comet tail pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J Cell Biol. 1997;137:155–167. doi: 10.1083/jcb.137.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 18.Bierne H, Sabet C, Personnic N, Cossart P. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infec. 2007;9:1156–66. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Salazar MA, et al. Tuba, a novel protein containing Bin-Amphysin-Rvs and Dbl homology domains, links Dynamin to regulation of the actin cytoskeleton. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 20.Kovacs EM, Makar RS, Gertler FB. Tuba stimulates N-WASP-dependent actin assembly. J Cell Sci. 2006;119:2715–26. doi: 10.1242/jcs.03005. [DOI] [PubMed] [Google Scholar]
- 21.Li SSC. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 23.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:456–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 24.Nusrat A, Turner JR, Madara JL. Molecular physiology and pathophysiology of tight junctions IV. Regulation of tight junctions by extracellular stimuli: nutrients, cytokines, and immune cells. Am J Physiol Gastrointest Liver Physiol. 2000;279:G851–G857. doi: 10.1152/ajpgi.2000.279.5.G851. [DOI] [PubMed] [Google Scholar]
- 25.Wuenscher MD, Kohler S, Goebel W, Chakraborty T. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol Gen Genet. 1991;228:177–182. doi: 10.1007/BF00282463. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, et al. Host adaptor proteins Gab1 and CrkII promote InlB-dependent entry of Listeria monocytogenes. Cell Microbiol. 2005;7:443–457. doi: 10.1111/j.1462-5822.2004.00475.x. [DOI] [PubMed] [Google Scholar]
- 27.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auerbuch V, Loureiro JJ, Gertler FB, Theriot JA, Portnoy DA. Ena-VASP proteins contribute to Listeria monocytogenes pathogenesis by controlling temporal and spatial persistence of bacterial actin-based motility. Mol Microbiol. 2003;49:1361–1375. doi: 10.1046/j.1365-2958.2003.03639.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.