Abstract
Methylation is a major biological process. It has been shown to be important in formation of compounds such as phosphatidylcholine, creatine, and many others and also participates in epigenetic effects through methylation of histones and DNA. The donor of methyl groups for almost all cellular methylation reactions is S-adenosylmethionine. It seems that the level of S-adenosylmethionine must be regulated in response to developmental stages and metabolic changes, and the enzyme glycine N-methyltransferase has been shown to play a major role in such regulation in mammals. This minireview will focus on the latest discoveries in the elucidation of the mechanism of that regulation.
Discovery of S-Adenosylmethionine and Its Versatility
AdoMet2 was discovered in 1951 by Cantoni as the “active methionine” used in the enzymatic transfer of the methyl group of methionine to nicotinamide to form N1-methylnicotinamide (1). With the exception of a few intracellular parasites that take up AdoMet from their hosts, AdoMet is formed from ATP and methionine by methionine adenosyltransferases present in all (or virtually all) cells of all organisms, including archaea, eubacteria, and eukaryotes. The reaction involves, initially, transfer of the adenosyl group of ATP to methionine, with the remainder of the ATP being converted to enzyme-bound tripolyphosphate. The latter compound is hydrolyzed to pyrophosphate and phosphate, which are then released (2). Being a sulfonium compound, AdoMet provides the large amounts of free energy (20 ∼kcal/mol) needed for methyl group transfers.
AdoMet is possibly the most (or, compared with ATP, the second most) versatile compound in Nature. It is a source not only of methyl groups but, in diverse reactions in various organisms, provides methylene groups, four-carbon moieties, ribosyl groups, amino groups, and, after decarboxylation, three-carbon moieties for polyamines and ethylene (3). It may be converted to a 5′-deoxyadenosyl free radical that participates in a great variety of “radical SAM” reactions (4). AdoMet also functions as a regulator of many metabolic pathways in mammals, plants, and bacteria.
In mammals, >90% of AdoMet is used for methylation reactions by at least 50 different methyltransferases (5). Methylation of both small molecules (e.g. phosphatidylethanolamine and guanidinoacetate) and macromolecules (DNA, RNA, histones, and other proteins) plays critical roles in cellular metabolism. Methylations of DNA and histones are major events in epigenetics. Therefore, the level of AdoMet must be carefully regulated to maintain cellular homeostasis. Recent evidence has established that GNMT plays a major role in maintaining normal AdoMet levels in mammals.
GNMT Genes and Proteins
In 1960, enzymatically catalyzed direct transfer of a methyl group from AdoMet to glycine (forming sarcosine) was demonstrated. The activity was found in liver extracts from guinea pig, rat, rabbit, and mouse, but the enzyme was not purified until 1972 when Heady and Kerr, upon finding that glycine was a better acceptor of AdoMet methyl groups than tRNA, proceeded to purify the GNMT activity from rabbit liver (6).
Tissue Distribution
GNMT is a tetrameric, cytosolic protein present in large amounts in liver (1–3% of cytosolic protein) and in pancreas and prostate (0.4% of cytosolic protein). Immunohistochemical studies confirmed these findings (7) and showed that GNMT protein is located in the exocrine tissue of the pancreas as well as in additional tissues active in secretion (proximal kidney tubules, submaxillary glands, intestinal mucosa, cortical neurons, and Purkinje cells of the brain).
Possible Nuclear Localization
Although GNMT is a cytosolic protein, its presence in liver nuclei has been reported by a number of laboratories based on both activity and immunological measurements (7, 8). The Bresnick group suggested that GNMT is the putative “4 S” aryl hydrocarbon receptor responsible for induction of cytochrome P-450 1A (CYP1A) by benzo(a)pyrene that is translocated to the nucleus (9). Other studies have questioned the role of GNMT as the 4 S aryl hydrocarbon receptor (8). Studies in which GNMT was incubated with isolated rat liver nuclei showed that native tetrameric GNMT was unable to enter the purified liver nuclei (10). The possible function of GNMT in the nucleus is still a matter of speculation, and if GNMT in the nucleus is not simply a result of cytoplasmic contamination, its function there is not clear because it does not appear to be involved in the induction of the cytochrome P-450 family of enzymes.
GNMT Genes and Expression
The first three GNMT genes to be cloned and sequenced were those from rat, mouse, and human. Each is relatively small with a simple structure. About 3200 nucleotides from the first ATG to the stop codons (11), the genes consist of six exons with an average size of 170 nucleotides and five introns. As genomes of other species were sequenced, open reading frames with homology to human, rat, and mouse GNMTs were found in zebrafish, red flour beetle, mosquito, and other organisms. Unpublished experiments3 have shown that GNMT enzyme activity is present in zebrafish liver, but for the other organisms, the presence of such activity has not been tested, and the function(s) of any such activities have not been elucidated. In mammals, the levels of mRNA are highest in liver, kidney, pancreas, and prostate, the organs with the greatest GNMT activities. GNMT either is not detected or is present in minimal amounts in livers of embryos of the experimental animals tested, but it is expressed strongly soon after birth.
GNMT expression is down-regulated or even completely blocked in liver and prostate tumor tissue (11, 12) and in most cultured cells. Although little is known about the properties of the GNMT gene promoters of experimental animals, it has been shown that GNMT activity is induced by vitamin A, glucocorticoids, and glucagon (13).
The greatly decreased levels of GNMT in prostate tumor tissues (11) might predict that the level of sarcosine in prostate should be relatively low (see Fig. 2). However, a recent study reported that sarcosine is found in elevated amounts in metastatic prostate tissue compared with normal human prostate and localized prostate cancer. Sarcosine is increased in the urine of men with metastatic prostate cancer, leading to the suggestion that this metabolite may be a marker for progression of the cancer (14). This remains to be confirmed.
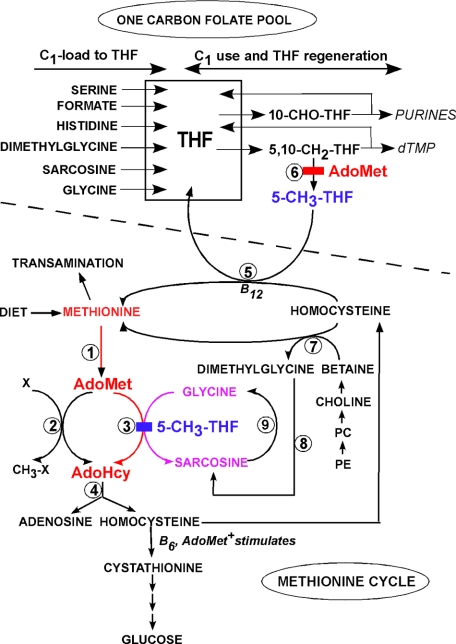
FIGURE 2.
Metabolic pathways related to the regulatory role of GNMT. Two metabolic pathways are shown separated by a dashed line; the ONE CARBON FOLATE POOL and the METHIONINE CYCLE. Indicated on the left of the one-carbon pool are the sources of the one-carbon groups carried by THF; on the right are indicated the metabolic uses of these groups. The methionine cycle includes reactions that transfer methyl groups from methionine to AdoMet and methylation of acceptors. These are followed by regeneration of methionine via methylation of homocysteine by either methionine synthase or betaine-homocysteine methyltransferase. Homocysteine can also be converted to cysteine and, ultimately, to glucose via cystathionine breakdown. The reactions are denoted by numbers: 1, methionine adenosyltransferase; 2, the majority of methyltransferases; 3, GNMT (inhibition by 5-methyl-THF is shown); 4, AdoHcy hydrolase; 5, methionine synthase; 6, 5,10-methylene-THF reductase (with inhibition by AdoMet); 7, betaine-homocysteine methyltransferase; 8, dimethylglycine dehydrogenase; 9, sarcosine dehydrogenase. X refers to the group of methyl acceptors, and CH3-X refers to the methylated products. Fig. 2 first appeared in Ref. 43. PE, phosphatidylethanolamine.
GNMT Protein
The enzymatic characteristics of GNMT and the properties of the protein from rabbit, rat, and human, either purified from liver or pancreas or expressed in Escherichia coli, have been studied most extensively. Protein sequences of human, rabbit, rat, pig, and mouse have ∼90% sequence identity (15). All GNMTs are 130-kDa tetramers consisting of four identical subunits. Each subunit consists of 292–296 amino acid residues (depending on the source) and possesses an active center for enzymatic reaction (Fig. 1). The initial methionine residue is removed, and GNMT proteins from liver or expressed in E. coli have an N-terminal valine. The N-terminal valines in rat and human GNMTs are acetylated, but when recombinant rat or human GNMT is expressed in E. coli, they are not acetylated (16, 17). The only known post-translational modification of liver GNMT is serine phosphorylation. Serines 9, 71, 139, 182, and 241 may be partially phosphorylated in rat liver and recombinant GNMTs, but in the purified rat liver enzyme, the amount of phosphorylation is very low (17). Ser9 of rat GNMT can be phosphorylated in vitro by the glucagon-activated cAMP-dependent protein kinase (18). Gluconeogenesis (facilitated by glucagon) from certain amino acids, including methionine, is activated in diabetes and fasting, and it has been suggested that GNMT may be a major enzyme involved in the ultimate utilization of the four-carbon moiety of methionine (Fig. 2) for gluconeogenesis.
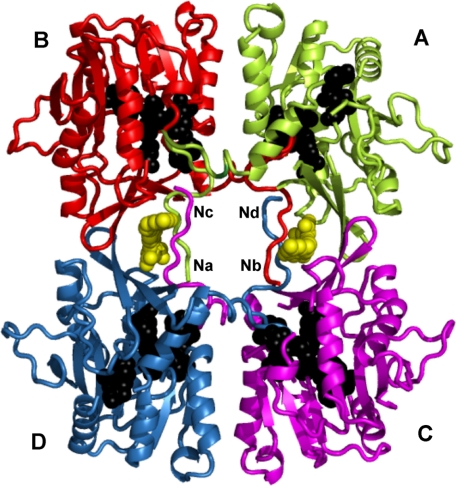
FIGURE 1.
Crystal structure of recombinant rat GNMT complexed with 5-methyl-THF. Coordinates of the structure from Protein Data Bank code 2IDK were used for preparation of this figure. Each subunit is denoted A, B, C, and D. Two molecules of folate are shown as yellow spheres. The overall structure was drawn in schematic mode of the PyMOL program. The residues participating in binding AdoMet (Trp39, Arg40, Ala64, Asp85, Asn116, Trp117, and Leu136) are drawn as black spheres. The N termini of subunits are denoted Na, Nb, Nc, and Nd. Interaction of the N-terminal fragment of subunit A (green) with the active center of subunit B (red) is clearly seen.
Crystal structures have been solved only for recombinant rat, mouse, and human GNMTs (19–22) crystallized either as apoproteins or complexed with AdoMet, AdoHcy, both AdoMet and acetate (a competitive inhibitor of glycine), or 5-CH3-H4PteGlu. In all crystal structures, GNMT is modeled as a flat-shaped tetramer with numerous interactions between the subunits (Fig. 1). The active sites lie deep within the globular portion of each subunit. Based on the crystal structures of GNMT complexed with AdoMet or AdoHcy and kinetic studies of numerous mutants of recombinant rat GNMT, a mechanism of enzyme reaction has been proposed (21): inactive GNMT exists in a “closed” conformation in which the N-terminal fragments of each subunit interact with the globular part of the adjacent subunit in a manner that closes access to the active centers (Fig. 1). In the presence of AdoMet, an “open” configuration is formed in which the substrate competes with the N termini for access to the active centers, and when AdoMet binds at the active centers, the N termini protrude from the binding sites, no longer interacting with the globular parts of the molecule.
Characteristics of Enzyme Activity
GNMT activity is optimum at pH 9.0. Depending upon the source of the enzyme, half-maximal activity is attained with 0.03–0.2 mm AdoMet and 2–20 mm glycine. The kcat values are in the range of 35–96 min−1. Early studies indicated that activity is cooperative with respect to AdoMet but not glycine (23). However, it was subsequently found that such cooperativity is seen only with GNMT purified from liver, not with recombinant enzyme expressed in E. coli, a finding perhaps explained either by the difference in N-terminal acetylation between the native and recombinant enzymes (24) or by the possibility that tightly bound folate is not completely removed during enzyme purification from liver, whereas there is no folate bound to the enzyme expressed in E. coli.
Glycine N-Methyltransferase Is a Major Folate-binding Protein
A new and novel role for GNMT was discovered in 1977 (25). Injection of rats with [3H]folic acid, followed in 24 h by extraction of liver proteins and gel filtration chromatography of cytosolic proteins, showed a major peak of tightly bound radioactivity in which the bound folate was primarily the pentaglutamate form of 5-CH3-H4PteGlu. It was eventually found that this cytosolic folate-binding protein was identical to GNMT (26). 5-CH3-H4PteGlu5 is also the form of folate bound preferentially in vitro by purified rat liver GNMT, a surprising observation because GNMT does not use any form of folate as a substrate. Subsequently, it was discovered that, when 5-CH3-H4PteGlu5 is bound, it behaves as an inhibitor of the enzyme. Thus, GNMT links changes in AdoMet concentration to the transfer and synthesis of one-carbon units by folate-metabolizing enzymes, thereby serving as a bridge between methionine and one-carbon metabolism.
Inhibition of GNMT by Folate
A unique aspect of GNMT activity is its inhibition by 5-CH3-H4PteGlu5 (27), studied most thoroughly with the enzymes from rat liver and pancreas (28), although unpublished observations3 have shown that the GNMT activity of zebrafish liver is also inhibited by 5-CH3-H4PteGlu5. The crystal structure of the complex of recombinant rat GNMT with the monoglutamate form of 5-CH3-H4PteGlu (Fig. 1) (29) elucidates the mechanism of the inhibition: two molecules of folate are bound by the tetrameric protein, and each interacts with the N termini of two subunits, making entrance of AdoMet into the active centers much more difficult. The bound folate is thought to impede the swinging out of position of the N termini that normally allows access of the substrates to the active sites. Despite similar binding constants, folate inhibition of the liver enzyme with its acetylated N-terminal valines is much stronger than inhibition of the enzyme expressed in E. coli, in which valine is not acetylated; 50% inhibition is seen at 0.0013 and 0.59 mm 5-CH3-H4PteGlu5 for the native and recombinant enzymes, respectively (30).
Role of GNMT in Regulation of AdoMet Levels
Fig. 2 shows how GNMT is involved in both hepatic methyl group and one-carbon metabolism. AdoMet is synthesized from methionine and ATP by methionine adenosyltransferases (reaction 1). The methyl group of AdoMet is transferred to a variety of acceptors by methyltransferases (reactions 2 and 3). AdoHcy is formed in each such reaction. With normal dietary intakes, there are usually not enough preformed methyl groups (contained chiefly in methionine- and choline-containing compounds) to meet the total need for transmethylation. Additional methyl groups are synthesized de novo via the one-carbon folate pool. Folate coenzymes carry one-carbon units (formyl, formaldehyde, and methyl groups) attached at either position 5 or 10 of THF. Intracellular forms of folate are all polyglutamates. The folate coenzymes bearing the one-carbon substituents are metabolically connected to one another and in Fig. 2 are collectively referred to as the ONE CARBON FOLATE POOL.
The reactions labeled 2 in Fig. 2 consist of a wide variety of 50 (or more) methyltransferases that catalyze the synthesis of essential products, including small molecules such as creatine and PC, as well as the methylation of macromolecules such as proteins, RNA, and DNA. Reaction 3 also involves a methyltransferase, GNMT, that is highly unusual in that the product it forms, sarcosine, has no known essential metabolic function. A mitochondrial enzyme, sarcosine dehydrogenase, catalyzes the conversion of sarcosine to glycine and methylene-THF (Fig. 2, reaction 8). Together, these facts are consistent with and give rise to the interpretation that the importance of GNMT lies not in its ability to form sarcosine but rather in its capacity to regulate utilization of AdoMet and thus affect the AdoMet/AdoHcy ratio (a ratio sometimes considered to be an index of the methylating ability of the cell).
How Regulation Takes Place
The metabolic control is pictured to operate as follows. When methionine levels in the diet are high, the AdoMet level is also high, and there is no need for de novo methyl group synthesis via the one-carbon folate pool. The high AdoMet levels inhibit 5,10-methylene-THF reductase (Fig. 2, reaction 6), and the levels of 5-CH3-H4PteGlu5 are decreased. The lower levels of 5-CH3-H4PteGlu5 lessen the inhibition of GNMT, and the excess AdoMet is converted to sarcosine and AdoHcy. On the other hand, when methionine is low, AdoMet will be low, and inhibition of 5,10-methylene-THF reductase is relieved, permitting more de novo synthesis of methyl groups by formation of 5-CH3-H4PteGlu5. The latter compound then inhibits GNMT, conserving AdoMet for physiologically important methylation reactions.
Early support for the above scheme was provided by measurements of sarcosine excretion in humans deficient in sarcosine dehydrogenase activity, who therefore excreted the majority of the sarcosine they formed rather than catabolizing it, as do normal subjects, to glycine and methylene-THF. On normal diets, these subjects excreted relatively little sarcosine, but when their dietary methyl group intakes exceeded the amounts required for necessary transmethylation reactions, the excess methyl groups appeared in the urine as sarcosine (31). Further support was provided by studies of rats fed a methyl group-deficient diet (choline omitted and methionine replaced by equimolar homocysteine) (32). The amount of GNMT protein measured immunologically did not change, but the specific activity of GNMT decreased, indicating inhibition of activity. This was presumably due to decreased dietary methionine having lowered AdoMet levels, thus removing the inhibition of 5,10-methylene-THF reductase and producing elevated amounts of 5-CH3-H4PteGlu5 and greater inhibition of GNMT. Additional experiments showed that folate deficiency results in elevated GNMT activity (33). Again, the amount of enzyme protein did not change. The level of 5-CH3-H4PteGlu5 was greatly reduced, and inhibition of GNMT was lessened. Thus, when more AdoMet is formed than is needed for the usual methylation reactions, it is catabolized by methylating glycine, forming sarcosine, which is catabolized in turn to recover the input glycine and the one-carbon moiety as methylene-THF.
Human GNMT Deficiency
Recently, definitive proof of the proposed model has been provided by studies of genetically determined GNMT deficiency in human children (34–36). The first two are Italian siblings who, on normal diets, have very high levels of plasma methionine and AdoMet with normal AdoHcy and total homocysteine and without elevation of sarcosine. Both have moderate liver disease (elevated plasma liver transaminases and slightly elevated alkaline phosphatase and triglycerides) and hepatomegaly. DNA sequencing showed that both siblings are compound heterozygotes for a GNMT mutation leading to replacement of Leu49 by proline and a second one producing H176N. A third case is a Greek child who has similar metabolic abnormalities. He has elevated liver transaminases but no hepatomegaly and is homozygous for an N140S substitution. Assay of the mutant GNMTs expressed in E. coli showed that each inactivates GNMT (37).
GNMT Knock-out Mice
A GNMT knock-out mouse was developed by Luka et al. (38) using gene targeting. Transgenic Gnmt−/− mice are fertile and able to reproduce. Their appearances and growth rates for 3–6 months are similar to those of wild-type mice. GNMT activity assay and Western blotting showed that the transgenic animals possess neither GNMT activity nor protein. The changes in the levels of key metabolites in liver were similar to those in the plasma of human patients: methionine increased from 100 nmol/g of liver in wild-type mice to ∼700 nmol/g in Gnmt−/− animals, and AdoMet increased from 37 nmol/g to ∼1334 nmol/g. In addition, AdoHcy slightly decreased from 12–15 to 3–5 nmol/g of liver. Thus, the concentration of AdoMet increased ∼36-fold, and the AdoMet/AdoHcy ratio increased ∼100-fold, suggesting significant increases in a variety of cellular methylation reactions.
The livers at 3 and 8 months of age show evidence of fatty accumulation and fibrosis that worsen progressively. At 8 months, Gnmt−/− mice have fibrosis and inflammation, and all develop HCC (39). HCC development in Gnmt−/− mice, as in humans, coincides with activation of the Ras and JAK/STAT (signal transducer and activator of transcription) signaling pathways. In Gnmt−/− mice, this is caused by inactivation of inhibitors of these pathways, RASSF1 and SOCS2, via hypermethylation of their promoters. In addition, histone methylation in Gnmt−/− mice is altered.
That the Gnmt−/− mice develop fatty livers is somewhat unexpected. Fatty liver in rodents has been ascribed to low levels of “lipotropes” (methionine, choline, folate, and vitamin B12), so-called because inadequate amounts in the diet lead to fatty liver. These metabolites are involved not only in the direct maintenance of methionine and choline but also in the regeneration of methionine (from homocysteine) (Fig. 2, Methionine Cycle) and choline through AdoMet-dependent methylation of phosphatidylethanolamine to form PC. The fatty livers of animals on such lipotrope-deficient diets have been thought to result from either an inability to transport triglycerides from the liver to extrahepatic tissues via very low density lipoprotein and/or a defect in the synthesis of phospholipids (40). Because a large portion of the synthesis of PC in liver is dependent upon AdoMet, it is puzzling that Gnmt−/− mice develop fatty livers when they have increased AdoMet.
A second GNMT knock-out mouse model has been developed by Liu et al. (41) using a similar approach to that developed by Luka et al. (38). Changes in metabolite concentrations were similar, but there were differences with regard to fat deposition and HCC. Initially, the Taiwan group found no HCC, and fatty liver did not develop in their Gnmt−/− mice. Instead, abnormal glycogen storage was found in livers at 9 months of age. Subsequently, HCC did occur but at significantly older ages (17 months) (42). These phenotypic differences between the two GNMT knock-out mouse models are presently unexplained. Nevertheless, the Gnmt−/− models provide strong evidence that the role of GNMT is to regulate the level of AdoMet in tissues. In liver, where GNMT is most abundant, AdoMet levels are very high, and profound abnormalities are seen. Tissues other than liver that have high levels of GNMT (exocrine pancreas, prostate, and kidney) have not developed abnormalities or malignancies at times up to 8 months of age.
Concluding Remarks
The studies described in this minireview show that the main biological role of GNMT in mammals is to regulate the level of AdoMeto. This role is now well documented by studies of humans with GNMT deficiency and mouse models with GNMT knocked out. Because AdoMet is needed for many biological processes, the regulation of GNMT activity appears to be critical for homeostasis. This is done by folate inhibition of the enzyme activity or by regulation of gene expression. Inhibition of GNMT by folate is relatively well studied, but detailed structural studies involving natural polyglutamated folates have not yet been performed. Although GNMT is phosphorylated to a limited extent, it is not known whether such phosphorylation plays a significant role. Another question raised by the GNMT knock-out mouse is the fatty liver that develops in the presence of an abundance of AdoMet. This may require a re-evaluation of current theories for the development of fatty liver. Important next efforts would be to clarify the mechanism of GNMT gene regulation to more fully understand its role in biological methylation, epigenetics, and cancer.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK15289 and DK080010 and by a merit revue award from the Department of Veterans Affairs. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Z. Luka and C. Wagner, unpublished data.
- AdoMet
- S-adenosylmethionine
- GNMT
- glycine N-methyltransferase
- AdoHcy
- S-adenosylhomocysteine
- THF
- tetrahydrofolate
- 5-CH3-H4PteGlu
- 5-methyltetrahydrofolic acid
- PC
- phosphatidylcholine
- HCC
- hepatocellular carcinoma.
REFERENCES
- 1.Cantoni G. L. (1951) J. Biol. Chem. 189,203–216 [PubMed] [Google Scholar]
- 2.Mudd S. H. (1963) J. Biol. Chem. 238,2156–2163 [PubMed] [Google Scholar]
- 3.Fontecave M., Atta M., Mulliez E. (2004) Trends Biochem. Sci. 29,243–249 [DOI] [PubMed] [Google Scholar]
- 4.Wang S. C., Frey P. A. (2007) Trends Biochem. Sci. 32,101–110 [DOI] [PubMed] [Google Scholar]
- 5.Clarke R., Lewington S., Landray M. (2003) Kidney Int. Suppl. 84,S131–133 [DOI] [PubMed] [Google Scholar]
- 6.Heady J. E., Kerr S. J. (1973) J. Biol. Chem. 248,69–72 [PubMed] [Google Scholar]
- 7.Yeo E. J., Wagner C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91,210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa H., Gomi T., Imamura T., Kobayashi M., Huh N. (1997) Biochem. Biophys. Res. Commun. 233,300–304 [DOI] [PubMed] [Google Scholar]
- 9.Bhat R., Weaver J. A., Wagner C., Bodwell J. E., Bresnick E. (1996) J. Biol. Chem. 271,32551–32556 [DOI] [PubMed] [Google Scholar]
- 10.Krupenko N. I., Wagner C. (1997) J. Biol. Chem. 272,27140–27146 [DOI] [PubMed] [Google Scholar]
- 11.Huang Y. C., Lee C. M., Chen M., Chung M. Y., Chang Y. H., Huang W. J., Ho D. M., Pan C. C., Wu T. T., Yang S., Lin M. W., Hsieh J. T., Chen Y. M. (2007) Clin. Cancer Res. 13,1412–1420 [DOI] [PubMed] [Google Scholar]
- 12.Heady J. E., Kerr S. J. (1975) Cancer Res. 35,640–643 [PubMed] [Google Scholar]
- 13.Williams K. T., Schalinske K. L. (2007) J. Nutr. 137,311–314 [DOI] [PubMed] [Google Scholar]
- 14.Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Nature 457,910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15.Ogawa H., Gomi T., Fujioka M. (1993) Comp. Biochem. Physiol. B 106,601–611 [DOI] [PubMed] [Google Scholar]
- 16.Ogawa H., Gomi T., Takata Y., Date T., Fujioka M. (1997) Biochem. J. 327,407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luka Z., Ham A. J., Norris J. L., Yeo E. J., Yermalitsky V., Glenn B., Caprioli R. M., Liebler D. C., Wagner C. (2006) Protein Sci. 15,785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner C., Decha-Umphai W., Corbin J. (1989) J. Biol. Chem. 264,9638–9642 [PubMed] [Google Scholar]
- 19.Pattanayek R., Newcomer M. E., Wagner C. (1998) Protein Sci. 7,1326–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y., Komoto J., Konishi K., Takata Y., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (2000) J. Mol. Biol. 298,149–162 [DOI] [PubMed] [Google Scholar]
- 21.Takata Y., Huang Y., Komoto J., Yamada T., Konishi K., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (2003) Biochemistry 42,8394–8402 [DOI] [PubMed] [Google Scholar]
- 22.Pakhomova S., Luka Z., Grohmann S., Wagner C., Newcomer M. E. (2004) Proteins 57,331–337 [DOI] [PubMed] [Google Scholar]
- 23.Ogawa H., Fujioka M. (1982) J. Biol. Chem. 257,3447–3452 [PubMed] [Google Scholar]
- 24.Konishi K., Fujioka M. (1988) J. Biol. Chem. 263,13381–13385 [PubMed] [Google Scholar]
- 25.Zamierowski M. M., Wagner C. (1977) J. Biol. Chem. 252,933–938 [PubMed] [Google Scholar]
- 26.Cook R. J., Wagner C. (1984) Proc. Natl. Acad. Sci. U.S.A. 81,3631–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner C., Briggs W. T., Cook R. J. (1985) Biochem. Biophys. Res. Commun. 127,746–752 [DOI] [PubMed] [Google Scholar]
- 28.Yeo E. J., Wagner C. (1992) J. Biol. Chem. 267,24669–24674 [PubMed] [Google Scholar]
- 29.Luka Z., Pakhomova S., Loukachevitch L. V., Egli M., Newcomer M. E., Wagner C. (2007) J. Biol. Chem. 282,4069–4075 [DOI] [PubMed] [Google Scholar]
- 30.Luka Z., Loukachevitch L. V., Wagner C. (2008) Biochim. Biophys. Acta 1784,1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mudd S. H., Ebert M. H., Scriver C. R. (1980) Metab. Clin. Exp. 29,707–720 [DOI] [PubMed] [Google Scholar]
- 32.Cook R. J., Horne D. W., Wagner C. (1989) J. Nutr. 119,612–617 [DOI] [PubMed] [Google Scholar]
- 33.Horne D. W., Cook R. J., Wagner C. (1989) J. Nutr. 119,618–621 [DOI] [PubMed] [Google Scholar]
- 34.Mudd S. H., Cerone R., Schiaffino M. C., Fantasia A. R., Minniti G., Caruso U., Lorini R., Watkins D., Matiaszuk N., Rosenblatt D. S., Schwahn B., Rozen R., LeGros L., Kotb M., Capdevila A., Luka Z., Finkelstein J. D., Tangerman A., Stabler S. P., Allen R. H., Wagner C. (2001) J. Inherit. Metab. Dis. 24,448–464 [DOI] [PubMed] [Google Scholar]
- 35.Luka Z., Cerone R., Phillips J. A., 3rd, Mudd H. S., Wagner C. (2002) Hum. Genet. 110,68–74 [DOI] [PubMed] [Google Scholar]
- 36.Augoustides-Savvopoulou P., Luka Z., Karyda S., Stabler S. P., Allen R. H., Patsiaoura K., Wagner C., Mudd S. H. (2003) J. Inherit. Metab. Dis. 26,745–759 [DOI] [PubMed] [Google Scholar]
- 37.Luka Z., Wagner C. (2003) Biochem. Biophys. Res. Commun. 312,1067–1072 [DOI] [PubMed] [Google Scholar]
- 38.Luka Z., Capdevila A., Mato J. M., Wagner C. (2006) Transgenic Res. 15,393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Chantar M. L., Vázquez-Chantada M., Ariz U., Martínez N., Varela M., Luka Z., Capdevila A., Rodríguez J., Aransay A. M., Matthiesen R., Yang H., Calvisi D. F., Esteller M., Fraga M., Lu S. C., Wagner C., Mato J. M. (2008) Hepatology 47,1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombardi B., Pani P., Schlunk F. F. (1968) J. Lipid Res. 9,437–446 [PubMed] [Google Scholar]
- 41.Liu S. P., Li Y. S., Chen Y. J., Chiang E. P., Li A. F., Lee Y. H., Tsai T. F., Hsiao M., Hwang S. F., Chen Y. M. (2007) Hepatology 46,1413–1425 [DOI] [PubMed] [Google Scholar]
- 42.Liao Y. J., Liu S. P., Lee C. M., Yen C. H., Chuang P. C., Chen C. Y., Tsai T. F., Huang S. F., Lee Y. H., Chen Y. M. (2009) Int. J. Cancer 124,816–826 [DOI] [PubMed] [Google Scholar]
- 43.Luka Z. (2008) Vitam. Horm. 79,325–345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.