Abstract
Methods for computationally predicting deleterious mutations have recently been investigated for proteins, mainly by probabilistic estimations in the context of genomic research for identifying single nucleotide polymorphisms that can potentially affect protein function. It has been demonstrated that in cases where a few homologs are available, ab initio predicted structures modeled by the Rosetta method can become useful for including structural information to improve the deleterious mutation prediction methods for proteins. In the field of RNAs where very few homologs are available at present, this analogy can serve as a precursor to investigate a deleterious mutation prediction approach that is based on RNA secondary structure. When attempting to develop models for the prediction of deleterious mutations in RNAs, useful structural information is available from folding algorithms that predict the secondary structure of RNAs, based on energy minimization. Detecting mutations with desired structural effects among all possible point mutations may then be valuable for the prediction of deleterious mutations that can be tested experimentally. Here, a method is introduced for the prediction of deleterious mutations in the secondary structure of RNAs. The mutation prediction method, based on subdivision of the initial structure into smaller substructures and construction of eigenvalue tables, is independent of the folding algorithms but relies on their success to predict the folding of small RNA structures. Application of this method to predict mutations that may cause structural rearrangements, thereby disrupting stable motifs, is given for prokaryotic transcription termination in the thiamin pyrophosphate and S-adenosyl-methionine induced riboswitches. Ribo switches are mRNA structures that have recently been found to regulate transcription termination or translation initiation in bacteria by conformation rearrangement in response to direct metabolite binding. Predicting deleterious mutations on riboswitches may succeed to systematically intervene in bacterial genetic control.
INTRODUCTION
In recent years, deleterious mutation prediction has been an area of active research in computational studies of proteins. Based on sequence homologs, probabilistic methods were developed that sort intolerant from tolerant amino acid substitutions (1) and predict whether an amino acid substitution at a particular position in a protein will have a disruptive effect. The success of these probabilistic methods depends on the availability of enough homologs in protein databases. To complement this work for cases where fewer than five or 10 homologs are available, ab initio protein structure prediction methods such as the Rosetta method (2) have been suggested. Thus, the potential benefit of involving structural information in methods for predicting deleterious mutations in proteins has been noticed. When transforming this prediction problem to RNAs, the importance of structural information is amplified since compared with proteins the number of available structures in databases for performing RNA homology predictions is significantly smaller.
As with proteins, mutations in RNAs have potential to alter their functionality in important biological processes. Recently, much progress has been achieved in understanding the role of riboswitches during transcription termination (3) and translation initiation (4) in bacteria (5–7). A riboswitch is a small segment of the mRNA (∼100–200 nt) that can sense small molecules and, by responding with a structural change, alter gene expression accordingly. In transcription termination during the thiamin (vitamin B1) biosynthesis in Bacillus subtilis, the riboswitch senses the thiamin pyrophosphate (TPP) ligand that binds to it and retains a secondary structure that contains a terminator hairpin which is highly stable (3). The terminator structure is responsible for the termination of transcription. With the absence of the TPP ligand, according to the model, the terminator structure is replaced by an antiterminator structure that interferes with transcription termination. The riboswitch structure is responsible for the regulation of transcription termination in Gram-positive bacteria, and translation initiation in Gram-negative bacteria.
Here, a new method for deleterious mutation prediction based on structural information for RNAs is presented. Suspected deleterious mutations found by structure predictions can easily be tested in laboratory experiments, and if found to inhibit important processes such as translation and transcription, the structural rearrangement as a cause of introducing the mutation can alter the regulatory mechanism. At present, such predictive calculations are limited to secondary structure in RNAs, whereas in the future it may be possible to extend these predictions to include tertiary structure information. Nevertheless, due to the hierarchical nature of RNA folding (8), the secondary structure is indicative of the folding to tertiary structure. Moreover, a substantial amount of information can be inferred about the riboswitch mechanism by examining its secondary structure, since certain highly important secondary structure motifs (e.g. terminators/anti-terminators and sequestors/anti-sequestors) have been located (9) in addition to the conserved box (10,11) that can identify riboswitch configurations.
The effect of single point mutations in RNA secondary structure has been addressed before using RNA tree-graph representation (12). Here, a novel similarity measure is introduced that facilitates computational predictions by using spectral decomposition for the RNA tree graphs, enabling one to examine higher order extensions to a single point mutation in a computational tractable manner. Therefore, the prediction problem becomes 2-fold, to find the minimal number of nucleotide mutations required to disrupt a stable motif and to specify their locations within the sequence. An illustrative example of the prediction method is given for attempting to disrupt the L5b tetraloop GAAA of the P5abc subdomain in the group I intron ribozyme of Tetrahymena thermophila, followed by deleterious mutation prediction for disrupting the prokaryotic transcription termination in the TPP (3) and S-adenosyl-methionine (SAM) (6) induced riboswitches.
MATERIALS AND METHODS
The tree graph representation of the secondary structure of RNAs was introduced by Shapiro (13) and Le et al. (14) and has consequently been tried by Margalit et al. (12) to examine the prediction of single point mutations in the L11 mRNA of Escherichia coli, without the eigenvalue methodology presented here. Riboswitch mechanisms were not known at the time and the mathematics of spectral graph theory was less developed. Here, the introduction of spectral decomposition methods suggested in Barash and Comaniciu (15), and the clustering of tree graphs into eigenvalues, allows an efficient organization of the data that enables the prediction of multiple mutations to disrupt desired motifs in riboswitches.
The Laplacian matrix
Given a tree-graph representation of an RNA, an equivalent way to represent tree graphs is by a special matrix called the Laplacian. The Laplacian matrix L(G) corresponding to a graph G is a symmetric matrix, with one row and column for each node on the graph. It is constructed as follows: in the diagonal of L, the degree of the vertex (number of incident edges) is specified, while in the off-diagonals the value ‘–1’ is inserted if there is an edge at that location, or ‘0’ if there is no connecting edge. As an example, for the tree-graph representation of the RNA structure in Figure 1A, the corresponding Laplacian matrix becomes:
Figure 1.
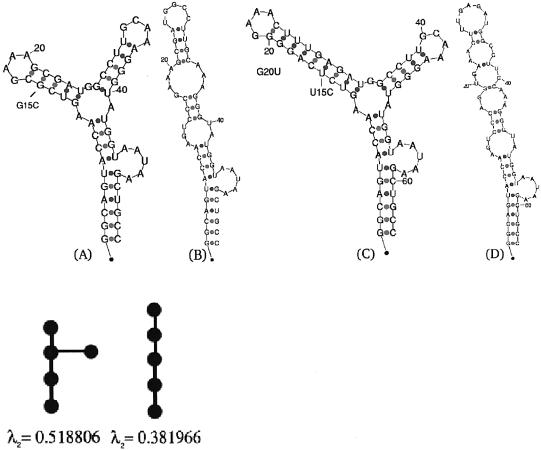
The secondary structure of the P5abc subdomain of the T.thermophila group I intron ribozyme. (A) The wild-type folded structure, along with its tree graph representation and the corresponding second eigenvalue below it. The tree graph possesses five vertices, since a ‘pseudo-loop’ representing the 5′-3′ is uniformly accounted for as a node, in addition to the loops, bulges and hairpins. An artificial structure without 4 bp near the L5b GAAA tetraloop has been shown by NMR experiments (21) to yield the same secondary structure predicted by mfold without the addition of magnesium ions. (B) Mutated structure; predicted mutation G15C with respect to the wild-type structure in (A), along with its tree graph representation and the corresponding second eigenvalue below it. (C) The predicted wild-type folded structure of the natural P5abc subdomain, with the 4 bp included in the P5b stem, as in Cate et al. (22). (D) Mutated structure; predicted mutation U15C and G20U with respect to the wild-type structure in (C). A two point mutation is required to disrupt the L5b GAAA tetraloop in case the 4 bp are not removed, when using the deleterious mutation prediction method described in the text.
Note that the Laplacian matrix is symmetric, and its rows and columns sum up to zero. The complete set of eigenvalues of the Laplacian matrix is called the spectrum of the graph, and is independent of how graph vertices are labeled. The following properties characterize the Laplacian matrix eigenvalues: (i) the eigenvalues of L(G) are non-negative and the first eigenvalue is zero; (ii) the second smallest eigenvalue is the algebraic connectivity of G (16); (iii) for a ‘star-shaped’ tree graph, the second smallest eigenvalue is unity (17).
Second eigenvalue of the Laplacian matrix
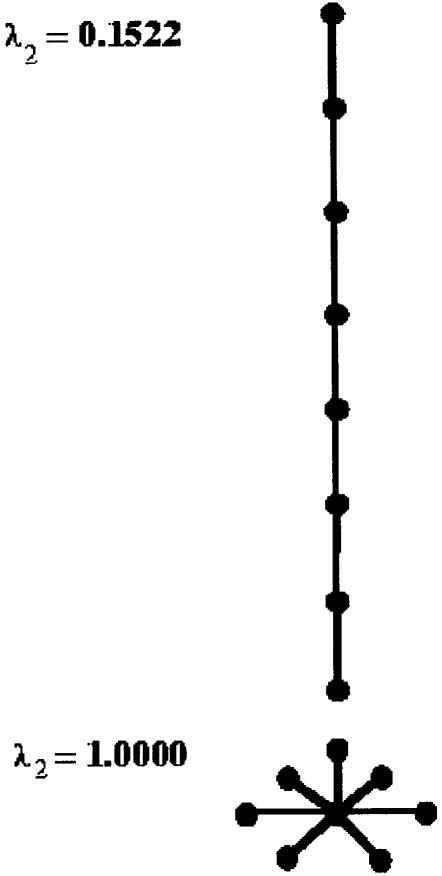
It has been originally proposed in Barash and Comaniciu (15), based on analogies with domain decomposition in parallel computing, to consider the second eigenvalue of the Laplacian matrix as a measure of the RNA tree-graph compactness. The second eigenvalue of the Laplacian matrix, corresponding to a general graph, is the measure of its graph connectivity (16). Intuitively, it is monotonically increasing from its lowest value for a linear tree-graph structure to its highest value for a ‘star shaped’ tree-graph structure, as illustrated in Figure 2. In the example of Figure 1, it can be noticed how the eigenvalues appear in descending order from a higher value for the initial tree graph structure depicted in Figure 1A to a lower value corresponding to a linear tree-graph structure depicted in Figure 1B.
Figure 2.
Second eigenvalue of the Laplacian matrix for a graph of eight vertices is lowest for a linear tree-graph structure (λ2 = 0.1522) and highest for a ‘star shaped’ tree-graph structure (λ2 = 1.0).
Other graph motif measures besides the eigenvalues can be used for deleterious mutation prediction, but the theorem by Fiedler (16), associating the second smallest eigenvalue of the Laplacian matrix to the algebraic connectivity of a graph, guarantees that the second eigenvalue measure is the most efficient single number to be used for capturing the geometry of the secondary structure in practical RNA calculations. This allows an economical procedure that is especially effective in time-consuming deleterious mutation prediction tasks, as in the case of the SAM riboswitch described in the applications. Rare cases of ambiguity can arise when two structures correspond to the same degree of compactness, therefore possessing the same second eigenvalue. In those cases, the energies of the folded structures can be used to differentiate between the two and resolve the ambiguity.
The proposed method is independent of the folding algorithms, but relies in the predictions presented here on Zuker’s mfold (18) and the Vienna package (19), both using the energy parameters described in Mathews et al. (20).
RESULTS AND DISCUSSION
Illustrative example
The secondary structure of the P5abc subdomain in the group I intron ribozyme of T.thermophila, derived by NMR experiments (21) without the addition of magnesium ions, is well predicted by mfold (18) and the Vienna package (19). The predicted structure is depicted in Figure 1A and assumes the tree-graph representation below it, which is quantified by the eigenvalue λ2 = 0.518806 as a measure of its graph compactness (see Materials and Methods). Given an initial structure, it is now possible to computationally predict deleterious mutations that may cause structural rearrangements, by performing an efficient exhaustive search over all possible mutations without the need to manually examine each fold as a consequence of introducing a mutation. In the example in Figure 1A, there are (56 nt × 3) = 168 possible single point mutations that can be introduced to the structure, (56 nt × 3)2 = 28 224 possible two point mutations, and so on. The goal is to predict a deleterious mutation that causes a structural rearrangement with the least amount of mutations. Thus, the examination of each of the possible 168 folds as a consequence of a single point mutation is a formidable task, and becomes non-realistic for two point mutations and more. Therefore, the possibility of effectively calculating only the second eigenvalue of the Laplacian matrix representation of each fold offers a computationally tractable means by which deleterious mutations can be predicted in a systematic procedure. This procedure can be referred to as the eigenvalue method.
Using the proposed method, it was found that almost all single point mutations will lead to a folded structure with the same eigenvalue as the wild-type structure in Figure 1A (λ2 = 0.518806). Only the single point mutation G15C, labeled in Figure 1A, will lead to the linear predicted folded structure drawn in Figure 1B with an eigenvalue of λ2 = 0.381966 corresponding to a linear tree-graph representation. The calculated mutation may cause a structural rearrangement that eliminates the L5b GAAA tetraloop (21), resulting in Figure 1B as the new predicted global minimum energy solution. In addition, mutations C22G and G15U are found to generate a folding prediction that consists of two suboptimal solutions: the first solution retains the same structure as the wild type corresponding to the eigenvalue λ2 = 0.518806, while the second solution possesses the linear structure corresponding to the eigenvalue λ2 = 0.381966. Thus, locating the three predicted deleterious point mutations mentioned above (G15C, C22G and G15U) illustrates the use of the eigenvalue search method proposed. The method can be extended to other nucleic acid structures of interest. The search can also be expanded to encompass more than single point mutations, although these may eventually become expensive since (N × 3)m folding calculations are required, where N is the number of nucleotides and m is the number of mutations. Nevertheless, using prior information, there is no need to calculate all possible mutations as in the example above. It is possible to interactively subdivide the structure as more layers of mutation are added, which will be demonstrated in the riboswitch applications.
It should be noted that the wild-type structure of the P5abc subdomain used in Wu and Tinoco (21) does not correspond to a natural ribozyme molecule [i.e. the P4-P6 crystal structure derived in Cate et al. (22)] because of a deletion of 4 bp performed in Wu and Tinoco (21). When the missing 4 bp are included in the stem belonging to the P5b subdomain, the predicted structure by energy minimization depicted in Figure 1C becomes less resistant to mutations than its artificial counterpart in Figure 1A. No single point mutation will succeed to rearrange the structure in Figure 1C performing energy minimization predictions by either mfold or the Vienna package, using the energy parameters as described in Mathews et al. (20). For all single point mutations, it is found that the folded structure, as a consequence of introducing the mutation, will result in the same tree-graph structure, hence the same eigenvalue. Thus, an exhaustive search of all two point mutations was performed using the eigenvalue method. This led to a predicted mutation pair U15C and G20U, labeled in Figure 1C, causing a structural rearrangement that results in the linear folded structure of Figure 1D. Therefore, by attempting to disrupt the L5b GAAA tetraloop in the P5abc subdomain, the illustrative example serves as a proof of concept for the deleterious mutation prediction method proposed. Even a highly stable motif that has likely consolidated to become structurally insensitive to mutations in the course of evolution, as will be observed in the case of the terminator hairpin in riboswitches, can be disrupted by adding more mutations. However, some motifs seem to be more resilient to mutations than others. Thus, in addition to the prediction of the mutation location, the minimal number of mutations that is necessary to disrupt a stable motif can be viewed as an indirect stability measure from the predictive standpoint, before experiments are conducted. It should be noted that this stability measure is of the type discussed in Lambert and Moran (23), which is different from the common terminology that refers to thermodynamic stability. This assessment of stability is closely related to the notions of neutral mutations and robustness to genetic perturbations. It can be used to expand theoretical studies about the nature of transitions in the domain of RNA secondary structures (24). Thus, the stability that conforms to how many mutations are needed to disrupt a motif can be helpful in studying mutation accumulation (25).
Applications
The prediction of deleterious mutations that lead to a structural rearrangement by energy minimization methods, thereby disrupting selected motifs, can be applied to riboswitches as a primary application with biological relevance. The important role of riboswitches in controlling transcription termination (3) and translation initiation (4) in the vitamin B1 biosynthesis in bacteria by sensing small molecules has been studied extensively. It is one of several examples of the unique feedback mechanism possessed by these small mRNA segments that are believed to be of ancient origin. Their secondary structure contains essential information that relates to their functional role.
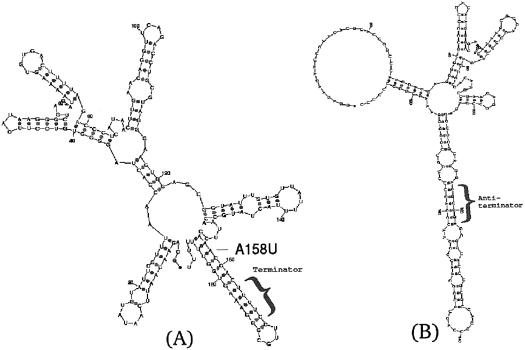
In transcription termination during the thiamin (vitamin B1) biosynthesis in B.subtilis, the riboswitch in Figure 3A (3) senses the TPP ligand that binds to it and maintains a secondary structure that accommodates a terminator hairpin responsible for termination of transcription. With the absence of TPP, an antiterminator structure emerges, suppressing transcription termination. The question arises whether mutations in the riboswitch structure can affect this regulatory process and how. Several mutations have been performed and investigated in Mironov et al. (3), experimentally measuring the termination efficiency as a consequence of introducing the mutations. Here, we wish to predict the location of the minimal number of mutations that can affect transcription termination. Using mfold to initially predict the secondary structure depicted in Figure 3A and then to computationally examine the riboswitch alterations as a response to several randomly selected mutations, it is clear that while other parts of the riboswitch are responding with structural changes, the terminator hairpin remains unaltered. A challenging problem is to predict where to introduce a mutation over the whole structure that will cause the antiterminator to emerge, as with the absence of TPP.
Figure 3.
TPP riboswitch in transcription termination of B.subtilis. (A) The depicted secondary structure of the wild type according to the riboswitch model (3) possesses a stable terminator hairpin, downstream of the evolutionary conserved bases in the thi-box. The anti-antiterminator structure (3) that prohibits the emergence of an antiterminator structure as in (B), guarding the terminator hairpin in the presence of TPP, is located upstream of the terminator. (B) Mutated riboswitch; predicted mutation A158U with respect to the wild-type structure in (A) leads to the emergence of the same antiterminator structure as in the absence of TPP. An antiterminator structure is challenging to obtain as a consequence of introducing a single point mutation. It is possible to efficiently locate this predicted peculiar mutation using the eigenvalue method described in the text, after generating Table 1 and noticing the eigenvalue λ2 = 0.223239. All other single point mutations that have been computationally tried will retain the stable terminator hairpin.
There are (189 nt × 3) = 567 possible single point mutations that need to be examined in order to cover all space of single point nucleotide mutations, (189 nt × 3)2 = 321 489 two point mutations, and so on. The manual visualization of each secondary structure fold as a consequence of introducing a mutation is not feasible. Instead, if the second eigenvalue of the Laplacian matrix corresponding to the tree-graph representation (see Materials and Methods) of each mutated fold is calculated and organized in a table, it is possible to easily discard the vast majority of folds that do not affect the terminator structure. Table 1 summarizes the result of these calculations on all possible single point mutations. In principle, after generating an eigenvalue table the folds corresponding to second eigenvalues which are furthest away from the wild-type eigenvalue should be examined first, especially the ones that occur rarely and possess unique traits. Caution should be exercised since the range of eigenvalues from lowest to highest, belonging to the different number of vertices, will vary. In general, when decreasing the number of vertices the eigenvalue range shifts upwards since a deletion of a node implies that a linear tree graph is more compact, and for a ‘star shaped’ tree graph of any number of vertices higher than two the second eigenvalue is unity [an upper limit for a tree (17)]. Thus, it is recommended to examine eigenvalues belonging to different number of vertices, especially where peculiarities appear. In this example, λ2 = 0.223239 corresponding to the only nine-vertex tree-graph structure and occurring only once, is an ideal candidate. It is now possible to easily check which mutation on the wild-type sequence caused the mutant sequence to fold to a structure corresponding to this eigenvalue, found to be mutation A158U. Indeed, inserting this mutation into the wild-type sequence leads by standard energy minimization methods to a predicted antiterminator structure with nine vertices in the mutant fold, as depicted in Figure 3B. We note that λ2 = 0.223239 is among the highest eigenvalues in Table 1, in accordance with the observation that the second eigenvalue of the Laplacian matrix will most likely be higher for a tree graph with fewer vertices (e.g. suppose we delete a node from a linear 4-vertex tree graph corresponding to a hypothetical wild-type structure, we obtain a star with three vertices corresponding to an eigenvalue of 1.0). Although specifically in the TPP riboswitch example the number of vertices by itself could in principle be used to detect mutation A158U, such an approach is likely to fail in other examples such as the P5abc subdomain described previously. The comprehensive procedure of generating an eigenvalue table with the Laplacian matrix second eigenvalues taken into account as initial guides, together with the number of vertices and possibly other numbers in concert, has the best chances of succeeding as a general recipe for the purpose of detecting vulnerable sequence spots in diverse structures. Besides mutation A158U, all other single point mutations fail to alter the stable terminator structure in the many folding prediction calculations that were manually examined. It is now of biological interest to check whether experimentally, this mutation can affect the riboswitch mechanism in suppressing transcription termination in B.subtilis.
Table 1. Eigenvalue table for the prediction of single point deleterious mutations in the TPP riboswitch, aimed at destroying the terminator hairpin structure in B.subtilis transcription termination.
| Second eigenvalue | Number of vertices | 11 vertices | Frequency |
|---|---|---|---|
| 0.079696 | 13 | 2 | |
| 0.079779 | 12 | 1 | |
| 0.084202 | 12 | 2 | |
| 0.088744 | 11 | * | 1 |
| 0.089199 | 12 | 1 | |
| 0.091622 | 12 | 2 | |
| 0.092741 | 13 | 14 | |
| 0.097460 | 13 | 5 | |
| 0.111249 | 12 | 10 | |
| 0.117230 | 12 | 23 | |
| 0.118492 | 12 | 1 | |
| 0.119840 | 11 | * | 1 |
| 0.125732 | 10 | 1 | |
| 0.133802 | 10 | 2 | |
| 0.140689 | 12 | 2 | |
| 0.156646 | 10 | 6 | |
| 0.162321 | 12 | 1 | |
| 0.162608 | 10 | 1 | |
| 0.164714 | 12 | 2 | |
| 0.178480 | 12 | 6 | |
| 0.178481 | 12 | 3 | |
| 0.185596 | 11 | * | 5 |
| 0.195933 | 11 | WT | 228 |
| 0.201543 | 10 | 4 | |
| 0.207595 | 10 | 13 | |
| 0.214769 | 11 | * | 5 |
| 0.220556 | 10 | 16 | |
| 0.223239 | 9 | ! | 1 |
| 0.233037 | 10 | 3 |
The clustering to discrete eigenvalues enables one to concentrate on examining only the most probable candidates for a deleterious mutation that may cause a structural rearrangement. Second eigenvalues which are furthest away from the wild-type eigenvalue, especially those that are unique with respect to their corresponding number of vertices and frequency of occurrence, should be examined first. Eigenvalues with the same corresponding number of vertices as the wild type are labeled with ‘*’ (note that a loop with only a single isolated nucleotide on each side is not considered a node, therefore only 11 vertices in the wild-type structure are accounted for). Based on this table, it is possible to identify the eigenvalue 0.223239 that is labeled by ‘!’ as a probable candidate for a structural rearrangement, then to locate the mutation A158U that leads to this eigenvalue. Using mfold or the Vienna package, it is verified that the mutation A158U found by the eigenvalue table assumes the predicted structural transition from Figure 3A to B.
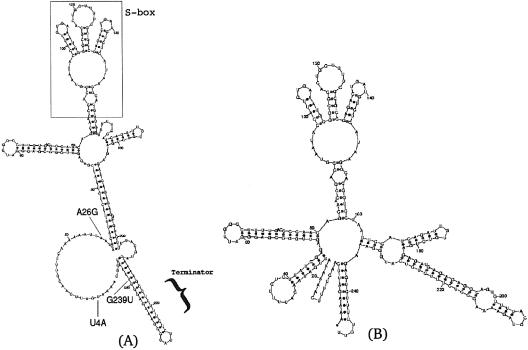
An even more challenging application is to predict the disruption of the terminator hairpin in the SAM riboswitch (6,7,10). In this structure, depicted in Figure 4A, the generation of an initial eigenvalue table and extensive trials of single point mutations did not succeed to predict a structural change in the stable terminator hairpin of the SAM riboswitch. Moreover, the deleterious mutation prediction was extended to two point mutations, since an automated exhaustive search of all such mutations is feasible using the eigenvalue method. The number of 248 nt secondary structure foldings (for the SAM riboswitch) that are calculated with an exhaustive search is (248 nt × 3)2 = 553 536. On a single processor laptop with an Intel Pentium III 1.2 GHz, this extended calculation took ∼3 days. No two point mutations were found that disrupt the terminator hairpin. Therefore, an efficient pruning procedure was devised. First, the S-box (6) was taken out before the folding prediction calculation, after verifying that the remaining artificial sequence with 188 nt conforms to the same folding prediction as the 248 nt wild-type sequence within the region of interest. Secondly, an eigenvalue table was generated for single point mutations on the artificial structure with 188 nt, and the mutation candidate that is most likely to begin the creation of an antiterminator structure [a structure known from metabolite binding experiments (6), or in other systems, any prior information if available] was picked. Thirdly, a two point mutation eigenvalue table was generated, starting from the structure caused by the single point mutation that was chosen in the previous step. In the newly constructed eigenvalue table, it was finally possible to identify a two point mutation that will lead to the disruption of the terminator hairpin. Thus, concatenating all the mutations introduced in the second and third steps, the three point mutation U4A, A26G, G239U, leading to a predicted disruption of the SAM riboswitch terminator hairpin, is depicted in Figure 4B. The reported nucleotide positions are referring to the wild-type sequence, after the S-box was added back to the reduced sequence used in the calculation. The predictions can now be tested in an experiment, adding insights in understanding riboswitch mechanisms and how they can be disrupted or repaired by introducing mutations. Furthermore, the eigenvalue method for predicting clever mutations that may cause structural rearrangements can potentially become useful in designing genetically engineered riboswitches (5) in the future.
Figure 4.
SAM riboswitch in transcription termination of B.subtilis. (A) The depicted secondary structure of the wild type according to the riboswitch model (6) possesses a highly stable terminator hairpin, downstream of the evolutionary conserved bases in the S-box. (B) Mutated riboswitch; predicted mutation U4A, A26G, G239U with respect to the wild-type structure in (A) succeeds to disrupt the terminator structure, using the pruning procedure in conjunction with repetitive generation of eigenvalue tables (data not shown) as outlined in the text.
Acknowledgments
ACKNOWLEDGEMENTS
The author would like to thank the anonymous referees for their useful feedback and valuable comments. He is indebted to the suggestions he received within the community throughout this work.
REFERENCES
- 1.Ng P.C. and Henikoff,S. (2001) Predicting deleterious amino acid substitutions. Genome Res., 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders C.T. and Baker,D. (2002) Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol., 322, 891–901. [DOI] [PubMed] [Google Scholar]
- 3.Mironov A.S., Gusarov,I., Rafikov,R., Lopez,L.E., Shatalin,K., Kreneva,R.A., Perumov,D.A. and Nudler,E. (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell, 111, 747–756. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W., Nahvi,A. and Breaker,R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. [DOI] [PubMed] [Google Scholar]
- 5.Winkler W.C. and Breaker,R.R. (2003) Genetic control by metabolite-binding riboswitches. Chem. Bio. Chem., 4, 1024–1032. [DOI] [PubMed] [Google Scholar]
- 6.Epshtein V., Mironov,A.S. and Nudler,E. (2003) The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA, 100, 5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler W.C., Nahvi,A., Sudarsan,N., Barrick,J.E. and Breaker,R.R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct. Biol., 10, 701–707. [DOI] [PubMed] [Google Scholar]
- 8.Tinoco I Jr. and Bustamante,C. (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 9.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2002) Comparative genomics of thiamin biosynthesis in procaryotes. J. Biol. Chem., 277, 48949–48959. [DOI] [PubMed] [Google Scholar]
- 10.Grundy F.J. and Henkin,T.M. (1998) The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol., 30, 737–749. [DOI] [PubMed] [Google Scholar]
- 11.Miranda-Rios J., Navarro,M. and Soberon,M. (2001) A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl Acad. Sci. USA, 98, 9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margalit H., Shapiro,B.A., Oppenheim,A.B. and Maizel,J.V. (1989) Detection of common motifs in RNA secondary structures. Nucleic Acids Res., 17, 4829–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro B.A. (1988) An algorithm for comparing multiple RNA secondary structures. Comput. Appl. Biosci., 4, 387–393. [DOI] [PubMed] [Google Scholar]
- 14.Le S.Y., Nussinov,R. and Maizel,J.V. (1989) Tree graphs of RNA secondary structures and their comparisons. Comput. Biomed. Res., 22, 461–473. [DOI] [PubMed] [Google Scholar]
- 15.Barash D. and Comaniciu,D. (2003) A common viewpoint on broad kernel filtering and nonlinear diffusion. In Griffin,L.D. and Lillholm,M. (eds), Proceedings of the 4th International Conference on Scale-Space Theories in Computer Vision. Springer-Verlag, Heidelberg, Germany, pp. 683–698. [Google Scholar]
- 16.Fiedler M. (1973) Algebraic connectivity of graphs. Czechoslovak Math. J., 23, 298–305. [Google Scholar]
- 17.Merris R. (1987) Characteristic vertices of trees. Lin. Multi. Alg., 22, 115–131. [Google Scholar]
- 18.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofacker I.L. (2003) Vienna RNA secondary structure server. Nucleic Acids Res., 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews D.H., Sabina,J., Zuker,M. and Turner D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 21.Wu M. and Tinoco,I.Jr (1998) RNA folding causes secondary structure rearrangement. Proc. Natl Acad. Sci. USA, 95, 11555–11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 23.Lambert D.L. and Moran,N.A. (1998) Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA, 95, 4458–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontana W. and Schuster,P. (1998) Continuity in evolution: on the nature of transitions. Science, 280, 1451–1455. [DOI] [PubMed] [Google Scholar]
- 25.Lewin R. (1997) Patterns in Evolution: The New Molecular View. Scientific American Library, New York, NY. [Google Scholar]