Abstract
Transcription of genes coding for the small nuclear RNAs (snRNAs) is dependent upon a unique transcription factor known as the small nuclear RNA-activating protein complex (SNAPc). SNAPc binds to an essential proximal sequence element located about 40–65 base pairs upstream of the snRNA transcription start site. In the fruit fly Drosophila melanogaster, DmSNAPc contains three distinct polypeptides (DmSNAP190, DmSNAP50, and DmSNAP43) that are stably associated with each other and bind to the DNA as a complex. We have used mutational analysis to identify domains within each subunit that are involved in complex formation with the other two subunits in vivo. We have also identified domains in each subunit required for sequence-specific DNA binding. With one exception, domains required for subunit-subunit interactions lie in the most evolutionarily conserved regions of the proteins. However, DNA binding by DmSNAPc is dependent not only upon the conserved regions but is also highly dependent upon domains outside the conserved regions. Comparison with functional domains identified in human SNAPc indicates many parallels but also reveals significant differences in this ancient yet rapidly evolving system.
The small nuclear RNA (snRNA)3-activating protein complex (SNAPc) is a multisubunit protein required for transcription of genes that code for the spliceosomal (and certain other) snRNAs (1–4). SNAPc recognizes and binds specifically to a proximal sequence element (PSE) located about 40–65 base pairs upstream of the transcription start site. SNAPc has also variously been called PSE-binding protein (5, 6) and PSE-binding transcription factor (1, 3, 7). In humans, SNAPc contains five distinct polypeptide chains (SNAP190, SNAP50, SNAP45, SNAP43, and SNAP19) named based upon the apparent molecular weights of these subunits (4, 7–12). For the remainder of this article, the human protein and its subunits will be indicated by the prefix “Hs.”
In the fruit fly Drosophila melanogaster, DmSNAPc contains three distinct polypeptide chains that are orthologous to HsSNAP190, HsSNAP50, and HsSNAP43 (13, 14). The three fly subunits, DmSNAP190, DmSNAP50, and DmSNAP43, are each present in a single copy in native DmSNAPc (15) and have calculated molecular masses of 84, 43, and 42 kDa, respectively. Interestingly, a homologous complex (tSNAPc) is required for transcription of the spliced leader snRNA in trypanosomes (16–18). This indicates that a SNAP-like complex arose very early in eukaryotic evolution and continues to be essential for snRNA transcription in widely divergent contemporary eukaryotes. However, even within insects, snRNA gene promoter sequences recognized by SNAPc have diverged fairly rapidly (19).
The subunits of eukaryotic SNAPc tightly associate with each other in solution even when the complex is not bound to DNA. The subunits co-purify through numerous chromatography columns (1–3, 16–18, 20). Moreover, each of the three metazoan core subunits is essential for sequence-specific binding to the PSE as none can bind individually or in pairwise combinations (21–23).4 However, an isolated region of HsSNAP190 that contains two Myb repeats binds weakly and with little sequence specificity to DNA (11, 22, 24).
We now report a mutational analysis of the three subunits of DmSNAPc from the fruit fly D. melanogaster to identify functional domains within each of the subunits. Truncated tagged versions of each of the proteins were expressed in D. melanogaster S2 cells together with the other two untagged subunits. The ability of the truncated subunits to participate in the assembly of DmSNAPc in the homologous in vivo system was monitored by coimmunoprecipitation assays from cellular extracts. In each case, we have identified regions within the polypeptides that are necessary or sufficient for in vivo assembly with the other two subunits. We have also identified domains within each DmSNAP polypeptide that are essential for sequence-specific binding to the PSEA (the fly equivalent of the PSE) but are dispensable for assembly with the other two subunits. Our mutational analysis of DmSNAPc expands on the knowledge obtained in the human system and in some cases reveals surprising differences in the localization of functional domains for complex assembly and DNA binding of the different metazoan SNAP complexes.
EXPERIMENTAL PROCEDURES
DmSNAP Expression Constructs
Constructs for expression of full-length untagged or V5 epitope-tagged DmSNAPs in D. melanogaster S2 cells have been previously described (14). Briefly, open reading frames encoding each of the DmSNAP proteins had been cloned into the pMT/V5-His-TOPO vector, a component of Invitrogen's Drosophila Expression System. To convert those to FLAG-tagged constructs, the appropriate plasmids were digested with BstBI and MluI to remove the V5-coding sequence; this was then replaced with synthetic DNA sequence encoding the FLAG epitope.
Truncation constructs were prepared by using the full-length constructs as templates for the PCR together with appropriate primers. For N-terminal truncations, forward primers were designed to contain the natural start codon of the protein together with about 7 nucleotides 5′ of the start codon as the first one-third of the forward primer (∼10 nucleotides). The second two-thirds of the forward primer (∼20 nucleotides) was designed to match the sequence at the starting point of the desired truncation. PCR products were then cloned initially into the pMT/V5-His-TOPO expression vector. Fragments were removed from these constructs and re-cloned into the FLAG-modified expression vector.
To prepare C-terminal truncations tagged at the N terminus, PCR was used to introduce a C-terminal stop codon at each of the desired positions within the DmSNAP genes. The amplified DNA was initially cloned into pMT-V5-His-TOPO. These constructs were used as templates with forward primers that contained an SpeI site just upstream of the codon for the second amino acid of the DmSNAP190, DmSNAP50, or DmSNAP43 genes. Following digestion with SpeI and NotI, the PCR fragments were cloned into a pMT-V5-His-TOPO-based expression vector that contained an N-terminal FLAG sequence. This N-terminal FLAG vector was prepared as follows: synthetic DNA containing a translation initiation codon and the FLAG encoding sequence was cloned between the KpnI and SpeI sites of pMT-V5-His-TOPO. This FLAG-containing vector was then used for cloning C-terminal-truncated DmSNAPs after digestion with SpeI and NotI.
Stably Transfected S2 Cells
Expression plasmids for each of the DmSNAPs were used to co-transfect S2 cells together with pCoBlast according to conditions recommended by Invitrogen. Stably transfected cell lines were established by selection in blasticidin-containing medium. Expression of DmSNAPs was induced from the metallothionein promoter by treatment of the cells for 24 h with 0.5 mm copper sulfate.
FLAG Purification/Coimmunoprecipitation
Following induction, cells were washed in phosphate-buffered saline and lysed in CelLytic M lysis buffer (Sigma catalog number C2978) containing protease inhibitor mixture (Sigma catalog number P8340). For low-salt FLAG immunopurifications, cell lysates were used directly in overnight incubations with anti-FLAG M2-agarose beads (Sigma catalog number A2220). The beads were then washed four times in 0.05 m Tris-HCl (pH 7.4), 0.15 m NaCl and then twice in HEMG wash buffer (81 mm KCl, 32.5 mm Hepes K+, 5.5 mm MgCl2, 5.0 mm dithiothreitol, 10% glycerol, and 0.1 mm EDTA (pH 7.6)). For high-salt immunopurifications, lysates were first adjusted to a NaCl concentration of 0.35 m prior to incubating overnight with the anti-FLAG beads. The beads were then washed twice in 0.01 m Tris-HCl (pH 7.4), 0.35 m NaCl and then three times with HEMG wash buffer. In both cases, the DmSNAP complexes were eluted from the affinity gel by competition with 3× FLAG peptide (Sigma catalog number F4799) at a concentration of 200 μg/ml in HEMG wash buffer.
Immunoblots
Whole cell lysate and FLAG-purified samples were run on 10–14% denaturing polyacrylamide gels and the proteins were transferred to polyvinylidene difluoride membranes. FLAG-tagged DmSNAP subunits were detected using anti-FLAG M2 monoclonal antibodies (Sigma) conjugated to either alkaline phosphatase or horseradish peroxidase. Untagged DmSNAP subunits were detected as previously described using primary antibodies generated in rabbits against amino acid sequences at or near the C termini of the DmSNAP polypeptides (14). Data shown in the figures are from gels in which the amount of FLAG-tagged protein loaded in each lane was normalized by running gels repetitively and each time adjusting the amounts of protein until the intensity of the signals from the tagged subunit in the different samples was as similar as reasonably possible. For all immunoblots shown, the lanes in each horizontal panel (top, middle, or bottom) are from the same gel blot developed for the same length of time. Thus the intensities of the bands within a horizontal panel reflect the relative amounts of protein present in the various cell extracts or FLAG affinity-purified fractions. This permitted an easy visual assessment of the relative amounts of the untagged subunits in each sample in comparison to a constant amount of the tagged subunit.
Electrophoretic Mobility Shift Assays
DNA mobility shift assays were carried out in 18-μl volumes in a final concentration of ∼80 mm KCl, 25 mm Hepes (pH 7.6), 5 mm MgCl2, 10 μm ZnCl2, 0.1 mm EDTA, 9 mm dithiothreitol, 9% glycerol. The radioactive DNA probe contained the PSEA sequence of the D. melanogaster U1:95Ca gene (formerly called the U1–95.1 gene (25)). FLAG affinity-purified DmSNAP complexes, normalized for the content of the FLAG-tagged subunit (as determined by immunoblotting, described above), were added to each reaction. Incubation was carried out for 30 min at 20 °C. For complexes exhibiting weak or no binding, 2–6 times as much protein (constrained by the maximum volume of the reaction) was sometimes used in additional lanes to maximize the possibility of observing binding. Complexes were run on 5% non-denaturing polyacrylamide gels and the bands were detected by autoradiography.
Chromatin Immunoprecipitations (ChIPs)
ChIP assays were carried out as recently described (26). Affinity-purified rabbit polyclonal antibodies produced against the FLAG peptide (Sigma catalog number F7425) were used for FLAG ChIPs. The anti-DmSNAP43 antibodies used as positive controls in the ChIP assays were produced in a rabbit immunized with bacterially expressed recombinant DmSNAP43 (26). The preimmune serum was from the same rabbit prior to immunization. The ChIP PCR forward primer (5′-GTGTGGCATACCTTATAGGGGTGCT-3′) and reverse primer (5′-GCTTTTCGATGCTCGGCAGCAG-3′) amplify the promoter region of the U1:95Ca gene (26). In all experiments, the amounts of DNA samples and PCR conditions were chosen such that the resultant signals were within a semi-quantitative range.
RESULTS
Functional Domains of DmSNAP190
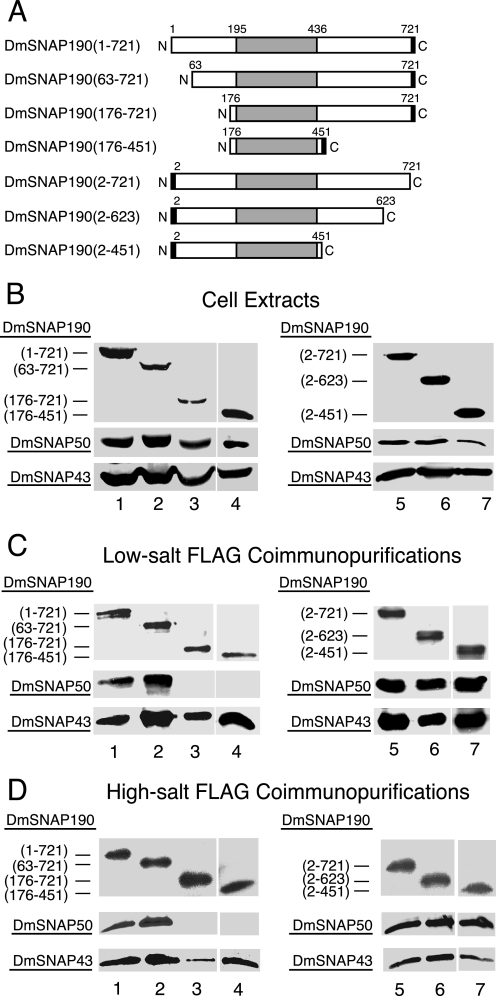
An unusual feature of the amino acid sequence of DmSNAP190 is the existence of 4.5 tandem Myb repeats located between residues 195 and 436 (14). This region (Fig. 1) is conserved between fly and human SNAP190 with an identity of 27% and overall similarity of 44% (11, 14). In human SNAPc, the Myb repeat domain of HsSNAP190 is involved in DNA binding (11, 22, 24). Because it seemed likely that deletion of this most conserved region of DmSNAP190 would be highly detrimental to DmSNAPc activity, we focused our studies on the non-conserved regions. Full-length and N- and C-terminal DmSNAP190 truncation constructs under control of the copper-inducible metallothionein promoter were prepared with FLAG-His6 tags at the C terminus or with His6-FLAG tags at the N terminus (Fig. 2A). Each tagged DmSNAP190 construct was used separately to transfect Drosophila S2 cells together with pCoBlast (Invitrogen) and the two complementary constructs to express the full-length DmSNAP50 and DmSNAP43 subunits without tags. Stably transfected cells were selected by growth on blasticidin.
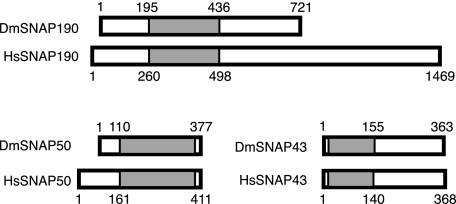
FIGURE 1.
Comparison of D. melanogaster (Dm) and human (Hs) SNAP subunits. The rectangles indicate the relative lengths of the proteins, and the shaded areas indicate the evolutionarily conserved regions of the orthologous protein pairs. The numbers above and below the rectangles designate the amino acid positions in the proteins.
FIGURE 2.
Domains of DmSNAP190 involved in assembly in vivo with DmSNAP50 and DmSNAP43. A, schematic representation of full-length and truncated DmSNAP190 constructs co-transfected into S2 cells together with constructs expressing full-length untagged DmSNAP50 and DmSNAP43. The shaded area represents the 4.5-Myb-repeat domain conserved between humans and fruit flies. The black rectangles represent FLAG tags at the C or N termini of DmSNAP190. The names of the constructs in the column at the left indicate the extent of the wild type amino acid residues present in the expressed constructs. B, co-overexpression of tagged DmSNAP190 with DmSNAP50 and DmSNAP43 in stably transfected S2 cells. Whole cell extracts from stably transfected cells co-overexpressing all three DmSNAP subunits were run on denaturing gels and DmSNAP subunits were detected by immunoblot analysis. The amount of extract loaded in each lane was normalized so that the intensity of the signal obtained from the tagged DmSNAP190 construct was similar in each lane. The top panels show full-length or truncated tagged DmSNAP190 detected using monoclonal antibodies against the FLAG epitope. The middle and bottom panels show detection of DmSNAP50 or DmSNAP43, respectively, by using polyclonal anti-peptide antibodies prepared against C-terminal peptides of the respective proteins (14). For all immunoblots shown in this and subsequent figures, the lanes in each horizontal panel (top, middle, or bottom) are from the same gel blot developed for the same length of time. Thus the intensities of the bands within a horizontal panel reflect the relative amounts of protein present in the various transfected cell lines. C, co-purification of DmSNAP50 and DmSNAP43 with full-length and truncated DmSNAP190 constructs following FLAG affinity purification under low-salt conditions. Complexes containing full-length or truncated tagged DmSNAP190 were purified using FLAG antibody beads, and the presence of the individual subunits in the elution fractions was evaluated by immunoblotting. The volume of elution fraction loaded in each lane was normalized so that the intensity of the signal for the DmSNAP190 construct was similar in each lane when detected with FLAG antibodies. DmSNAP50 and DmSNAP43 subunits that co-purified with the FLAG-tagged DmSNAP190 constructs were detected with antibodies prepared against C-terminal peptides of the respective proteins. D, same as C except FLAG affinity purification was carried out under high-salt conditions (0.35 m NaCl).
Cellular extracts were prepared following copper induction and immunoblots were carried out to examine the expression of the DmSNAP190, DmSNAP50, and DmSNAP43 proteins in each of the cell lines. Detection of the respective proteins employed monoclonal antibodies against the FLAG epitope (for the DmSNAP190 constructs) or polyclonal antibodies specific for C-terminal peptides of DmSNAP50 or DmSNAP43 (14). Fig. 2B shows that each of the DmSNAP190 truncation mutants was expressed in the corresponding stably transfected cells (lanes 1–7, top panels). Fig. 2B also shows that both DmSNAP50 and DmSNAP43 were expressed among the different cell lines at relatively similar levels (middle and bottom panels). Expression of these proteins from the endogenous genes in normal S2 cells was undetectable under these conditions (data not shown, but see Ref. 14).
To determine whether the truncated DmSNAP190 proteins were able to assemble in vivo with the co-expressed DmSNAP50 and DmSNAP43 subunits, the extracts were applied to FLAG antibody affinity resins under either low-salt (Fig. 2C) or high-salt (Fig. 2D) conditions (10 and 350 mm NaCl, respectively). Bound protein was eluted by competition with FLAG peptide. The top panels in Fig. 2, C and D, show that all seven full-length or truncated DmSNAP190 proteins could be detected in the elution fractions by immunoblot analysis using the FLAG antibody. As revealed in the bottom most panels of Fig. 2, C and D, DmSNAP43 co-purified with each of the DmSNAP190 constructs, including construct DmSNAP190-(176–451), which contained little more than the 4.5 Myb repeats. This indicates that the Myb domain of DmSNAP190 is sufficient for association with DmSNAP43.
The result with DmSNAP50, however, was different. As expected, DmSNAP50 co-purified with full-length DmSNAP190 tagged at either the C or N terminus (Fig. 2, C and D, lanes 1 and 5, middle panels). Moreover, DmSNAP50 associated with the truncation construct that lacked the first 62 amino acid residues of DmSNAP190 (lane 2 in Fig. 2, C and D); likewise, DmSNAP50 associated with the two constructs that lacked amino acids C-terminal to the Myb domain (lanes 6 and 7). However, DmSNAP50 failed to co-purify with the two different DmSNAP190 constructs that were missing 175 residues from the N terminus (Fig. 2, C and D, lanes 3 and 4, middle panels). These results indicate that residues located between positions 63 and 175 of DmSNAP190 are essential for its assembly with DmSNAP50.
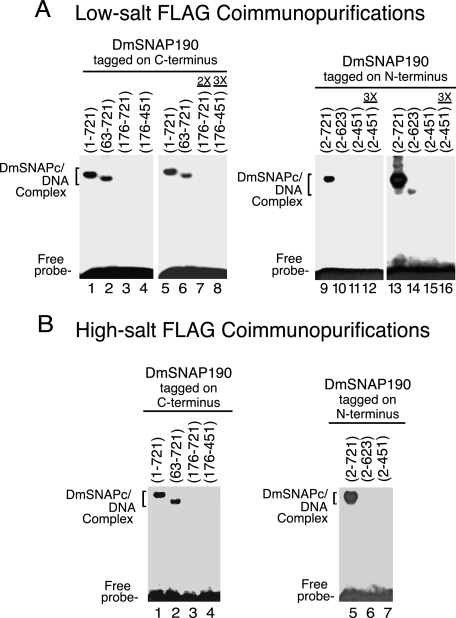
Next, to investigate which regions of DmSNAP190 contribute to DNA binding by DmSNAPc, the FLAG affinity-purified complexes were subjected to electrophoretic mobility shift analysis (Fig. 3). In Fig. 3, A, lanes 1–4, and B, lanes 1–4, protein amounts in each lane were normalized as determined from the immunoblots shown in Fig. 2, C and D, respectively. The complexes that contained DmSNAP190 that was either full-length or missing 62 amino acids from its N terminus bound efficiently to the PSEA (Fig. 3, A, lanes 1 and 2, and B, lanes 1 and 2). However, the complex that contained DmSNAP190 lacking 175 N-terminal residues was not able to bind DNA (Fig. 3, A, lane 3, and B, lane 3). DmSNAP190 that contained only the Myb domain gave a similar result (lane 4). Neither the use of 2 or 3 times the amount of normalized protein (Fig. 3A, lanes 7 and 8), nor longer exposure of the film (data not shown), was able to detect any DNA binding activity for these latter two truncation constructs. However, this was expected because these complexes lacked DmSNAP50 (Fig. 2, C and D), and previous results have indicated that all three subunits of DmSNAPc contact the DNA (13, 14) and are required for the sequence-specific DNA-binding activity of DmSNAPc.5
FIGURE 3.
Domains of DmSNAP190 required for sequence-specific DNA binding by DmSNAPc. A, DmSNAP complexes FLAG affinity purified under low-salt conditions (Fig. 2C) were used for DNA mobility shift analysis with a DNA probe containing a U1 PSEA sequence. Complexes containing DmSNAP190 constructs tagged at the C terminus (N-terminal truncations) are shown in lanes 1–8, and constructs tagged at the N terminus (C-terminal truncations) are shown in lanes 9–16. Lanes 1–6 were carried out with complexes containing a normalized amount of DmSNAP190 as determined in Fig. 2C, whereas lanes 7 and 8 contained 2 or 3 times the normalized amount of truncated protein. (The maximum excess of protein that could be added was limited by the final volume of the reaction and the relative concentrations of each sample.) Lanes 9–11 also contained normalized amounts of protein, whereas lane 12 contained 3 times this amount of protein. Lanes 13–16 show a longer exposure of the same gel shown in lanes 9–12. B, similar to A except DmSNAP complexes were affinity purified under high-salt conditions and protein amounts were normalized based upon the immunoblots shown in Fig. 2D.
DmSNAPc that contained full-length DmSNAP190 tagged at the N terminus also bound efficiently to DNA (Fig. 3, A, lane 9, and B, lane 5). However, DNA binding was severely compromised by truncations from the C terminus following residues 623 or 451 (Fig. 3, A, lanes 10–12, and B, lanes 6 and 7). Fig. 3A, lanes 13–16, show a much longer exposure of film to the same gel shown in lanes 9–12. This longer exposure revealed that DmSNAPc, which contained DmSNAP190 truncated following position 623 (lane 14), retained an extremely low level of DNA-binding activity, whereas truncation following position 451 completely abrogated detectable DNA binding (lane 15) even when 3 times the normalized amount of protein was used (lane 16).
These results indicated that the C-terminal domain of DmSNAP190 is essential for DmSNAPc to bind efficiently to the PSEA, even though it is not required for assembly with either DmSNAP43 or DmSNAP50 (Fig. 2, C and D). This result was surprising because human SNAP190 truncated immediately following the Myb domain can associate with HsSNAP43 and HsSNAP50 into a complex known as miniSNAPc that binds very efficiently to DNA (21, 22, 24). Thus, the C-terminal domain of DmSNAP190 is important for DNA binding by fly SNAPc despite the fact that it is not required, and is even inhibitory (21), for human SNAPc binding to DNA.
Functional Domains of DmSNAP50
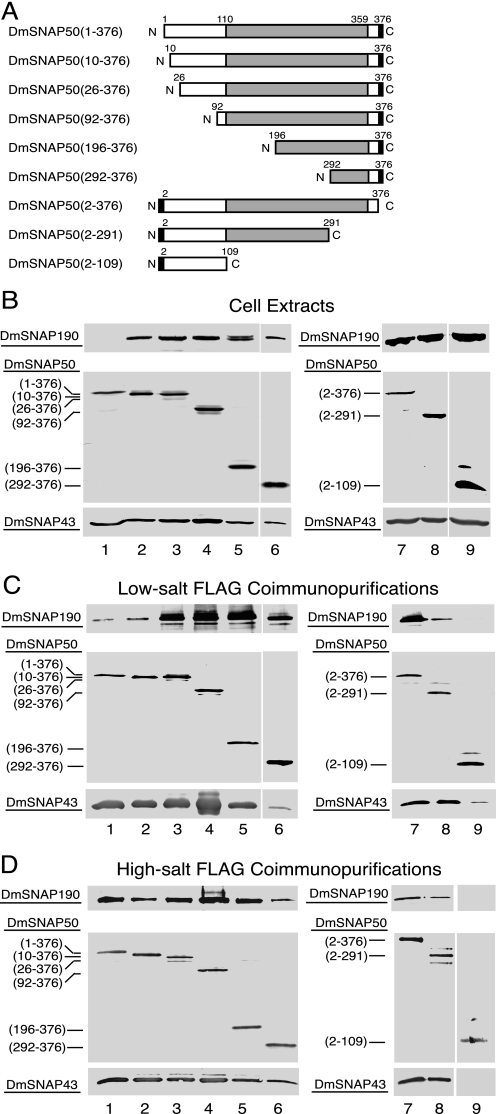
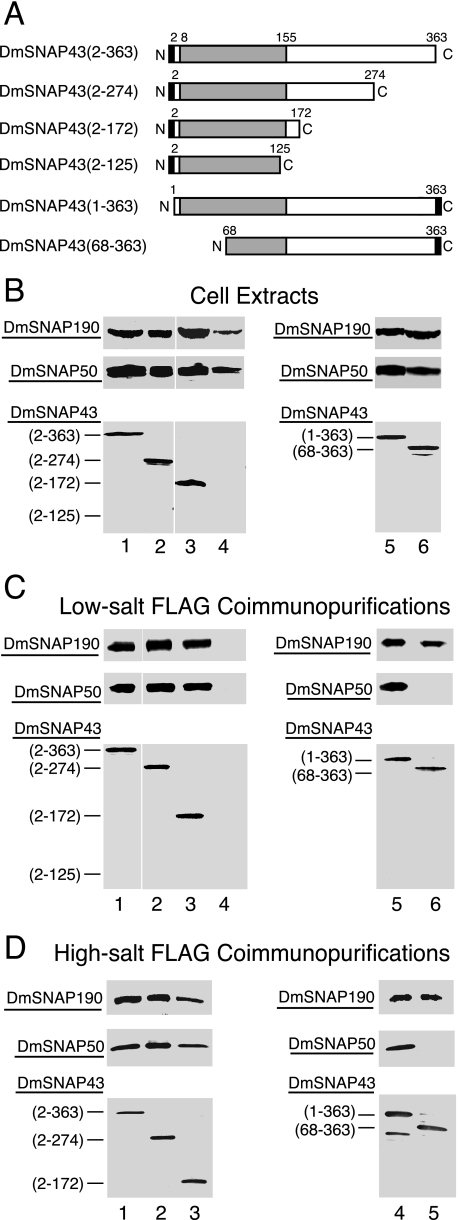
The SNAP50 orthologs are the most evolutionarily conserved of the SNAP proteins (14, 16). The most conserved region of DmSNAP50 is the C-terminal region where residues 110 to 359 have 33% identity and 51% similarity to the C-terminal region of HsSNAP50 (Fig. 1) (14). We concentrated most of our efforts on truncations from the N terminus, but two C-terminal truncations were also prepared (Fig. 4A). N-terminal truncations had FLAG-His6 tags at the C terminus, whereas the C-terminal truncations had His6-FLAG tags at the N terminus. Each DmSNAP50 construct was co-overexpressed in S2 cells together with full-length untagged DmSNAP190 and DmSNAP43.
FIGURE 4.
Domains of DmSNAP50 involved in assembly in vivo with DmSNAP190 and DmSNAP43. A, schematic representation of full-length and truncated DmSNAP50 constructs co-transfected into S2 cells together with constructs expressing full-length untagged DmSNAP190 and DmSNAP43. The shaded area represents the region most conserved between humans and fruit flies. The black rectangles represent FLAG tags at the C or N termini of DmSNAP50. The names of the constructs in the column at the left indicate the extent of the wild type amino acid residues present in the expressed constructs. B, co-overexpression of tagged DmSNAP50 with DmSNAP190 and DmSNAP43 in stably transfected S2 cells. Whole cell extracts from stably transfected cells co-overexpressing all three DmSNAP subunits were run on denaturing gels and DmSNAP subunits were detected by immunoblot analysis. The amount of extract loaded in each lane was normalized so that the intensity of the signal obtained from the tagged DmSNAP50 construct was similar in each lane. The middle panels show full-length or truncated tagged DmSNAP50 detected using monoclonal antibodies against the FLAG epitope. The top and bottom panels show detection of DmSNAP190 or DmSNAP43, respectively, by using polyclonal anti-peptide antibodies prepared against C-terminal peptides of the respective proteins (14). C, co-purification of DmSNAP190 and DmSNAP43 with full-length and truncated DmSNAP50 constructs following FLAG affinity purification under low-salt conditions. Complexes containing full-length or truncated tagged DmSNAP50 were purified using FLAG antibody beads, and the presence of the individual subunits in the elution fractions was evaluated by immunoblotting. The volume of elution fraction loaded in each lane was normalized so that the intensity of the signal for the DmSNAP50 construct was similar in each lane when detected with FLAG antibodies. DmSNAP190 and DmSNAP43 subunits that co-purified with the FLAG-tagged DmSNAP50 constructs were detected with antibodies prepared against C-terminal peptides of the respective proteins. D, same as C except FLAG affinity purification was carried out under high-salt conditions.
Successful expression of each of the tagged DmSNAP50 constructs was indicated by immunoblots of total cellular lysates prepared from each of these cell lines (Fig. 4B, lanes 1–9, middle panels). DmSNAP43 and DmSNAP190 were also readily detected in all cell lines with one exception: DmSNAP190 was difficult to detect in lysates from cells expressing full-length C-terminal tagged DmSNAP50 under conditions where it was readily detectable in the remaining cell lines (Fig. 4B, upper panel, lane 1 versus lanes 2–9). However, upon FLAG purification of DmSNAPc from those cells, DmSNAP190 became detectable in the FLAG-purified fraction (see Fig. 4, C and D, below).
The abilities of DmSNAP190 and DmSNAP43 to associate with the DmSNAP50 truncations were assayed by immunoblotting following FLAG affinity purification under either low-salt (Fig. 4C) or high-salt (Fig. 4D) conditions. Lanes 1–9 in the middle panels of Fig. 4, C and D, show that each of the FLAG-tagged DmSNAP50 variants was immunopurified in the elution fractions from the FLAG affinity resins. The top panels in Fig. 4, C and D, reveal the ability of DmSNAP190 to co-purify with the various full-length and truncated DmSNAP50 constructs. Under low-salt conditions of purification, DmSNAP50 truncation mutants that had 25 or more amino acid residues deleted from the N terminus co-precipitated greater amounts of DmSNAP190 compared with that precipitating with the full-length or nearly full-length constructs (Fig. 4C, top panels, lanes 3–6 versus lanes 1 and 2). However, when the co-immunopurifications were carried out in higher salt conditions (Fig. 4D), the amount of co-immunopurified DmSNAP190 exhibited much less variability. Thus, under certain conditions the N terminus of DmSNAP50 may play a role in preventing an artifactual association of extra copies of DmSNAP190 with DmSNAP50.
The last 85 residues of DmSNAP50 (residues 292–376) contain an evolutionarily conserved (14, 16) yet unorthodox zinc-binding domain termed the “SNAP finger” (23). Interestingly, these last 85 residues were sufficient to co-immunoprecipitate both DmSNAP190 and DmSNAP43 under either low- or high-salt conditions, although possibly at reduced levels in certain cases (Fig. 4, C and D, lane 6, top and bottom panels). Thus the SNAP finger region of DmSNAP50 appeared to interact with both DmSNAP190 and DmSNAP43; however, from those data alone it remained possible that the interaction with one of these subunits could be indirect as a result of mutual interactions between DmSNAP190 and DmSNAP43.
To further examine whether DmSNAP43 and DmSNAP190 can independently interact with the SNAP finger of DmSNAP50, we expressed the SNAP finger domain, DmSNAP50-(292–376), in pairwise combinations either with DmSNAP190 alone or with DmSNAP43 alone. In each case, we observed that DmSNAP190 and DmSNAP43 copurified with FLAG-tagged DmSNAP50-(292–376) (data not shown). Thus both DmSNAP190 and DmSNAP43 can interact independently with the SNAP finger domain of DmSNAP50.
Results from the C-terminal truncations of DmSNAP50 are shown in Fig. 4, C and D, lanes 7–9. Truncation of DmSNAP50 beyond residue 291 partially impaired the co-purification of DmSNAP190 (compare lanes 7 and 8 in the top panels of Fig. 4, C and D) although having no effect on the co-purification of DmSNAP43 (compare lanes 7 and 8 in the bottom panels of Fig. 4, C and D). These data suggest that residues of DmSNAP50 between 110 and 291 comprise a very significant site of interaction with DmSNAP43, even though the SNAP finger region undoubtedly also contributes to the interaction. Deletion of DmSNAP50 amino acids C-terminal to position 109 completely abrogated the binding of DmSNAP190 and severely compromised or eliminated the binding of DmSNAP43 (Fig. 4, C and D, top and bottom panels, lanes 9). Together with the results from the other truncations, this suggests that the non-conserved region of DmSNAP50 (residues 1–109) makes little if any contribution toward forming a complex with DmSNAP190 and DmSNAP43.
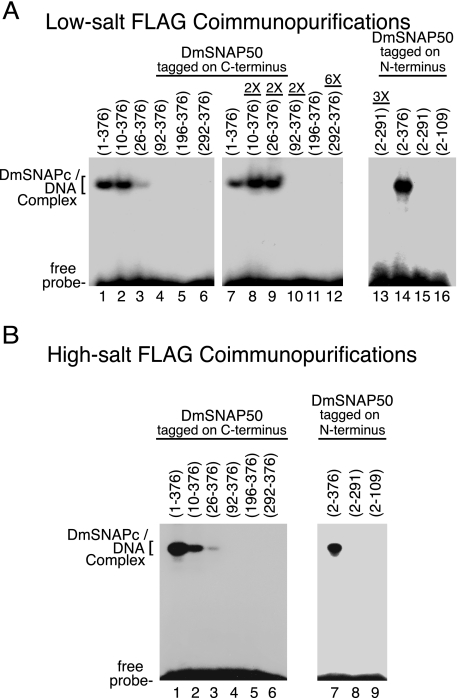
To map regions of DmSNAP50 important for the binding of DmSNAPc to the PSEA, the FLAG-purified DmSNAP complexes that contained full-length or truncated DmSNAP50 were used in electrophoretic mobility shift assays (Fig. 5). DmSNAPc that contained full-length DmSNAP50 tagged on the C terminus bound DNA efficiently (Fig. 5, A, lanes 1 and 7, and B, lane 1). SNAP complexes that contained DmSNAP50 lacking either the first 9 or 25 amino acid residues retained partial DNA-binding activity (Fig. 5, A, lanes 2, 3, 8, and 9, and B, lanes 2 and 3). However, deletion of the first 91 amino acid residues of DmSNAP50 resulted in the complete loss of detectable DNA-binding activity (Fig. 5, A, lanes 4 and 10, and B, lane 4) even though this DmSNAP50 construct associated very well with both DmSNAP190 and DmSNAP43 (Fig. 4, C and D, lane 4). Thus, amino acids between positions 26 to 91 of DmSNAP50 are critical for DNA binding even though these lie outside of the evolutionarily conserved region and are not essential for complex formation with DmSNAP190 and DmSNAP43.
FIGURE 5.
Domains of DmSNAP50 required for sequence-specific DNA binding by DmSNAPc. A, DmSNAP complexes FLAG affinity purified under low-salt conditions (Fig. 4C) were used for DNA mobility shift analysis with a DNA probe containing a U1 PSEA sequence. Complexes containing DmSNAP50 constructs tagged at the C terminus (N-terminal truncations) are shown in lanes 1–12, and constructs tagged at the N terminus (C-terminal truncations) are shown in lanes 13–16. Lanes 1–7 and 11 were carried out with complexes containing a normalized amount of DmSNAP50 as determined in Fig. 4C, whereas lanes 8–10 contained two times the normalized amount of truncated protein, and lane 12 contained six times the normalized amount of truncated protein. Lanes 14–16 also contained normalized amounts of protein, whereas lane 13 contained 3 times the normalized amount of protein. (The maximum amount of protein that could be used was limited by the volume of the reaction and the relative concentration of each sample.) B, similar to A except DmSNAP complexes were affinity purified under high-salt conditions and protein amounts were normalized based upon the immunoblots shown in Fig. 4D.
Although DmSNAPc that contained full-length DmSNAP50 tagged at the N terminus bound efficiently to the PSEA (Fig. 5, A, lane 14, and B, lane 7), deletion of the SNAP finger of DmSNAP50 eliminated DNA-binding activity (Fig. 5, A, lanes 13 and 15, and B, lane 8). These results were to be expected because the SNAP finger was clearly required for DNA binding in the human system (23).
Functional Domains of DmSNAP43
DmSNAP43 is 363 amino acid residues in length, and the evolutionarily most conserved region (residues 8 to 155, shaded in Fig. 1) has 31% identity and 48% similarity to the human ortholog (14). We concentrated our analysis on truncations that removed portions of the non-conserved C terminus; however, one N-terminal truncation was also prepared (Fig. 6A). Each construct was co-overexpressed in S2 cells with untagged DmSNAP190 and DmSNAP50.
FIGURE 6.
Domains of DmSNAP43 involved in assembly in vivo with DmSNAP190 and DmSNAP50. A, schematic representation of full-length and truncated DmSNAP43 constructs co-transfected into S2 cells together with constructs expressing full-length untagged DmSNAP190 and DmSNAP50. The shaded area represents the region most conserved between humans and fruit flies. The black rectangles represent FLAG tags at the C or N termini of DmSNAP43. The names of the constructs in the column at the left indicate the extent of the wild type amino acid residues present in the expressed constructs. B, co-overexpression of tagged DmSNAP43 with DmSNAP190 and DmSNAP50 in stably transfected S2 cells. Whole cell extracts from stably transfected cells co-overexpressing all three DmSNAP subunits were run on denaturing gels and DmSNAP subunits were detected by immunoblot analysis. The amount of extract loaded in each lane was normalized so that the intensity of the signal obtained from the tagged DmSNAP43 construct was similar in each lane. The bottom panels show full-length or truncated tagged DmSNAP43 detected using monoclonal antibodies against the FLAG epitope. The top and middle panels show detection of DmSNAP190 or DmSNAP50, respectively, by using polyclonal anti-peptide antibodies prepared against C-terminal peptides of the respective proteins (14). C, co-purification of DmSNAP190 and DmSNAP50 with full-length and truncated DmSNAP43 constructs following FLAG affinity purification under low-salt conditions. Complexes containing full-length or truncated tagged DmSNAP43 were purified using FLAG antibody beads, and the presence of the individual subunits in the elution fractions was evaluated by immunoblotting. The volume of elution fraction loaded in each lane was normalized so that the intensity of the signal for the DmSNAP43 construct was similar in each lane when detected with FLAG antibodies. DmSNAP190 and DmSNAP50 subunits that co-purified with the FLAG-tagged DmSNAP43 constructs were detected with antibodies prepared against C-terminal peptides of the respective proteins. D, same as C except FLAG affinity purification was carried out under high-salt conditions.
Expression of each of the full-length and truncated proteins was examined by immunoblot analysis of cellular extracts prepared from the individual stably transfected cell lines. The bottom panels in Fig. 6B show that expression of the two tagged full-length and three of the truncated DmSNAP43 constructs were readily detected. However, expression of the construct truncated following position 125 was nearly undetectable (lane 4), suggesting that this shortest construct was unstable. The upper panels in Fig. 6B illustrate that DmSNAP190 and DmSNAP50 were expressed in all six cell lines.
The DmSNAP complexes were then immunoprecipitated by FLAG affinity chromatography under either low-salt or high-salt conditions, and the ability of DmSNAP190 or DmSNAP50 to co-precipitate with the tagged DmSNAP43 was assayed by immunoblotting (Fig. 6, C and D). As expected, there was no detectable purification of the poorly expressed DmSNAP43-(2–125) construct (Fig. 6C, lane 4, bottom panel); furthermore, no co-eluting DmSNAP190 nor DmSNAP50 could be detected in the elution fractions from that cell line (Fig. 6C, lane 4, top and middle panels).
However, all other truncated and full-length DmSNAP43 constructs were readily detectable in the FLAG elution fractions (Fig. 6, C, lanes 1–3, 5–6, and D, lanes 1–5, in the bottom panels). The top most panels in Fig. 6, C and D, show that DmSNAP190 co-purified with each of the stably expressed DmSNAP43 constructs. This indicates that neither the last 191 residues (those beyond position 172) nor the first 67 amino acid residues of DmSNAP43 are essential for recruitment of DmSNAP190 into the complex. By inference, the data suggest that DmSNAP43 residues between 68 and 172 are likely involved in interaction with DmSNAP190.
Interestingly, DmSNAP50 co-purified with the two stably expressed DmSNAP43 C-terminal truncation constructs (Fig. 6, C and D, middle panels, lanes 2 and 3) but failed to co-purify with the N-terminal truncation construct DmSNAP43-(68–363) (Fig. 6, C, lane 6, and D, lane 5, middle panels). This latter result indicates that the first 67 amino acid residues of DmSNAP43 are required for stable complex formation with DmSNAP50.
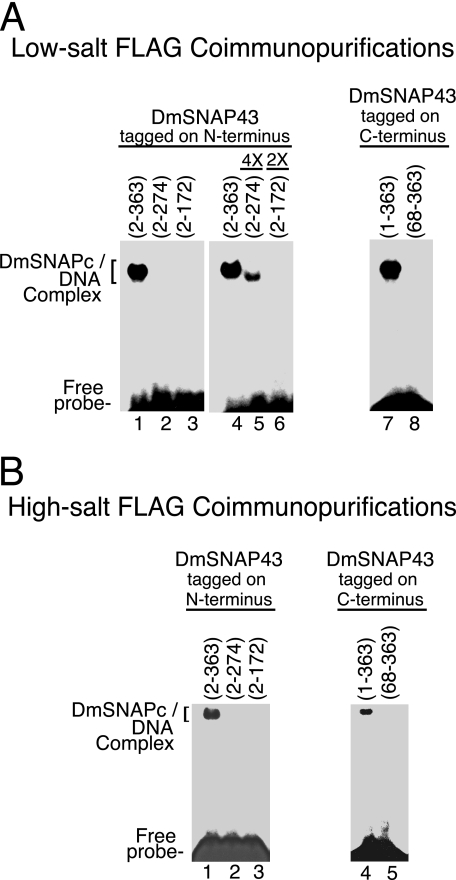
Finally, to investigate which domains of DmSNAP43 contribute to DNA binding by DmSNAPc, the FLAG affinity-purified complexes were subjected to electrophoretic mobility shift analysis (Fig. 7). Complexes that contained N-terminal tagged full-length DmSNAP43 bound efficiently to the PSEA (Fig. 7, A, lanes 1 and 4, and B, lane 1). However, DNA binding by the construct that was truncated following residue 274 was weakly detectable only when 4 times the normalized amount of protein was used (Fig. 7, A, lanes 2 and 5, and B, lane 2). No DNA-binding activity was detectable for the construct that was truncated following position 172 (Fig. 7, A, lanes 3 and 6, and B, lane 3). These results indicate that the non-conserved C-terminal domain of DmSNAP43 plays a critical role in DmSNAPc binding to the PSEA, even though this region is not required for DmSNAP43 to assemble into a complex with both DmSNAP190 and DmSNAP50.
FIGURE 7.
Domains of DmSNAP43 required for sequence-specific DNA binding by DmSNAPc. A, DmSNAP complexes FLAG affinity purified under low-salt conditions (Fig. 6C) were used for DNA mobility shift analysis with a DNA probe containing a U1 PSEA sequence. Complexes containing DmSNAP43 constructs tagged at the N terminus (C-terminal truncations) are shown in lanes 1–6, and constructs tagged at the C terminus (N-terminal truncations) are shown in lanes 7 and 8. Lanes 1–4 and 7 and 8 were carried out with complexes containing a normalized amount of DmSNAP43 as determined in Fig. 6C, whereas lanes 5 and 6 contained 4 or 2 times the normalized amount of truncated protein as indicated above each lane. (The maximum amount of protein that could be used was limited by the volume of the reaction and the relative concentration of each sample.) B, similar to A except DmSNAP complexes were affinity purified under high-salt conditions and protein amounts were normalized based upon the immunoblots shown in Fig. 6D.
DmSNAPc that contained full-length C-terminal tagged DmSNAP43 bound efficiently to DNA (Fig. 7, A, lane 7, and B, lane 4). However, no DNA-binding activity was detectable following the deletion of the N-terminal 67 amino acid residues of DmSNAP43 (Fig. 7, A, lane 8, and B, lane 5). This was to be expected because DmSNAP50 was not complexed with this N-terminal-truncated DmSNAP43 (Fig. 6, C, lane 6, and D, lane 5).
Requirements for the Binding of DmSNAPc to the U1 Gene Promoter in Vivo
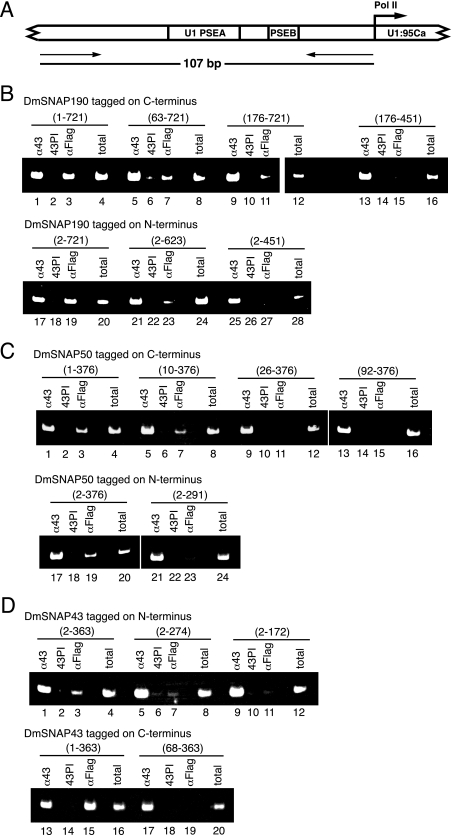
Electrophoretic mobility shift experiments described above (Figs. 3, 5, and 7) examined the in vitro sequence-specific DNA-binding activity of DmSNAPc that contained tagged full-length or truncated DmSNAP subunits. To examine the in vivo DNA-binding activity of these mutant DmSNAP complexes, chromatin immunoprecipitation (ChIP) assays were conducted by using the same stably transfected cell lines prepared to make extracts for the immunoaffinity purification experiments.
ChIPs were carried out by using antibodies against the FLAG epitope to examine the in vivo occupancy of the well characterized endogenous U1:95Ca gene promoter by the FLAG-tagged constructs. The promoter of the U1:95Ca gene is active in vivo and has been previously employed for ChIP analysis (26). The PCR primers specific for this promoter amplify a 107-base pair DNA fragment, and their genomic locations in the 5′-flanking DNA of the U1:95Ca gene are illustrated in Fig. 8A. Polyclonal antibodies prepared against full-length DmSNAP43 were used as positive ChIP controls because DmSNAP43 should consistently be present at this promoter either as a component of the overexpressed FLAG-tagged DmSNAPc, or in its absence, as a component of endogenous DmSNAPc. Preimmune antibodies from the same rabbits were used as negative controls in the ChIP assays.
FIGURE 8.
Effect of DmSNAP mutations on the in vivo occupancy of a U1 snRNA gene promoter by DmSNAPc. A, schematic diagram of the endogenous D. melanogaster U1:95Ca promoter. Positions of primers utilized for ChIP are indicated by arrows. B, ChIP from S2 cell lines that co-express full-length or truncated FLAG-tagged DmSNAP190 constructs (as shown in Fig. 2A) with full-length untagged DmSNAP50 and DmSNAP43. Antibody against the FLAG epitope (αFLAG) was used to detect the presence of tagged DmSNAPc at the U1:95Ca promoter. Antibody against DmSNAP43 (α43), which recognizes both endogenous and tagged DmSNAPc was used as a positive control. DmSNAP43 preimmune serum (43PI) was used as a negative control. Positive PCR controls that utilized 20% of the unselected total input DNA (total) were also included. The upper panel shows ChIP results from S2 cell lines expressing DmSNAP190 constructs tagged at the C terminus (N-terminal truncations). The lower panel shows ChIP results from S2 cell lines expressing DmSNAP190 constructs tagged at the N terminus (C-terminal truncations). C, ChIP from S2 cell lines that co-express full-length or truncated FLAG-tagged DmSNAP50 constructs (as shown in Fig. 4A) with full-length untagged DmSNAP190 and DmSNAP43. Antibodies and other conditions were the same as in B. The upper panel shows ChIP results from S2 cell lines expressing DmSNAP50 constructs tagged at the C terminus (N-terminal truncations). The lower panel shows ChIP results from S2 cell lines expressing DmSNAP50 constructs tagged at the N terminus (C-terminal truncations). D, ChIP from S2 cell lines that co-express full-length or truncated FLAG-tagged DmSNAP43 constructs (as shown in Fig. 6A) with full-length untagged DmSNAP190 and DmSNAP50. Antibodies and other conditions were the same as in B. The upper panel shows ChIP results from S2 cell lines expressing DmSNAP43 constructs tagged at the N terminus (C-terminal truncations). The lower panel shows ChIP results from S2 cell lines expressing DmSNAP43 constructs tagged at the C terminus (N-terminal truncations).
In the transfected cell lines that overexpressed tagged full-length DmSNAP190, the anti-FLAG antibodies efficiently precipitated the U1 promoter. This was true whether the tag was at the C or N terminus of the protein (Fig. 8B, lanes 3 and 19). Truncation of amino acid residues from either terminus progressively decreased the intensity of the PCR signal (Fig. 8B, lanes 7, 11, 15, 23, and 27). This indicates that residues both N- and C-terminal to the conserved Myb domain of DmSNAP190 are important for the efficient binding of DmSNAPc to the U1 promoter in vivo. Moreover, these results were similar to those obtained with the in vitro binding assays of the full-length and truncated DmSNAP190 constructs shown in Fig. 3.
ChIP results from cells expressing FLAG-tagged DmSNAP50 and DmSNAP43 truncation constructs were also quite consistent with the band shift data. DmSNAP complexes that contained tagged full-length DmSNAP50 and DmSNAP43 were readily detected on the U1 promoter by ChIP (Fig. 8, C, lanes 3 and 19, and D, lanes 3 and 15). Removal of the first nine amino acids from DmSNAP50 had little effect on DmSNAPc-binding activity in vivo (Fig. 8C, lane 7), whereas U1 promoter occupancy was greatly reduced or no longer detectable as a result of additional truncations from the N or C terminus of DmSNAP50 (Fig. 8C, lanes 11, 15, and 23). Similarly, each of the tested truncations of DmSNAP43 from the N or C terminus severely damaged the in vivo DNA-binding activity of DmSNAPc (Fig. 8D, lanes 7, 11, and 19).
DISCUSSION
Functional Domains of DmSNAPc Subunits
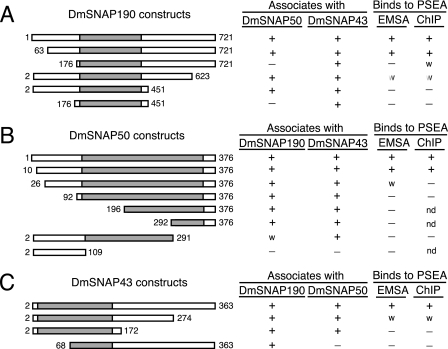
Fig. 9 summarizes the results of our truncation analyses to identify protein domains involved in DmSNAPc assembly and in binding to the PSEA. With one exception, the evolutionarily most conserved region of each DmSNAP subunit is sufficient for its assembly with the other two subunits. For example, DmSNAP43-(2–172) can assemble with both DmSNAP190 and DmSNAP50. DmSNAP50- (92–376) can assemble with both DmSNAP190 and DmSNAP43. Furthermore, the conserved Myb domain of DmSNAP190-(176–451) can associate with DmSNAP43. On the other hand, this conserved region of DmSNAP190 is insufficient for its assembly with DmSNAP50. Additional amino acid residues located just N-terminal to the Myb domain of DmSNAP190 (between positions 63 and 176) are required for DmSNAP50 assembly into the complex. In all other cases, however, the regions of the proteins that are the most evolutionarily conserved mediate assembly of the DmSNAPc subunits.
FIGURE 9.
Summary of the effects of DmSNAP truncations on DmSNAPc assembly and DNA-binding activity. The various DmSNAP truncations analyzed are shown on the left side of the figure. The columns on the right indicate whether the specific truncation construct can associate with the other two DmSNAP subunits and whether the DmSNAP complex that contains the truncation construct can bind sequence specifically to the PSEA as assayed either by electrophoretic mobility shift assay or ChIP. A w indicates weak association or binding. nd indicates “not done,” reflecting the fact that these represent more extensive truncations beyond constructs that already lacked DNA-binding activity.
Furthermore, all DmSNAP mutations that impaired complex assembly also impaired DNA-binding activity both in vitro and in vivo (Fig. 9). This was to be expected because all three subunits are required for sequence-specific DNA binding by metazoan SNAPc (21–23).4
Somewhat surprisingly, mutations in the non-conserved regions of the DmSNAP subunits often resulted in the loss of DNA-binding activity, even when subunit assembly was not affected (Fig. 9). For example, deletion of DmSNAP190 residues beyond position 623 severely compromised DNA binding by the complex, and deletion of residues beyond position 451 destroyed the ability of the complex to bind to DNA. Similarly, deletion of non-conserved DmSNAP50 residues between positions 10 and 92 resulted in a loss of DNA-binding activity. Finally, deletion of amino acid residues following position 274 of DmSNAP43 significantly weakened the DNA binding activity of the complex, and deletion beyond position 172 of DmSNAP43 eliminated DNA binding. Thus, the evolutionarily non-conserved regions of the DmSNAP subunits are in many cases essential for the DNA binding activity of DmSNAPc. Whether these domains directly contact DNA, or whether they are required for DmSNAPc to adopt a conformation required for DNA-binding activity, remains an open question. However, considering that DmSNAPc contacts over 40 base pairs of DNA on the U1 promoter (14) and that all three subunits contact DNA (13), it is entirely possible that the less-conserved domains are involved in making direct contacts to the DNA. This suggests that the requirements for subunit-subunit interactions within metazoan SNAPc are more constrained evolutionarily than the protein-DNA interactions. This agrees with our recent observations that the DNA sequences of the 3′ half of the PSEAs have changed fairly rapidly during insect evolution (19), suggesting that interactions between the protein and the DNA are more free to co-evolve.
Comparison to Findings with Human SNAPc
A number of studies have been published that address the roles of domains within the human SNAP subunits (21–24, 27). Sometimes our results with fly SNAPc have lead to conclusions similar to those reached in the human system; in other cases the fly data expand our knowledge and provide new insights; at still other times unexpected differences have been revealed. Some of the main similarities and differences are summarized in the following paragraphs.
Fly SNAP190 Versus Human SNAP190
Figs. 2 and 9 show that the conserved Myb domain of DmSNAP190 interacts with DmSNAP43. In contrast, no interaction was observed between HsSNAP43 and the Myb domain of HsSNAP190 (27). In the human system, an additional subunit, HsSNAP19, is required for stable association between HsSNAP190 and HsSNAP43, and this interaction requires a small region (residues 84–138) in the non-conserved N terminus of HsSNAP190 (27). Our finding that DmSNAP43 interacts with the Myb domain of DmSNAP190 likely reveals an actual difference between the fly and human systems as no ortholog of HsSNAP19 has been identified in the fruit fly. The lack of strong interactions of HsSNAP43 with the Myb domain of HsSNAP190 may be compensated in the human system by interactions involving HsSNAP19 (and HsSNAP43) with residues 84–138 of HsSNAP190 (27).
We have determined in the fly that amino acids of DmSNAP190 located between positions 63 and 176, just upstream of the Myb domain, are required for association with DmSNAP50 (Figs. 2 and 9). We are not aware of any comparable experiments reported in the human system that localize a region of HsSNAP190 required for interaction with HsSNAP50. In fact available evidence in the human system indicates that HsSNAP190 and HsSNAP50 do not directly interact (11, 21, 23, 27).
Even more surprisingly, we found that the non-conserved C-terminal domain of DmSNAP190 is essential for the binding of DmSNAPc to the PSEA (Figs. 3 and 8). This was unexpected because the C-terminal domain of HsSNAP190 is completely dispensable for sequence-specific DNA binding in the human system (21, 22, 24, 27, 28). In fact, the C-terminal domain of HsSNAP190 serves to dampen DNA binding by HsSNAPc (21). In contrast, in flies it appears that the C-terminal domain of DmSNAP190 is required for the stable binding of DmSNAPc to the DNA.
Fly SNAP50 Versus Human SNAP50
Domains of HsSNAP50 involved in interacting with HsSNAP190 have not been reported in the human system. In fact, as mentioned above, there is no published data supporting a direct interaction between HsSNAP190 and HsSNAP50. However, our results (Figs. 4 and 9 and data not shown) indicate that sequences at the C terminus of DmSNAP50 (residues 292–376 that form part of the highly conserved SNAP finger domain) are sufficient for interaction with DmSNAP190. Jawdekar et al. (23) did not report any interaction of this region of HsSNAP50 with HsSNAP190. Instead, their work emphasized the DNA-binding activity of the SNAP finger domain. More than likely, the SNAP finger domain of DmSNAP50 is involved both in DNA binding and in interaction with DmSNAP190. The SNAP finger of DmSNAP50 also interacts with DmSNAP43 (Figs. 4 and 9 and data not shown). However, the central region of DmSNAP50 (residues 110–291) also participates in complex assembly with DmSNAP43 (Figs. 4 and 9). This is the same region of SNAP50 that was found to interact with SNAP43 in the human system (23).
It was not surprising that the C-terminal truncation DmSNAP50-(2–291) that deleted a portion of the SNAP finger domain eliminated DNA-binding activity. However, DNA-binding activity was also ablated by a DmSNAP50 N-terminal truncation that deleted amino acids 1–91 even though this construct assembled efficiently with DmSNAP190 and DmSNAP43 (Fig. 9). Thus a portion of the non-conserved N terminus (as well as the C terminus) of DmSNAP50 is required for DNA binding. Comparable studies targeting the N-terminal function of HsSNAP50 have to our knowledge not been reported.
Fly SNAP43 Versus Human SNAP43
Results shown in Figs. 6 and 9 implicate DmSNAP43 residues located between positions 68 and 172 as those most likely involved in interaction with DmSNAP190. On the other hand, Ma and Hernandez (27) found that residues 164–268 of HsSNAP43 (which are equivalent to residues 179–283 of DmSNAP43) were sufficient for association with HsSNAP190 together with HsSNAP19. As mentioned above, HsSNAP43 does not interact strongly with HsSNAP190 in the absence of HsSNAP19. Therefore, the mapping of different regions in fly and human SNAP43 involved in association with SNAP190 could reflect the additional requirement and role for HsSNAP19 for stable association in the human system.
Our results indicate that the first 172 amino acids of DmSNAP43 are sufficient to recruit DmSNAP50 into the complex and that the first 68 amino acids of DmSNAP43 are required for this interaction. Ma and Hernandez (27), who employed very similar truncations of HsSNAP43, obtained nearly identical results in the human system. On the other hand, Hinkley et al. (22), also working in the human system, found that the C-terminal as well as the N-terminal half of HsSNAP43 could interact with HsSNAP50 in a glutathione S-transferase pulldown assay. It is possible that the conditions utilized by us and by Ma and Hernandez (27) are more stringent and detect only stronger interactions among the SNAP subunits.
Deletion of DmSNAP43 amino acids beyond position 274 reduced but did not completely eliminate sequence-specific binding of DmSNAPc to the PSEA. However, DNA-binding activity was undetectable following further truncation of DmSNAP43 to position 172 (Figs. 7–9). Results essentially identical to these were obtained by Ma and Hernandez (27) when they used truncations that were analogous to our truncations of DmSNAP43. However, Hinkley et al. (22) found that either the N-terminal half or the C-terminal half of HsSNAP43 could be incorporated into mini-SNAPc complexes that were capable of binding to DNA. The reason for the different findings within the human system is not clear. However, our data with fly SNAP43 parallel more closely the findings of Ma and Hernandez (27).
Conclusion
We have co-expressed full-length and truncated forms of all three DmSNAPc subunits in homologous S2 cells to determine the domains required for in vivo assembly of DmSNAPc and for DNA binding both in vitro and in vivo. With one exception, the regions evolutionarily conserved among animals are sufficient for complex assembly. However, non-conserved regions of the proteins are required for sequence-specific DNA binding. The ability to compare results in the human and fly systems has in several cases allowed us to identify domains that function similarly in both systems. However, in other cases novel insights into metazoan SNAPc functional domains have been obtained, and differences between the two systems suggest possible alternative mechanisms for achieving complex assembly and stable DNA binding.
Acknowledgments
We thank Anna D'Agostini, Kathleen McNamara- Schroeder, and David Almeida for assistance in plasmid construction and preparation.
This work was supported by National Science Foundation Grants MCB-0641350 and MCB-0842770 and the California Metabolic Research Foundation.
K.-H. Hung, M. Titus, S.-C. Chiang, and W. E. Stumph, unpublished observations.
K.-H. Hung, M. Titus, S.-C. Chiang, and W. E. Stumph, unpublished data.
- snRNA
- small nuclear RNA
- SNAPc
- small nuclear RNA-activating protein complex
- PSE
- proximal sequence element
- PSEA
- proximal sequence element A
- ChIP
- chromatin immunoprecipitation
- Hs
- human
- Dm
- D. melanogaster.
REFERENCES
- 1.Murphy S., Yoon J. B., Gerster T., Roeder R. G. (1992) Mol. Cell. Biol. 12,3247–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadowski C. L., Henry R. W., Lobo S. M., Hernandez N. (1993) Genes Dev. 7,1535–1548 [DOI] [PubMed] [Google Scholar]
- 3.Yoon J. B., Murphy S., Bai L., Wang Z., Roeder R. G. (1995) Mol. Cell. Biol. 15,2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry R. W., Sadowski C. L., Kobayashi R., Hernandez N. (1995) Nature 374,653–656 [DOI] [PubMed] [Google Scholar]
- 5.Waldschmidt R., Wanandi I., Seifart K. H. (1991) EMBO J. 10,2595–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanandi I., Waldschmidt R., Seifart K. H. (1993) J. Biol. Chem. 268,6629–6640 [PubMed] [Google Scholar]
- 7.Yoon J. B., Roeder R. G. (1996) Mol. Cell. Biol. 16,1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadowski C. L., Henry R. W., Kobayashi R., Hernandez N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93,4289–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry R. W., Ma B., Sadowski C. L., Kobayashi R., Hernandez N. (1996) EMBO J. 15,7129–7136 [PMC free article] [PubMed] [Google Scholar]
- 10.Bai L., Wang Z., Yoon J. B., Roeder R. G. (1996) Mol. Cell. Biol. 16,5419–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong M. W., Henry R. W., Ma B., Kobayashi R., Klages N., Matthias P., Strubin M., Hernandez N. (1998) Mol. Cell. Biol. 18,368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry R. W., Mittal V., Ma B., Kobayashi R., Hernandez N. (1998) Genes Dev. 12,2664–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Stumph W. E. (1998) Mol. Cell. Biol. 18,1570–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C., Harding G. A., Parise J., McNamara-Schroeder K. J., Stumph W. E. (2004) Mol. Cell. Biol. 24,1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai H. T., Kang Y. S., Stumph W. E. (2008) FEBS Lett. 582,3734–3738 [DOI] [PubMed] [Google Scholar]
- 16.Das A., Bellofatto V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A., Zhang Q., Palenchar J. B., Chatterjee B., Cross G. A., Bellofatto V. (2005) Mol. Cell. Biol. 25,7314–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schimanski B., Nguyen T. N., Günzl A. (2005) Mol. Cell. Biol. 25,7303–7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez G., Jr., Valafar F., Stumph W. E. (2007) Nucleic Acids Res. 35,21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y., Song Y., Wang Y., Jessop L., Zhan L., Stumph W. E. (1997) Eur. J. Biochem. 248,231–237 [DOI] [PubMed] [Google Scholar]
- 21.Mittal V., Ma B., Hernandez N. (1999) Genes Dev. 13,1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinkley C. S., Hirsch H. A., Gu L., LaMere B., Henry R. W. (2003) J. Biol. Chem. 278,18649–18657 [DOI] [PubMed] [Google Scholar]
- 23.Jawdekar G. W., Hanzlowsky A., Hovde S. L., Jelencic B., Feig M., Geiger J. H., Henry R. W. (2006) J. Biol. Chem. 281,31050–31060 [DOI] [PubMed] [Google Scholar]
- 24.Ma B., Hernandez N. (2002) Mol. Cell. Biol. 22,8067–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamrod Z., Tyree C. M., Song Y., Stumph W. E. (1993) Mol. Cell. Biol. 13,5918–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barakat N. H., Stumph W. E. (2008) FEBS Lett. 582,2413–2416 [DOI] [PubMed] [Google Scholar]
- 27.Ma B., Hernandez N. (2001) J. Biol. Chem. 276,5027–5035 [DOI] [PubMed] [Google Scholar]
- 28.Hanzlowsky A., Jelencic B., Jawdekar G., Hinkley C. S., Geiger J. H., Henry R. W. (2006) Protein Expr. Purif. 48,215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]