Abstract
The plastid terminal oxidase (PTOX) is distantly related to the mitochondrial alternative oxidase (AOX). Both are members of the diiron carboxylate quinol oxidase (DOX) class of proteins. PTOX and AOX contain 20 highly conserved amino acids, six of which are Fe-binding ligands. We have previously used in vitro and in planta activity assays to examine the functional importance of the Fe-binding sites. In this report, we conduct alanine-scanning mutagenesis on the 14 other conserved sites using our in vitro and in planta assay procedures. We found that the 14 sites fall into three classes: (i) Ala-139, Pro-142, Glu-171, Asn-174, Leu-179, Pro-216, Ala-230, Asp-287, and Arg-293 are dispensable for activity; (ii) Tyr-234 and Asp-295 are essential for activity; and (iii) Leu-135, His-151, and Tyr-212 are important but not essential for activity. Our data are consistent with the proposed role of some of these residues in active site conformation, substrate binding, and/or catalysis. Titration experiments showed that down-regulation of PTOX to ∼3% of wild-type levels did not compromise plant growth, at least under ambient growth conditions. This suggests that PTOX is normally in excess, especially early in thylakoid membrane biogenesis.
Alternative oxidase (AOX)2 is a terminal oxidase that functions in the alternative pathway of mitochondrial respiration (1). It catalyzes the four-electron reduction of oxygen to water and branches from the cytochrome pathway at the level of the quinone pool (1–5). The alternative oxidase is found in two of the three domains of life, Bacteria and Eucarya, but not in Archaea; among Eucarya it is found in all kingdoms (i.e. plants, animals, fungi, protists) (4, 5). It is thought that AOX is activated when the cytochrome pathway becomes saturated, for instance, during oxidative stress when the inner membrane is highly energized and prone to the production of reactive oxygen species (ROS) (3, 6–9).
The IMMUTANS locus of Arabidopsis codes for a plastid homolog of AOX (10, 11). IM-like proteins have subsequently been found in a diverse array of plant, algal, and cyanobacterial species; IM does not appear to be present in animals (12). Also similar to AOX, IM functions as a terminal oxidase, transferring electrons from the plastoquinol (PQ) pool to molecular oxygen (13–17). IM is thus frequently designated “PTOX” (plastid terminal oxidase). (To avoid confusion, we will use the term PTOX in this report and IMMUTANS when describing the Arabidopsis gene for PTOX.) Current thinking is that PTOX is an important alternative electron sink in plastid membranes and that it lies at the intersection of many redox pathways. These include the desaturation reactions of carotenoid biosynthesis and chlororespiration (18, 19). Reminiscent of AOX, PTOX has been hypothesized to serve as a “safety valve” for the dissipation of excess electron flow, e.g. during stress (20–23). Consistent with this view, studies in tomato ghost (the ortholog of the Arabidopsis im mutant) reveal that lack of PTOX in young seedlings as well as in mature tomato leaves results in increased sensitivity to high light stress due to disturbances in the redox status of the plastoquinone pool (23). In contrast, in vivo chlorophyll fluorescence measurements reveal that modulating IMMUTANS expression and/or protein accumulation does not alter the flux of intersystem electrons during steady state photosynthesis in Arabidopsis nor does it afford photoprotection (17). However, IMMUTANS expression is strongly regulated by developmental factors in Arabidopsis, and the phenotype of im strongly argues that PTOX function in Arabidopsis is required early in chloroplast biogenesis. Together, these studies suggest that PTOX functions in a developmental- and species-specific manner.
AOX and PTOX are members of the DOX (non-heme diiron carboxylate quinol oxidase) family of proteins (4, 5, 24–29). By analogy to crystal structure determinations of non-plant members of this family, it has been proposed that the diiron centers of AOX and PTOX are coordinated by four carboxylate and two His residues on a four-helix bundle (26–28). Sequence alignments have revealed that the six putative Fe-binding residues are highly conserved in the sequences of all AOX and PTOX proteins examined to date. In addition, nearly all PTOX enzymes contain a 16-amino acid domain near the C terminus that is highly conserved, but that is not found in AOX (13). Curiously, this sequence corresponds precisely to exon 8 of the gene.
We previously used PTOX as a model to test the functional significance of the conserved Fe-binding and exon 8 sequences (13). These experiments were facilitated by the availability of two important tools: 1) an in vitro activity assay, developed by Josse et al. (15, 18); and 2) null alleles of the Arabidopsis immutans (im) mutant, which made in planta mutagenesis experiments possible. Arabidopsis im mutants have a light-sensitive green- and white-variegation due to action of a nuclear recessive gene -enhanced light intensities promote white sector formation (30–32). The white sectors contain abnormal, photooxidized plastids as a consequence of a lack of colored carotenoid production, while the green sectors contain morphologically normal chloroplasts. When transformed with a wild-type IM sequence, im plants revert to an all-green phenotype. Mutagenized copies of IM can thus be tested for their ability to normalize the im variegation and restore a wild type appearance. Our previous in vitro and in planta experiments showed that the six amino acids that bind iron do not tolerate change, even conservative ones, and that the exon 8 domain is required for PTOX activity and stability (13).
In this report, we examine 14 other amino acids that are perfectly conserved, or nearly so, in the sequences of all AOX and PTOX reported to date: Leu-135, Pro-142, Ala-139, His-151, Glu-171, Asn-174, Leu-179, Tyr-212, Pro-216, Ala-230, Tyr-234, Asp-287, Arg-293, Asp-295 (numbering refers to the Arabidopsis PTOX sequence). Most of these residues are predicted to reside in close proximity to the six Fe-binding sites. In this report, we test the functional significance of these sites by in vitro and in planta alanine-scanning mutagenesis, and report that very few of these sites are absolutely essential for activity. In addition, some mutant enzymes are defective in vitro, but are able to complement im. RNAi, antisense, and co-suppression experiments showed that transgenic plants with less than 3% of wild-type PTOX levels produce normal appearing plants. Considered together, the data suggest that PTOX is normally in excess, especially during the process of thylakoid formation early in leaf development when sector formation is established (33).
MATERIALS AND METHODS
Plant Growth
Plants in this report included wild-type Arabidopsis (Columbia ecotype), the spotty allele of im (also in Columbia) (10), and transgenic im plants (described below). The plants were germinated and grown at 22 °C under conditions of continuous illumination. The wild-type plants were maintained at 100 μmol m−2 s−1. However, spotty is light sensitive and therefore was germinated under low light (15 μmol m−2 s−1 for 5 days) before transfer to normal light (100 μmol m−2 s−1).
Site-directed Mutagenesis in Vitro
An IM cDNA lacking the N-terminal chloroplast-targeting sequence (10) was cloned into the BamH1 and Nhe1 sites of the Escherichia coli expression vector pET-11a (Novagen, Madison, WI). The QuikChangeTM site-directed mutagenesis protocol (Stratagene, La Jolla, CA) was used to generate mutations; the mutations were confirmed by sequencing. Following standard notation, the mutant clones are designated by the amino acid that was altered, its location in the A. thaliana IM sequence, and the amino acid to which it was changed (e.g. L135A means Leu-135 was changed to Ala).
The following primers were used to introduce mutations into 14 conserved amino acids residues of AOX and IM: L135A: 5′-gcaaggttctttgttgctgagacagc-3′, 5′-gctgtctcagcaacaaagaaccttgc-3′. A139G: 5′-cttgagacaattggtagagtgccttattttgc-3′, 5′-gcaaaataaggcactctaccaattgtctcaag-3′. P142A: 5′-caattgctagagtggcttattttgcg-3′, 5′-cgcaaaataagccactctagcaattg-3′. H151A: 5′-gtctgtgctagctatgtatgagacctttgg-3′, 5′-ccaaaggtctcatacatagctagcacagac-3′. E171A: 5′-gtacactttgctgcgagctggaatgaaatg-3′, 5′-catttcattccagctcgcagcaaagtgtac-3′. N174A: 5′-gagagctgggctgaaatgcatcacttg-3′, 5′-caagtgatgcatttcagcccagctctc-3′. L179A: 5′-gaaatgcatcacgcgctcataatggaag-3′, 5′-cttccattatgagcgcgtgatgcatttc-3′. Y212A: 5′-gtgttcttggctatcttaagccctagaatg-3′, 5′-cattctagggcttaagatagccaagaacac-3′. P216A: 5′-gtatatcttaagcgctagaatggcatatcac-3′, 5′-gtgatatgccattctagcgcttaagatatac-3′. A230G: 5′-gtggagagtcatggatatgagacttat-3′, 5′-ataagtctcatatccatgactctccac-3′. Y234A: 5′-gcatatgagactgctgataaatttctc-3′, 5′-gagaaatttatcagcagtctcatatgc-3′. D287A: 5′-gaaaatctatacgctgtgtttgtgaac-3′, 5′-gttcacaaacacagcgtatagattttc-3′. R293A: 5′-gtgaacatagcagatgatgaagcagaacac-3′, 5′-gtgttctgcttcatcatctgctatgttcac-3′. D295A: 5′-gtgaacataagagatgctgaagcagaacac-3′, 5-gtgttctgcttcagcatctcttatgttcac-3′.
Site-directed Mutagenesis in Planta
Procedures for in planta mutagenesis have been described in Ref. 13. In brief, mutations were introduced (as above) into full-length IM cDNAs, and the mutant cDNAs were cloned behind the CaMV 35 S promoter in the binary vector pBI121. The constructs were transferred into Agrobacterium tumefaciens and introduced into im plants (spotty allele) by the floral dip method (34). Kanamycin-resistant seedlings were selected at the T1 generation on plates containing 1 × MS salts (pH 5.7), 1% sucrose, and 50 μg ml−1 kanamycin. PCR and Southern blotting methods were performed to verify that the plants were transformed. Phenotypic analyses and protein analysis were conducted on T2 generation plants.
Generation of RNAi, Antisense, and Co-suppression Plants
To generate RNAi plants, gene-specific sense and antisense fragments of IM were amplified by RT-PCR using primers: sense strand (5′-CCGGATATCGCAGTAAAATACTATACG-3′ and 5′-GGCTCTAGACCAAGGCTAGTAGTATGG-3′), antisense strand (5′-GCGGAATTCGCAGTAAAATACTATACG-3′ and 5′-CGCGGATCCCCAAGGCTAGTAGTATGG-3′). The resulting sense and antisense fragments were linked by a 1-kb spacer, which is a partial DNA fragment of the β-glucuonidase (GUS) gene. The whole DNA fragment containing sense, spacer, and antisense regions was cloned behind the CaMV 35 S promoter in the binary vector pBI121. To generate antisense and co-suppression plants, full-length wild-type IM cDNAs (the same as used for the in planta site-directed mutagenesis experiments) were cloned in the forward and reverse orientations behind the CaMV 35 S promoter in the binary vector pBI121. For all three constructs, the same procedures were used as described above for plant transformation. Western blotting was performed to identify antisense and co-suppression lines with reduced levels of the PTOX protein in T2 generation plants; these procedures have been described (13).
PTOX Activity Assays
We have previously described methods to estimate in vitro PTOX activity based on procedures described by Josse et al. (15, 18). In brief, mutant constructs were transformed into E. coli strain BL21-DE3, and protein expression was induced by the addition of 1.6 mm isopropyl-1-thio-β-d-galactopyranoside (final concentration). The cells were then lysed in a buffer containing 20 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, and 0.3% protease inhibitor mixture (PIC). The membranes were collected by ultracentrifugation (200,000 × g for 2 h at 4 °C), and the concentration of proteins in the pellet was measured using the BioRad protocol with bovine serum albumin as a standard. Oxygen consumption was monitored at 25 °C with a Clark O2 electrode (Hansatech, Norfolk, England) using 100–200 μg of membrane protein in a 1.5-ml reaction mixture that contained a final concentration of 50 mm Tris-maleate (pH 7.5), 10 mm KCl, 5 mm MgCl2, 1 mm EDTA, and 0.2 mm decyl-plastoquinone (2,3-methyl-5decyl-1,4-benzoquinone) (Sigma). The reactions were initiated by the addition of NADH (1 mm final concentration), followed by KCN (3 mm final concentration) and n-propyl gallate (3,4,5-tri-hydroxy-benzoic acid-n-propyl ester) (0.5 mm final concentration), all from Sigma. Reactions were followed for a total of 8 min: KCN was added 1 min after initiating the reaction with NADH; n-PG was added after 6 min. The activity measured in these assays represents the CN-resistant and nPG-sensitive oxygen uptake by PTOX.
Isolation and Detection of Proteins
Proteins were isolated from partially purified chloroplasts by previously described procedures (35). In brief, leaves from 4-week-old Arabidopsis seedlings were homogenized in 0.33 m sorbitol, 10 mm EDTA, and 50 mm HEPES (pH 8.0). The homogenate was filtered through two layers of Miracloth (Calbiochem, La Jolla, CA), and the filtrate was centrifuged at 2600 × g for 5 min. The pellet was resuspended in 10 mm MOPS (pH 8.0) then centrifuged at 10,000 × g for 10 min. The resulting pellet was resuspended in 0.33 m sorbitol, 5 mm MgCl2, 50 mm HEPES (pH 8.0), and the chlorophyll concentration of the suspension was determined (35).
Procedures for Western blotting have been described (13). In brief, 5 μg of protein (for E. coli samples) or an amount of protein corresponding to 5 μg of chlorophyll (for chloroplast samples) were electrophoresed through 12.5% SDS-polyacrylamide gels, and the proteins were transferred to a nitrocellulose membrane. The membrane was incubated with a polyclonal antibody generated to the Arabidopsis PTOX protein (1:3000 dilution) (20), and the proteins were visualized using the ECL immunodetection procedure (Pierce).
RESULTS
Identification of Conserved Residues in PTOX and AOX
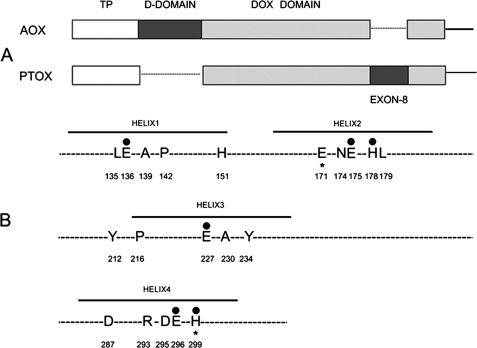
Fig. 1A shows that AOX and PTOX have two structural features in common: both have an N-terminal transit peptide (TP) that targets the protein to the organelle (mitochondria in the case of AOX and chloroplasts in the case of PTOX); and both have a “DOX” domain, which comprises a four-helix bundle that includes the catalytic site (27–29). The active site is composed, in part, of six amino acids that serve as ligands for two iron atoms (Glu-136, Glu-175, His-178, Glu-227, Glu-296, and His-299) (Fig. 1B). AOX and PTOX also have unique structural domains: AOX contains a “dimerization domain” (D-domain) that has been implicated in AOX-AOX dimer formation (36), and PTOX contains a 16 amino acid (exon 8) domain near the active site that is essential for protein stability and activity (13).
FIGURE 1.
Conserved residues in AOX and PTOX. A, structures of AOX and PTOX. Both proteins have an N-terminal organelle-targeting sequence (TP, transit peptide). AOX has a dimerization domain (D-domain) (not present in PTOX), whereas PTOX has an exon 8 sequence (absent in AOX). The DOX domain is shown in gray. B, early sequence alignments revealed the presence of 20 perfectly conserved residues in comparisons of the 27 AOX and PTOX sequences that were available at that time. The figure shows the 20 amino acids: 18 of the residues are perfectly conserved in all plant AOX and PTOX sequences. The exceptions are the following: Glu-171 (Gln in maize) and His-299 (Ile in wheat) (indicated by *).The six iron binding sites are indicated by darkened circles, and the location of the four helices is shown.
When the present experiments were initiated, 7 PTOX and 20 AOX protein sequences were available for comparison. 20 residues were found to be perfectly conserved among these sequences, and these 20 became the focus of our structure/function studies. Many more AOX and PTOX protein sequences have become available since that time, and a recent comparison of these sequences using ClustalW reveals that the 20 amino acid residues are still the most highly conserved, with exceptions being largely confined to those taxa that are distantly related to higher plants such as protists, cyanobacteria, and fungi. For example, an alignment of over 100 plant AOX and PTOX sequences from NCBI reveals that 18 of the 20 amino acids are perfectly conserved in plants and that the other two have single exceptions (Glu-171 is Gln in Zea mays PTOX whereas His-299 is Ile in wheat PTOX). Several of these residues have been shown to be essential for catalytic activity and/or are predicted to play a role in AOX regulation and stability (4, 5). Six of the 20 residues are putative Fe-binding sites, and our results of in vitro and in planta mutagenesis of these six have been reported (13). The present report describes mutagenesis studies on the other 14 residues (Fig. 1B).
Mutagenesis of 14 Conserved Residues in Vitro
To test the functionality of the 14 residues, we conducted site-directed mutagenesis using an in vitro PTOX activity assay developed by Josse et al. (15, 18); this assay was used by us in our functional analyses of the six Fe-binding sites (13). In brief, this assay involves the measurement of CN-resistant and nPG-sensitive O2 consumption using membranes from E. coli transformed with various mutant IM constructs. Prior to measurement, the membranes are treated sequentially with NADH, KCN, and n-propyl gallate (n-PG). The rationale is that NADH serves as an electron donor to quinone (via a membrane-bound NADH dehydrogenase), and electrons are then transferred to molecular oxygen via either PTOX or the cytochrome pathway. Flux is regulated by KCN, which inhibits the cytochrome pathway (but not PTOX activity), or by n-propyl gallate (n-PG), which inhibits PTOX but not the cytochrome pathway. In short, O2 consumption occurs by the cytochrome pathway when n-PG is added to the membranes, but by PTOX when KCN is added. Addition of both inhibitors abolishes O2 consumption.
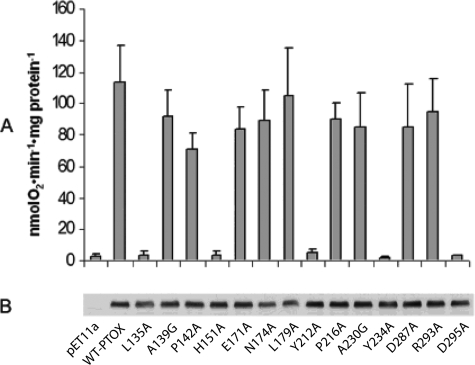
For our experiments, we performed alanine-scanning mutagenesis on 12 of 14 residues; the two conserved alanine residues (Ala-139 and Ala-230) were mutated to glycine. Fig. 2A shows that wild-type PTOX has an average KCN-resistant O2 consumption activity of 115 nmol O2·min−1·mg-protein−1; this activity is negligible in an empty vector control (∼3 nmol O2·min−1·mg-protein−1). Nine of the mutants have high KCN-resistant O2 consumption rates that in most cases closely resemble the wild-type protein (A139G, P142A, E171A, N174A, L179A, P216A, A230G, D287A, R293A), but O2 consumption is nearly abolished in five of the mutants (L135A, H151A, Y212A, Y234A, and D295A). We conclude that five of the conserved amino acids are essential for in vitro activity.
FIGURE 2.
Mutagenesis of 14 conserved residues in vitro. A, oxygen consumption rates were determined from the slopes of O2 traces following the addition of 1 mm NADH, 3 mm KCN, and 0.5 mm propyl gallate (PG) (as in Ref. 13). PTOX activity is defined as the oxygen consumption rate in the presence of 3 mm KCN minus the oxygen consumption rate in the presence of 0.5 mm n-PG. Each rate is the average of four independent measurements (±S.D.). B, total membrane proteins were isolated from transformed E. coli, and equal amounts were electrophoresed through 12.5% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. The membranes were treated with an antibody to Arabidopsis PTOX and visualized by the ECL system.
To examine whether the severe decreases in KCN-resistant O2 consumption are due to poor expression and/or instability of the mutant proteins, PTOX abundance was assayed by Western blot analysis of membrane proteins from the various transformants using a polyclonal IM antibody (13, 20). Fig. 2B shows that mutant and wild type PTOX accumulate to similar levels in E. coli; the empty expression vector, as anticipated, lacks PTOX. We also found that PTOX is localized in the membrane (versus soluble) fractions in all the samples, consistent with the idea that the various mutations do not compromise protein incorporation into the E. coli membrane (data not shown). One other possibility for the lack of activity of some of the constructs is that the proteins fail to fold properly. We have previously discussed the evidence that makes this unlikely (13). One is that the three-dimensional structures of DOX proteins appear to be quite stable and resistant to alterations in charge or mobility of side chains. Taken together, the data indicate that the mutant PTOX proteins are expressed and stable at wild-type enzyme levels, and that reductions in O2 consumption are due to reductions in PTOX activity.
Mutagenesis of 14 Conserved Amino Acid Residues in Planta
Mutants that lack PTOX (e.g im) are a powerful tool to gain insight into structure/function relationships of PTOX and AOX (13). This is because of the high degree of structural similarity of their DOX domains. Taking advantage of null alleles of im, we tested the impact of each of the 14 mutations described above on PTOX activity in planta. Wild-type IM cDNAs and IM cDNAs with single mutations in each of the 14 residues were generated and transformed into im. Full-length cDNAs were used to assure proper targeting to the plastid; the constructs were driven by the CaMV 35 S promoter. 12 independent transgenic lines were selected in the T1 generation for each mutant construct (i.e. a total of 12 × 14 = 168 independent transgenic lines were examined at the molecular level). Each of these lines had intact inserts, as determined by PCR and Southern blotting using primers or a probe specific for the Streptomyces hygroscopicus phosphinothricin acetyltransferase (bar) gene. The T1 plants were selfed, and Western blotting was carried out to determine the abundance of PTOX in the 168 T2 generation lines. Mutants with the most similar levels of PTOX compared with wild-type plants (Fig. 3B) were selected for phenotypic comparisons (Fig. 3A).
FIGURE 3.
Mutagenesis of 14 conserved residues in planta. A, same mutations as in the in vitro mutagenesis experiments were introduced into im plants. Kanamycin-resistant seedlings were scored by PCR and Southern blotting for the presence of the kanamycin gene (NPTII). At least eight transformation events per construct were examined; T2 generation seedlings are shown. All plants were grown for 3–4 weeks under continuous light (100 μmol m−2 s−1) after initial low light growth. B, Western blotting analyses were conducted using 5 μg of chlorophyll per lane from chloroplast membranes of partially purified plastids using an antibody to Arabidopsis PTOX. The proteins were visualized by the ECL system.
Fig. 3A shows that im plants transformed with the empty BI121 binary vector resemble non-transformed im (“pBI121”), whereas transformation with wild type IM produces all-green leaves (“WTIM”). The 14 mutants fell into three categories: (i) Transformation of im with nine of the mutant DNAs generated all-green transgenics: A139G, P142A, E171A, N174A, L179A, P216A, A230G, D287A, and R293A. These mutants were able to complement the mutant phenotype. All of these mutants had high in vitro activities, most very similar to wild-type. This suggests that these sites are not important for PTOX activity. (ii) Transformation of im with three of the mutant constructs (L135A, H151A, and Y212A) gave rise to variegated plants, but the extent of variegation was significantly less than found in im, i.e. these plants were primarily green. These mutants had negligible in vitro activities. This suggests that these sites are important for PTOX activity in vitro, but that not much activity is required for normal function in planta. (iii) Y234A and D295A produced transgenic im plants that resembled im, and they had negligible in vitro activities. This suggests that these sites are essential for PTOX activity.
RNAi, Antisense, and Co-suppression Plants
The data in Figs. 2 and 3 were based on samples (E. coli or Arabidopsis) with similar amounts of PTOX protein. We were next interested in determining the phenotypes of plants with different amounts of the wild-type enzyme. For this purpose we carried out a titration experiment in which we modulated PTOX levels using RNAi, antisense, and co-suppression technologies. Fig. 4A shows a representative sample of the transgenics; wild type and a null allele of im are controls. Fig. 4B shows that lines with greater than ∼3% of wild-type PTOX levels have a normal phenotype and growth habit under conditions of 100 μmol m−2 s−1 of continuous illumination. By contrast, the RNAi line (in which PTOX is not detectable) is virescent (delayed greening). All the leaves of the RNAi plants are yellow-green during expansion, but turn all-green after expansion is complete. At flowering the leaves and plants are nearly normal-sized. Interestingly, we failed to identify variegated plants among the various transgenic populations, despite the fact that plants could be found with no detectable PTOX protein accumulation (such as the RNAi line). We also did not observe any differences in the phenotype of these plants grown under higher light conditions (300 μmol m−2 s−1) i.e. the plants did not show a variegated phenotype (data not shown). We conclude that PTOX is normally in excess of the amount required for normal chloroplast development and function, at least under the growth conditions used.
FIGURE 4.
PTOX RNAi, antisense, and co-suppression plants. A, representative RNAi, antisense and co-suppression plants (T2 generation) are shown. The seeds were germinated at the same time and grown as described in Fig. 3A. Kanamycin-resistant seedlings were scored by PCR and Southern blotting for the presence of the kanamycin gene (NPTII). B, Western immunoblot analyses were conducted using 5 μg of chlorophyll per lane of partially-purified chloroplast membranes from wild type leaves and leaves of RNAi, antisense, and co-suppression plants. The filters were probed with an antibody to Arabidopsis PTOX. The numbers are the relative amount of PTOX compared with wild type (100%). ND, not detectable.
DISCUSSION
Mechanism of im Variegation
IM is a single copy gene in Arabidopsis (10), and thus an intriguing question is why some cells of im are green and others white, against a mutant background. We hypothesize that a lack of PTOX (as in immutans) results in over-reduction of the PQ pool under restrictive conditions (e.g. high light); this has been demonstrated (21, 23). This over-reduction prevents further transfer of electrons to the PQ pool and, consequently, an inhibition of the carotenoid pathway at the PDS step. This results in an accumulation of phytoene and a lack of production of downstream, colored (photoprotective) carotenoids. Carotenoids quench triplet chlorophyll and toxic oxygen radicals, and in their absence, photooxidized, white plastids are generated under restrictive light conditions.
Consistent with the variegated phenotype of im, we have hypothesized that PTOX might be particularly important as an electron sink during early chloroplast biogenesis when undifferentiated proplastids in the leaf meristem develop into photosynthetically competent chloroplasts, and components of the photosynthetic apparatus are synthesized and assembled. During this stage, some plastids lack a threshold of photoprotective activities (white plastids), while others, even in the absence of PTOX, have a threshold and develop into functional chloroplasts (10). In accord with this notion, IMMUTANS expression is significantly regulated by the developmental stage of the plant. However, in contrast to early development, studies with im and with Arabidopsis PTOX overexpressor lines show that alterations in IMMUTANS expression and/or protein accumulation have no effect on electron flow during steady state photosynthesis in mature leaves (17). Interestingly, this does not appear to be the case in tomato, where recent studies in the ghost variegation mutant have demonstrated that PTOX activity is necessary in mature leaves, especially under high light conditions (23).
Comparison of AOX and PTOX Sequences
Phylogenetic studies have revealed that genes for AOX and PTOX have a prokaryotic origin and, intriguingly, that they diverged prior to the endosymbiotic events that gave rise to mitochondria and chloroplasts (4, 5, 12, 29, 37, 38). Given the evolutionary distance between AOX and PTOX, it is striking that certain residues are perfectly conserved, or nearly so, among the PTOX and AOX sequences in the databases, with most differences arising in those taxa most distantly related to higher plants - e.g. protists, cyanobacteria, and fungi (4, 5). Among the 20 amino acid residues examined in this report, recent sequence comparisons have revealed that 18 are perfectly conserved among all plant AOX and PTOX sequences, thus underscoring the potential importance of these amino acids in PTOX activity and/or structure.
Classes of Mutants
In the present experiments, we tested the functionality of 14 of the 20 conserved residues using both in vitro and in planta mutagenesis procedures. In an earlier study, we examined the other 6 of these 20 sites (the putative Fe-binding ligands) and found that they are essential for activity in vitro and in planta; they do not tolerate change, even conservative ones. In this report, we found that the 14 residues fall into three classes: (I) essential for activity in vitro and in planta, Tyr-234 and Asp-295; (II) not important for activity in vitro and in planta, Ala-139, Pro-142, Glu-171, Asn-174, Leu-179, Pro-216, Ala-230, Asp-287, and Arg-293; (III) important for activity in vitro but not essential for activity in planta, Leu-135, His-151, and Tyr-212.
Classes I and II are relatively straightforward to explain. Mutant enzymes with negligible activity in vitro are not able to complement im and are thus essential for activity, whereas mutant enzymes that have high levels of activity in vitro (most of which closely resemble wild type) are able to complement im, and are thus not essential for activity. On the other hand, it is more difficult to explain the class III sites. Class III mutant enzymes have defective activities in vitro but are able to complement im, at least in part. Comparisons of activities in the in vitro and in planta experiments were based on E. coli and Arabidopsis with similar amounts of PTOX protein, so the class III mutants cannot be easily explained by a higher abundance of the mutant enzyme in some samples versus the others. Yet, the in planta studies were conducted with mature leaves, and since the variegated phenotype is due to a lack of PTOX activity early in chloroplast biogenesis (i.e. in young expanding leaves), it is possible that ectopic overexpression of a defective enzyme might yield enough activity at this early developmental stage, especially if only low amounts are required to allow the generation of plants that are all green or nearly so. The fact that plants need very little PTOX is indicated by our IM titration experiments.
An alternative hypothesis for a seeming lack of concordance between in planta and in vitro activities is that PTOX activities might be influenced by factors specific to each of the assay systems. For instance, differences in the protein and lipid components of E. coli versus thylakoid membranes could influence activities, as could differences in substrates that are present in the two systems (endogenous quinols and added plastoquinol in the E. coli system versus plastoquinol in thylakoids). Last, there could be other molecules that are synthesized in one system but not the other that influence enzyme activities.
Function of Conserved Active Site Amino Acids
Given the three classes of mutant enzymes, the question arises about the functions of the 20 conserved residues. In the AOX/PTOX structural model of Andersson and Nordlund (26) and Berthold et al. (27), the diiron site resides within the hydrophobic crevice of a four-helix bundle (Fig. 5), and the protein is anchored in the membrane by a fifth, short α-helical domain (i.e. AOX and PTOX are interfacial membrane proteins). It has been further proposed that the membrane-binding region between helix 2 and 3 in AOX functions as a substrate binding site; six amino acid residues (Gln-242, Asn-247, Tyr-253, Ser-256, His-261, Arg-262) in this region were proposed to form part of the ubiquinol-binding site (5). Consistent with this proposal, recent site-directed mutagenesis studies indicate that mutations in four of the amino acids (Gln-242, Tyr-253, Ser-256, and Arg-262) result in significant inhibition of AOX activity (5). However, only two of these sites are conserved in PTOX (Tyr-212 and Ser-256); of these two, only Tyr-212 was examined in this study. Furthermore, inhibitor binding studies revealed that amino acid residues near His-261 and Arg-262, such as Phe-259 and Met-263, are resistant to SHAM suggesting that they might be involved in AOX-quinol interactions (4). These residues are not conserved between AOX and PTOX. In the following paragraphs, the functions of the 14 conserved sites in PTOX will be discussed by each helix in turn.
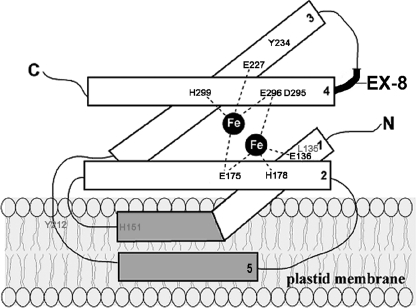
FIGURE 5.
Structural model of PTOX. PTOX is proposed to have four α-helices and a di-iron center. Exon 8, which is present only in PTOX, is indicated in bold. Amino acid residues important for PTOX function in vitro (Leu-135, His-151, and Tyr-212) are shown in gray and those essential for in vitro and in vivo function (Tyr-234, Asp-295, and the six Glu and His residues in the catalytic site) are shown in black.
Helix 1: Leu-135, Glu-136, Ala-139, Pro-142, and His-151
Helix 1 contains one of the six Fe ligands, the perfectly conserved Glu-136 (see Figs. 1B and 5). The importance of this site has been tested experimentally by mutagenesis (13). Immediately flanking Glu-136 is the perfectly conserved Leu-135 and somewhat further away are Ala-139, Pro-142, and His-151.
It has been proposed that Leu-135, Ala-139, and Pro-142 (non-polar, hydrophobic R groups) are important for the structure of the hydrophobic crevice, while the positively charged His-151 might interact with phosphate groups on the membrane (26) or, alternatively, be involved in substrate binding or electron transfer (27). Given the high degree of conservation of these sites, it is surprising that our functional analyses revealed that these sites are either not important for activity (Ala-139 and Pro-142) or are important but not essential for activity (Leu-135 and His-151), at least when these amino acids are converted to Ala or Gly (for Ala-139). We surmise that if Ala-139 and Pro-142 play a role in the structure of the hydrophobic crevice, substitution with Ala or Gly (respectively) might not be dramatic enough to compromise function, since both normally have small, uncharged R groups.
As discussed above, it is possible that the activities of Class III enzymes, such as Leu-135 and His-151, are influenced by differences between the two assay systems, or, alternatively, that only low levels of PTOX activity are necessary for normal growth under the light conditions used. This might account for the observation that alterations of the perfectly conserved Leu-135 and His-151 to Ala (positively charged to uncharged R group), while substantially inhibiting in vitro activity was still able to partially complement im.
Helix 2: Glu-171, Asn-174, Glu-175, His-178, and Leu-179
Helix 2 contains two of the six Fe ligands (Glu-175 and His-178) (see Figs. 1B and 5). The importance of Glu-175 and His-178 has been demonstrated experimentally by mutagenesis in PTOX and AOX (13, 39–42). Immediately flanking the EXXH sequence are the residues Glu-171, Asn-174, and Leu-179. In the three-dimensional structure of Berthold et al. (27) and Andersson and Nordlund (26), Asn-174 is close to the Fe-binding motif in helix 4 (EXXH), and they proposed that Asn-174 interacts with Glu-296 or His-299 via a hydrogen bond to stabilize the four-helix bundle and/or to fine-tune Fe coordination. This notion has been supported by the phylogenetic studies of Finnegan et al. (29). Leu-179 has a hydrophobic R group, and thus its conservation might suggest that it is important in defining the structure of the hydrophobic crevice. The role of Glu-171 is not clear in the model, but the negatively charged group might facilitate electron transfer or stabilization of the pocket structure.
Despite the sequence conservation of Glu-171, Asn-174, and Leu-179, our in vitro and in planta experiments showed that they are not important for activity (class II sites). This might have been because the changes were not dramatic enough, at least in the case of L179A, since Leu and Ala both have hydrophobic R groups. The E171A substitution seems substantial, at least a priori (a large, negatively charged R group for a small, nonpolar one). However, the fact that Glu-171 is substituted with an amino acid containing uncharged side chains (Gln) in Zea mays PTOX suggests that even major substitutions at this position may not be deleterious. Nevertheless, the reason for the conservation of these three residues remains unclear.
Helix 3A: Glu-227 and Tyr-234, the Nakamura E(X)6Y Motif
In their structure/function studies of the Trypanosoma vivax AOX, Nakamura et al. (43) noted that the helix 3 motif, E(X)6Y, is perfectly conserved in all AOX and PTOX proteins (43). This motif extends from Glu-227 to Tyr-234 in the PTOX sequence (Fig. 1B). Glu-227 is one of the Fe-ligands (13), and the Berthold et al. (27) and Andersson and Nordlund (26) models predict that Tyr-234 is near the di-iron site.
Our in vitro and in planta studies confirm the essential nature of Glu-227 and Tyr-234 (Figs. 2 and 3 and Ref. 13). They are also in accord with the studies of Nakamura et al. (43) showing that conversion of Glu-227 and Tyr-234 to Ala abolishes activity of the trypanosome AOX in an E. coli/ΔhemA activity assay. The reason for the importance of Glu-227 is clear: it serves as an Fe-ligand (13, 26, 27). The reason for the essential nature of Tyr-234 is less clear, but it might be because the tyrosine radical plays a critical role in electron transfer, as demonstrated by Albury et al. (42).
Helix 3B: Tyr-212, Pro-216, and Ala-230, Other Conserved Residues in or near Helix 3
In addition to the conserved E(X)6Y motif, Nakamura et al. (43) noted that Tyr-199 of the T. vivax sequence is perfectly conserved (corresponds to Tyr-212, Fig. 1B); Nakamura et al. (43) found that substitution of Tyr-199 with Ala reduced the activity of the trypanosome AOX ∼50%, using the E. coli/ΔhemA activity assay. Recent site-directed mutagenesis studies of Sauromatum guttatum AOX also show that a mutation of the Tyr-253 (Tyr-212 in PTOX) results in significant inhibition of AOX activity (5). Tyr-253 is positioned in a hydrophobic pocket (membrane binding region) of AOX and is hypothesized to be one of the 6 amino acids involved in quinone binding (see Fig. 5). We found that the in vitro activity of the Y212A mutants was nearly abolished. This is consistent with results from the AOX studies and suggests that Tyr-212 may be involved in substrate binding.
Pro-216 and Ala-230 are highly conserved (Fig. 1B) in plant AOX and PTOX sequences but have not previously been examined functionally. We found that Pro-216 and Ala-230 are unimportant for activity when converted to Ala and Gly, respectively. As with Pro-142 (Helix 1), a Pro to Ala change (P216A) might be expected to perturb protein conformation, but if this is important, we could not detect it in our assays. Regarding Ala-230, altering this residue to Gly might not have been a significant enough substitution to observe a change. The reasons for the high degree of Pro-216 and Ala-230 conservation remain to be resolved.
Helix 4: Asp-287, Arg-293, Asp-295, Glu-296, and His-299
Helix 4 contains two of the six iron ligands (Glu-296 and His-299). As with the EXXH sequence on helix 2, the helix 4 EXXH sequence is perfectly conserved except that His-299 is an Ile in wheat PTOX. The functional importance of Glu-296 and His-299 has been demonstrated by mutagenesis both in PTOX and AOX (13, 40, 41, 44), but Asp-287, Arg-293, Asp-295 have not yet been examined. The alterations of these latter three sites are predicted to be substantial, since charged R groups were substituted for Ala. Despite this, we found that these changes do not affect the in vitro and in planta activities of the D287A and R293A mutants, making the reason for the conservation of these amino acids unclear. In contrast, the perfectly conserved Asp-295 next to Glu-296 is one of only two residues we identified in this study that is absolutely essential for activity both in vitro and in planta. The three-dimensional structural model predicts that Asp-295 is close to Glu-175 and His-178 (Fe ligands in helix 2) suggesting that Asp-295 interacts with Glu-175 and/or His-178 via hydrogen bonding to stabilize the active site structure or Fe atom coordination. If so, it is curious that mutations in the analogous Asn-174 in helix 2 (predicted to bind the EXXH motif in helix 4) are not required for activity. Further substitutions might resolve this question.
Summary
AOX contains a catalytic site comprising four Glu and two His residues. In addition, a putative quinone binding site between helix 2 and helix 3 has been proposed (5). Our previous mutagenesis studies showed that all six of the Fe-ligands in the catalytic site (Glu-136, Glu-175, His-178, Glu-227, Glu-296, and His-299) are essential for PTOX activity in vitro and in planta (Ref. 13, Fig. 5). In this report, we determined the functional significance of 14 other amino acid residues that are highly conserved between AOX and PTOX, and we identified five additional amino acid residues (Leu-135, His-151, Tyr-212, Tyr-234, and Asp-295) that are essential for in vitro PTOX activity; two of these (Tyr-234 and Asp-295) are also essential for activity in planta (Fig. 5). These five amino acid residues reside in areas of AOX that have been proposed to be important for catalysis, protein stability, and/or substrate binding. While our studies are consistent with the hypothesis that Tyr-212 is involved in substrate binding in PTOX (as suggested for the analogous Tyr-253 in AOX), it is interesting that only a few of the amino acid residues in the proposed quinone binding site in AOX are conserved in PTOX. This suggests that there might be flexibility in the substrate binding site. Differences between AOX and PTOX may not be surprising, as we have previously shown that minor changes in structure do occur between AOX and PTOX; for example, the presence of exon 8 in PTOX (Fig. 1A and Ref. 13). Furthermore, the substrates for these two proteins are different; AOX binds ubiquinol whereas PTOX acts on plastoquinol. The present studies thus set the stage for further structure/function studies on PTOX.
Footnotes
- AOX
- alternative oxidase
- PTOX
- plastid terminal oxidase
- DOX
- diiron carboxylate quinol oxidase
- IM
- IMMUTANS locus of Arabidopsis
- MOPS
- 3-(N-morpholino) propanesulfonic acid
- PQ
- plastoquinol
- RNAi
- RNA interference.
REFERENCES
- 1.Vanlerberghe G. C., McIntosh L. (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48,703–734 [DOI] [PubMed] [Google Scholar]
- 2.Siedow J. N., Umbach A. L. (2000) Biochim. Biophys. Acta 1459,432–439 [DOI] [PubMed] [Google Scholar]
- 3.Affourtit C., Albury M. S., Crichton P. G., Moore A. L. (2002) FEBS Lett. 510,121–126 [DOI] [PubMed] [Google Scholar]
- 4.McDonald A. (2008) Funct. Plant Biol. 35,535–552 [DOI] [PubMed] [Google Scholar]
- 5.Moore A. L., Albury M. S. (2008) Biochem. Soc. Trans. 36,1022–1026 [DOI] [PubMed] [Google Scholar]
- 6.Purvis A. C. (2001) J. Plant Physiol. 158,159–165 [Google Scholar]
- 7.Purvis A. C., Shewfelt R. L. (1993) Physiol. Plant 88,712–718 [DOI] [PubMed] [Google Scholar]
- 8.Maxwell D. P., Wang Y., McIntosh L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yip J. Y., Vanlerberghe G. C. (2001) Physiol. Plant. 112,327–333 [DOI] [PubMed] [Google Scholar]
- 10.Wu D., Wright D. A., Wetzel C., Voytas D. F., Rodermel S. (1999) Plant Cell 11,43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carol P., Stevenson D., Bisanz C., Breitenbach J., Sandmann G., Mache R., Coupland G., Kuntz M. (1999) Plant Cell 11,57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald A. E., Amirsadeghi S., Vanlerberghe G. C. (2003) Plant Mol. Biol. 53,865–876 [DOI] [PubMed] [Google Scholar]
- 13.Fu A., Park S., Rodermel S. (2005) J. Biol. Chem. 280,42489–42496 [DOI] [PubMed] [Google Scholar]
- 14.Cournac L., Josse E. M., Joët T., Rumeau D., Redding K., Kuntz M., Peltier G. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355,1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josse E. M., Simkin A. J., Gaffé J., Labouré A. M., Kuntz M., Carol P. (2000) Plant Physiol. 123,1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joët T., Genty B., Josse E. M., Kuntz M., Cournac L., Peltier G. (2002) J. Biol. Chem. 277,31623–31630 [DOI] [PubMed] [Google Scholar]
- 17.Rosso D., Ivanov A. G., Fu A., Giesler-Lee J., Hendrickson L., Geisler M., Stewart G., Krol M., Hurry V., Rodermel S. R., Maxwell D. P., Hüner N. P. (2006) Plant Physiol. 142,574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josse E. M., Alcaraz J. P., Labouré A. M., Kuntz M. (2003) Eur. J. Biochem. 270,3787–3794 [DOI] [PubMed] [Google Scholar]
- 19.Peltier G., Cournac L. (2002) Annu. Rev. Plant Biol. 53,523–550 [DOI] [PubMed] [Google Scholar]
- 20.Rizhsky L., Hallak-Herr E., Van Breusegem F., Rachmilevitch S., Barr J. E., Rodermel S., Inzé D., Mittler R. (2002) Plant J. 32,329–342 [DOI] [PubMed] [Google Scholar]
- 21.Baerr J. N., Thomas J. D., Taylor B. G., Rodermel S. R., Gray G. R. (2005) Physiol. Plant. 124,390–402 [Google Scholar]
- 22.Niyogi K. K. (2000) Curr. Opin. Plant Biol. 3,455–460 [DOI] [PubMed] [Google Scholar]
- 23.Shahbazi M., Gilbert M., Labouré A. M., Kuntz M. (2007) Plant Physiol. 145,691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore A. L., Umbach A. L., Siedow J. N. (1995) J. Bioenerg. Biomembr. 27,367–377 [DOI] [PubMed] [Google Scholar]
- 25.Siedow J. N., Umbach A. L., Morre A. L. (1995) FEBS Lett. 362,10–14 [DOI] [PubMed] [Google Scholar]
- 26.Andersson M. E., Nordlund P. (1999) FEBS Lett. 449,17–22 [DOI] [PubMed] [Google Scholar]
- 27.Berthold D. A., Andersson M. E., Nordlund P. (2000) Biochim. Biophys. Acta. 1460,241–254 [DOI] [PubMed] [Google Scholar]
- 28.Berthold D. A., Stenmark P. (2003) Annu. Rev. Plant Biol. 54,497–517 [DOI] [PubMed] [Google Scholar]
- 29.Finnegan P. M., Umbach A. L., Wilce J. A. (2003) FEBS Lett. 555,425–430 [DOI] [PubMed] [Google Scholar]
- 30.Rédei G. P. (1963) Science 139,767–769 [DOI] [PubMed] [Google Scholar]
- 31.Wetzel C. M., Jiang C. Z., Meehan L. J., Voytas D. F., Rodermel S. R. (1994) Plant J. 6,161–175 [DOI] [PubMed] [Google Scholar]
- 32.Aluru M. R., Bae H., Wu D., Rodermel S. R. (2001) Plant Physiol. 127,67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aluru M. R., Yu F., Fu A., Rodermel S. (2006) J. Exp. Bot. 57,1871–1881 [DOI] [PubMed] [Google Scholar]
- 34.Clough S. J., Bent A. F. (1998) Plant J. 16,735–743 [DOI] [PubMed] [Google Scholar]
- 35.Perry S. E., Li H. M., Keegstra K. (1991) Methods Cell Biol. 34,327–344 [DOI] [PubMed] [Google Scholar]
- 36.Umbach A. L., Siedow J. N. (2000) Arch. Biochem. Biophys. 378,234–245 [DOI] [PubMed] [Google Scholar]
- 37.Stenmark P., Nordlund P. (2003) FEBS Lett. 552,189–192 [DOI] [PubMed] [Google Scholar]
- 38.Atteia A., van Lis R., van Hellemond J. J., Tielens A. G., Martin W., Henze K. (2004) Gene 330,143–148 [DOI] [PubMed] [Google Scholar]
- 39.Chaudhuri M., Ajayi W., Hill G. C. (1998) Mol. Biochem. Parasitol. 95,53–68 [DOI] [PubMed] [Google Scholar]
- 40.Berthold D. A., Voevodskaya N., Stenmark P., Gräslund A., Nordlund P. (2002) J. Biol. Chem. 277,43608–43614 [DOI] [PubMed] [Google Scholar]
- 41.Ajayi W. U., Chaudhuri M., Hill G. C. (2002) J. Biol. Chem. 277,8187–8193 [DOI] [PubMed] [Google Scholar]
- 42.Albury M. S., Affourtit C., Crichton P. G., Moore A. L. (2002) J. Biol. Chem. 277,1190–1194 [DOI] [PubMed] [Google Scholar]
- 43.Nakamura K., Sakamoto K., Kido Y., Fujimoto Y., Suzuki T., Suzuki M., Yabu Y., Ohta N., Tsuda A., Onuma M., Kita K. (2005) Biochem. Biophys. Res. Commun. 334,593–600 [DOI] [PubMed] [Google Scholar]
- 44.Albury M. S., Affourtit C., Moore A. L. (1998) J. Biol. Chem. 273,30301–30305 [DOI] [PubMed] [Google Scholar]