Abstract
Superoxide dismutase 2 (SOD2) is one of the rare mitochondrial enzymes evolved to use manganese as a cofactor over the more abundant element iron. Although mitochondrial iron does not normally bind SOD2, iron will misincorporate into Saccharomyces cerevisiae Sod2p when cells are starved for manganese or when mitochondrial iron homeostasis is disrupted by mutations in yeast grx5, ssq1, and mtm1. We report here that such changes in mitochondrial manganese and iron similarly affect cofactor selection in a heterologously expressed Escherichia coli Mn-SOD, but not a highly homologous Fe-SOD. By x-ray absorption near edge structure and extended x-ray absorption fine structure analyses of isolated mitochondria, we find that misincorporation of iron into yeast Sod2p does not correlate with significant changes in the average oxidation state or coordination chemistry of bulk mitochondrial iron. Instead, small changes in mitochondrial iron are likely to promote iron-SOD2 interactions. Iron binds Sod2p in yeast mutants blocking late stages of iron-sulfur cluster biogenesis (grx5, ssq1, and atm1), but not in mutants defective in the upstream Isu proteins that serve as scaffolds for iron-sulfur biosynthesis. In fact, we observed a requirement for the Isu proteins in iron inactivation of yeast Sod2p. Sod2p activity was restored in mtm1 and grx5 mutants by depleting cells of Isu proteins or using a dominant negative Isu1p predicted to stabilize iron binding to Isu1p. In all cases where disruptions in iron homeostasis inactivated Sod2p, we observed an increase in mitochondrial Isu proteins. These studies indicate that the Isu proteins and the iron-sulfur pathway can donate iron to Sod2p.
Metal-containing enzymes are generally quite specific for their cognate cofactor. Misincorporation of the wrong metal ion can be deleterious and tends to be a rare occurrence in biology. A prime example of metal ion selectivity is illustrated by the family of manganese- and iron-containing superoxide dismutases (SODs)3. This large family of enzymes utilizes either manganese or iron as cofactors to scavenge superoxide anion. The iron- and manganese-containing forms are highly homologous to one another at primary, secondary, and tertiary levels and have virtually identical metal binding and catalytic sites (1–3). Despite this extensive homology, Mn- and Fe-SODs are only active with their cognate metal. Misincorporation of iron into Mn-SOD or vice versa alters the redox potential of the enzyme's active site and prohibits superoxide disproportionation (4, 5). Nevertheless, misincorporation of iron into Mn-SOD does occur in vivo (6, 7). The isolated Mn-SOD from Escherichia coli is found as a mixture of manganese- and iron-bound forms (7); binding of manganese is favored under oxidative stress, whereas iron binding is increased under anaerobic conditions (3, 8). It has been proposed that changes in bioavailability of manganese versus iron determine the metal selectivity of Mn-SOD in bacterial cells (3, 8). But is this also true for Fe-SOD? Currently, there is no documentation of manganese misincorporation into Fe-SOD in vivo.
Unlike bacteria that co-express Mn- and Fe-SOD molecules in the same cell, eukaryotic mitochondria generally harbor only one member of the Fe/Mn-SOD family, a tetrameric Mn-SOD typically known as SOD2 (9). In some organisms, SOD2 is essential for survival (10–12), and mitochondria have therefore evolved to prevent iron-SOD2 interactions despite high levels of mitochondrial iron relative to manganese. Using a yeast model system, we have shown previously that metal ion mis-incorporation can occur with Saccharomyces cerevisiae Sod2p (7). Specifically, iron binds and inactivates yeast Sod2p when cells are either starved for manganese or have certain disruptions in mitochondrial iron homeostasis. These disruptions include mutations in MTM1, a mitochondrial carrier protein that functions in iron metabolism (7, 13), and mutations in GRX5 or SSQ1, involved in iron-sulfur biogenesis (14). We proposed that these disruptions lead to expansion of a mitochondrial pool of so-called SOD2-reactive iron (7). Currently, it is unknown whether SOD2-reactive iron represents a major shift in the chemistry of bulk mitochondrial iron or whether it is just a small pool of the metal emerging from one or more specific sites.
The grx5 and ssq1 mutants that promote iron-SOD2 interactions encode just two of many components of a complex pathway for iron-sulfur biogenesis (15, 16). One of the key components is a well conserved iron-sulfur scaffold protein originally described for bacteria as IscU, also known as mammalian ISCU and S. cerevisiae Isu1p and Isu2p, referred collectively herein as “Isu proteins” (17–22). The iron-sulfur clusters on Isu proteins are labile and can be transferred to target iron-sulfur proteins through the aid of mitochondrial factors including Grx5p and Ssq1p (15, 16). It is not clear whether disruption of the iron-sulfur pathway per se is sufficient to promote iron interactions with yeast Sod2p or whether this effect is specific to grx5, ssq1, and mtm1 mutants.
In the current study, we explore the nature of mitochondrial iron that can interact with Sod2p. We find that the changes in mitochondrial metal homeostasis that shift metal binding in yeast Sod2p likewise alter metal cofactor selection in a heterologously expressed Mn-SOD, but not in a Fe-SOD molecule. Through x-ray absorption near edge structure (XANES) and extended x-ray absorption fine structure (EXAFS) analyses of mitochondrial iron, we detected no major change in bulk mitochondrial iron under conditions that promote iron-SOD2 interactions. SOD2-reactive iron appears to represent a small pool of the metal, and we provide evidence that the iron-sulfur scaffold Isu1p can act as an important source of this reactive iron.
EXPERIMENTAL PROCEDURES
Yeast Strains and Culture Conditions
The strains employed in this study are isogenic to either BY4741 (MATa, leu2Δ0, met15Δ0, ura3Δ0, and his3Δ1) or BY4742 (MATα, leu2Δ0, lys2Δ0, ura3Δ0, and his3Δ1). Strains include the previously reported MY019 (mtm1Δ::LEU2) (7), MY020 (mtm1Δ::LEU2 sod2Δ::kanMX4) (7), and LJ109 (rho−) (23). Single gene deletions strains in the BY4741 background 5278 (ssq1Δ::kanMX4), 2769 (grx5Δ::kanMX4), 1878 (smf2Δ::kanMX4), and 12482 (BY4742 isu2Δ::kanMX4) were obtained from Research Genetics, Inc. Strain AN008 (yfh1Δ::LEU2) is a derivative of the yfh1Δ::URA3 strain MY036 (7), where URA3 was replaced with LEU2. Strain LJ206 (atm1Δ::URA3) was created with the atm1Δ disruption plasmid pLJ170 (described below). The pGAL-ISU1 knock down strain LJ403 (isu1Δ::kanMX4, isu2Δ::HIS3, pGAL-ISU1) was created by deleting isu2 with pLJ318 (described below) in the isu1Δ::kanMX4 strain expressing ISU1 under GAL1 (plasmid pLJ386; described below). The pMET3-ISU1 knockdown strain LJ436 (BY4742 isu2Δ::kanMX4, pMET3-ISU1::HIS3) was created by integrating the MET3 promoter just prior to the ISU1 coding sequence using plasmid pLJ472 (described below) in a strain also harboring a isu2Δ deletion. MTM1 and GRX5 were deleted in BY4742 and LJ436 using plasmids pVC257 (13) and pLJ190 (described below) respectively generating strains LJ443 (mtm1Δ::LEU2), LJ444 (mtm1Δ::LEU2 isu2Δ::kanMX4, pMET3-ISU1::HIS3), LJ445 (grx5Δ::URA3), and LJ446 (grx5Δ::URA3, isu2Δ::kanMX4, pMET3-ISU1::HIS3).
Yeast cells were grown either in enriched yeast extract, peptone-based medium supplemented with 2% glucose (YPD), 2% galactose (YPGal), 3% glycerol (YPG), or in synthetic defined medium. To minimize oxidative damage in iron accumulating yeast strains, stocks of cells were maintained in oxygen-depleted culture jars (GasPak, Becton Dickinson), and liquid cultures were grown without aeration.
Plasmids
The S. cerevisiae SOD2 expressing vector pAN001 was created by mobilizing SOD2 sequences −552 to +889 from pEL111 (13) through SalI and BamHI digestion and by ligation into the CEN URA3 vector pRS316 (24). pAN002 expresses E. coli sodA fused to the SOD2 mitochondrial targeting signal and gene promoter. For this, sodA sequences +1 to +625 were PCR-amplified with NdeI and BglII sites and ligated to a derivative of pEL111 with a NdeI site engineered at +78 in yeast SOD2 in which SOD2 sequences +78 to +700 were removed by NdeI/BglII digestion. The pKL-SOD plasmid for expressing FLAG-tagged E. coli Fe-SOD was a generous gift from Dr. Jerry Kaplan, University of Utah (25, 26). To create the atm1Δ::URA3 plasmid pLJ170 for deleting ATM1 sequences +60 to +1,541, ATM1 fragments −493 to +60 and +1,541 to +2,566 mobilized through EcoRI/BglII and EcoRI/KpnI digestion were inserted into KpnI/BamHI-digested pRS306 (URA3). The grx5Δ::URA3 plasmid pLJ190 was generated by PCR-amplifying upstream (−703 to −232) and downstream sequences (+416 to +855) of GRX5 introducing EcoRI and SpeI (upstream) or KpnI and EcoRI (downstream) restriction sites. Following digestion the PCR products were ligated into KpnI/SpeI digested pRS306 (URA3). To generate pLJ129 expressing ISU1 under PGK1, the coding sequence for ISU1 was PCR-amplified with BamHI and SalI sites and inserted into these sites of pLJ150 (URA3 2 μm containing the PGK1 promoter and CYC1 terminator). Plasmid pLJ357 is a derivative of pLJ129 expressing D71A ISU1 generated by QuikChange mutagenesis (Stratagene). To create the GAL1-driven ISU1 vector pLJ386, the BamHI-SalI ISU1-containing fragment of pLJ129 was inserted into these same sites of pYC2/CT harboring the GAL1 promoter (URA3 CEN) (Invitrogen). The isu2Δ::HIS3 disruption plasmid pLJ388 was created by inserting the isu2 disruption cassette from pSG101 (18) into the BamHI-SalI sites of pRS403. The pMET3-ISU1 promoter replacement plasmid was generated by PCR-amplifying ISU1 upstream (−929 to −343) and downstream (+1 to +687) sequences introducing NotI and SacI (upstream) or SphI and NotI (downstream) restriction sites and MET3 sequences (−425 to −3) introducing BamHI and SphI sites. Following digestion the MET3 promoter and ISU1 downstream sequences were ligated into pRS313 (HIS3) digested with BamHI/NotI producing plasmid pLJ469. The pMET3-ISU1 (downstream) fusion was then excised with BamHI/NotI and ligated together with the ISU1 upstream sequences digested with NotI/SacI into pRS403 (HIS3) digested with BamHI/SacI. The resulting plasmid pLJ472 was linearized with NotI and used to replace the ISU1 promoter with the methionine-repressible MET3 promoter.
Analysis of SOD Activity and Immunoblots
Whole cell lysates of S. cerevisiae strains were prepared by glass bead homogenization (27) from cells that were grown in liquid cultures of either YPD or synthetic defined medium to late log. For glucose repression of GAL-ISU1 and methionine repression of pMET3-ISU1, cells were maintained on solid YPD for at least 48 h prior to 14–16 h in liquid YPD cultures. The lysates were analyzed for SOD activity by native gel electrophoresis and nitroblue tetrazolium staining (13) and for immunoblot analysis by denaturing gel electrophoresis using antibodies directed against SOD2 (27) or E. coli IscU that recognizes yeast Isu1p and Isu2p (18). Quantitation of SOD activity bands utilized ImageQuant TL (version 2005) software (GE Healthcare). Immunoblots were visualized by ECL detection (Amersham Pharmacia Biotech) or by the Odyssey infrared imaging system (Licor Biosciences). Immunoblots were quantitated by either ImageQuant TL software (version 2005) for ECL or Odyssey software (version 1.2).
Miscellaneous Biochemical Analyses
For measurements of total mitochondrial iron, crude mitochondria were prepared as described previously (23) from 100-ml YPD cultures, and iron measurements were carried out on a PerkinElmer Life Sciences AAnalyst 600 graphite furnace atomic absorption spectrometer according to published methods (23). For XANES and EXAFS analysis, cells were cultured for 16 h in YPD to an A600 4.0–4.5 and mitochondria were isolated from duplicate 2.0 liter cultures as above in buffers containing 1 mm dithiothreitol. The yield of protein was typically 10–20 μg/μl. 40 μl of each mitochondrial sample was transferred to Lucite cuvettes for x-ray absorption spectra data collection as described in the supplemental “Experimental Procedures”.
RESULTS
Mitochondrial Iron Homeostasis Effects on Heterologous SODs
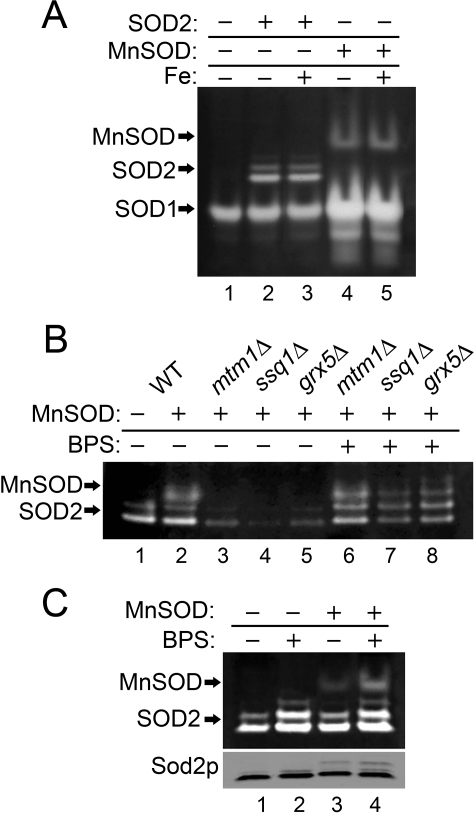
We have shown previously that cofactor selectivity in mitochondrial Sod2p is greatly influenced by mitochondrial manganese and iron homeostasis (7). We tested whether such effects of the mitochondrial environment are limited to eukaryotic SOD2 or can be extended to other members of the Fe/Mn-SOD family. In the experiment of Fig. 1, E. coli Mn-SOD was expressed in yeast mitochondria using the S. cerevisiae SOD2 gene promoter and mitochondrial targeting signal. E. coli Mn-SOD shows activity in wild type (WT) yeast mitochondria that was not inhibited by increasing mitochondrial iron through iron supplements (Fig. 1A, lanes 4 and 5). Thus, as with yeast Sod2p, E. coli Mn-SOD seems largely nonreactive to high mitochondrial iron from WT cells. Yet, iron does react with E. coli Mn-SOD in the mtm1, ssq1, and grx5 mutants that also inactivate yeast Sod2p. As seen in Fig. 1B, E. coli Mn-SOD activity is lost in mtm1, ssq1, and grx5 mutants (lanes 3–5), and activity is restored by depleting iron through chelation with BPS (bathophenanthrolinedisulfonate) (lanes 6–8). BPS treatments also somewhat increased activity of E. coli Mn-SOD and yeast Sod2p expressed in WT cells (Fig. 1C, lanes 2 and 4), consistent with a low level of iron reactivity with these SODs even in WT cells (7). The mitochondrial environment has the same impact on cofactor selection in a heterologous Mn-SOD from bacteria as is seen with the endogenous yeast Sod2p.
FIGURE 1.
Effects of mitochondrial iron homeostasis on a heterologous E. coli Mn-SOD. The designated strains were grown in YPD medium supplemented where indicated with 80 μm BPS or 2 mm ammonium iron(III) citrate (Fe:+). Whole-cell lysates were analyzed for SOD activity by native gel electrophoresis and nitroblue tetrazolium staining. Mn-SOD, SOD1, and SOD2 indicate positions of active Mn-SOD from E. coli and Cu/Zn-Sod1p and manganese Sod2p from S. cerevisiae, respectively. Sod2p polypeptide from the whole-cell lysates was analyzed by immunoblot (C, bottom panel). Strains utilized: the sod2Δ strain transformed where indicated with the yeast SOD2 plasmid pAN001 (A) or the E. coli Mn-SOD expression plasmid, pAN002 (B and C). The BY4741 WT (B, lanes 1 and 2, and C) or indicated mutant derivatives (B) transformed where designated (+) with plasmid pAN002 for expressing E. coli Mn-SOD.
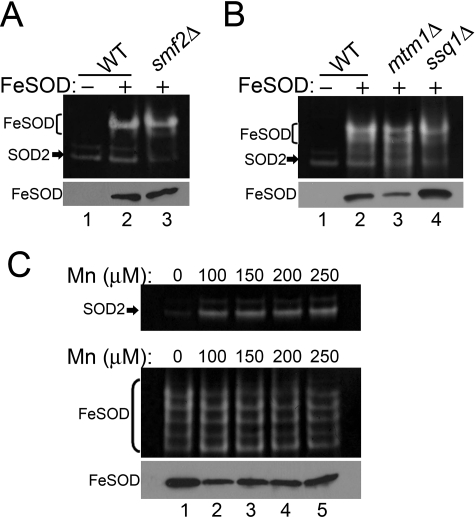
We tested whether the mitochondrial environment can likewise influence cofactor selection in an Fe-SOD. When expressed in WT yeast mitochondria, Fe-SOD from E. coli is active, consistent with iron binding (Fig. 2A, lane 2). To determine whether a fraction of the enzyme is inactivated by manganese binding, Fe-SOD was expressed in the mitochondria of smf2Δ mutants that accumulate very low manganese (7, 27). Fe-SOD activity was not significantly elevated in this manganese-starved cell (Fig. 2A, lane 3), indicating that mitochondrial manganese is not binding the enzyme. Fe-SOD activity was also largely unchanged by the same mtm1 and ssq1 mutants that favor iron binding to Mn-SOD over manganese (Fig. 2B, lane 3 and 4). Even when yeast cells are overloaded with manganese, Fe-SOD retains its activity. As seen in Fig. 2C, the same manganese treatments that promote manganese binding and manganese activation of yeast Sod2p (top) had no effect on Fe-SOD activity (bottom). Fe-SOD is largely refractory to the changes in metal homeostasis that affect cofactor selection in Mn-SOD molecules. Although mitochondrial iron is normally quite bioavailable for Fe-SOD, this is not the case for Mn-SOD. Only a specialized pool of mitochondrial iron, such as that expanded in mtm1, grx5, and ssq1 mutants, can gain access to manganese SOD molecules.
FIGURE 2.
Enzymatically active Fe-SOD expressed in yeast mitochondria. Strains that were transformed where indicated (Fe-SOD:+, A and B, and C, bottom panel) with the plasmid for expressing a FLAG-tagged version of E. coli Fe-SOD were analyzed for SOD activity (top) and for levels of Fe-SOD by immunoblotting with anti-FLAG (bottom). FLAG-tagged Fe-SOD typically runs as multiple bands on native gels; the major bands are shown in brackets. S. cerevisiae SOD2 activity is shown by arrows. Strains utilized: WT, BY4741; smf2Δ, 1878 (A); WT, BY4741; mtm1Δ, MY019; ssq1Δ, 5278 (B); mtm1Δ, MY019 (top panel); sod2Δ mtm1Δ, MY020 (C, bottom panel) transformed with the E. coli Fe-SOD expression plasmid.
Bulk Mitochondrial Iron Is Not Affected in Yeast Mutants Where Iron Binds Sod2p
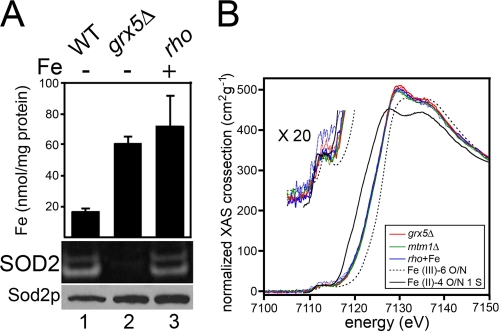
The mitochondrial iron that accumulates in grx5, ssq1, and mtm1 mutants would appear to be of a unique nature in that it readily reacts with Mn-SOD molecules. To address whether these mutants are associated with a bulk change in the average oxidative state and/or coordination environment of mitochondrial iron, we employed XANES and EXAFS. Such spectroscopy has previously been used to analyze mitochondrial iron of mammalian cells defective for frataxin (28), which, in our analogous yeast yfh1 mutants, does not react with SOD2. Because our interests lie in “SOD2-reactive iron,” we analyzed the mitochondrial iron of yeast grx5Δ and mtm1Δ mutants, as well as a rho mutant control (grx5 and mtm1 cells are rho−; (29)). When treated with iron, the rho strain accumulates high mitochondrial iron that does not react with or inactivate Sod2p, whereas the isogenic grx5Δ mutant does exhibit iron activation of Sod2p (Fig. 3A). EXAFS spectra were measured from duplicate samples as described previously (30) to k = 12 Å−1 (supplemental Tables S1 and S2 and Fig. S1). Spectra were fit using Feff 7.02 parameters over the k range 2–9 Å−1. As seen in Fig. 3B, the normalized (31) XANES spectra for the grx5Δ and mtm1Δ mutants and the rho control are identical, indicating that there is no detectable change in either the average iron oxidation state or geometry. The edge energy is consistent with an approximately equal mixture of Fe(II) and Fe(III) and the weak 1s-3d intensity indicates that the bulk of the iron has ∼octahedral symmetry. Although the absolute Fe(II):Fe(III) ratio cannot be discerned because the apparent ratio depends on the reference spectra used for Fe(II) and Fe(III), it is nevertheless clear that there is no significant change in oxidation state in mitochondrial iron from grx5 and mtm1Δ versus the rho control. A conversion of >5% of the iron from Fe(II) to Fe(III) or vice versa would be readily detectable. The EXAFS spectra (supplemental Fig. S1) were fit using a single iron-oxygen/nitrogen shell. Again, there was no significant change in spectra of grx5, mtm1, and the rho control; in all three cases, the apparent iron-oxygen distance was 1.97–2.00 Å, consistent with the expected RFe-O for 6-coordinate Fe(III). Overall, iron inactivation of Sod2p does not correlate with major changes in total mitochondrial iron. The pool of mitochondrial iron that inactivates SOD2 may represent a small fraction.
FIGURE 3.
Bulk mitochondrial iron is unchanged in cells accumulating SOD2-reactive iron. A, the designated strains grown where indicated in the presence of 2 mm ammonium iron(III) citrate were subjected to analysis of mitochondrial iron by atomic absorption spectrometry (values represent averages of four readings from two independent cultures, error bars indicate S.D.) (top) and to yeast Sod2 activity analysis (middle) and Sod2p protein levels (bottom) as in Fig. 1. B, normalized XANES spectra of mitochondria samples (grx5Δ, mtm1Δ, and rho supplemented with 2 mm ammonium iron(III) citrate; rho+Fe). Duplicate sample sets from independent cultures are color-coded per the inset legend, with each duplicate represented by a solid and dotted line. The expansion showing the 1s → 3d transition is enlarged 20-fold vertically. Strains utilized: WT, BY4741; grx5Δ, 2769; mtm1Δ, MY019; rho−, LJ109.
Effects of Blocking Specific Steps in Iron-Sulfur Biogenesis on Sod2p
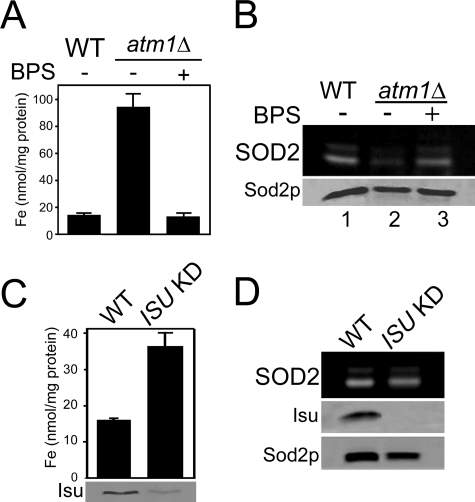
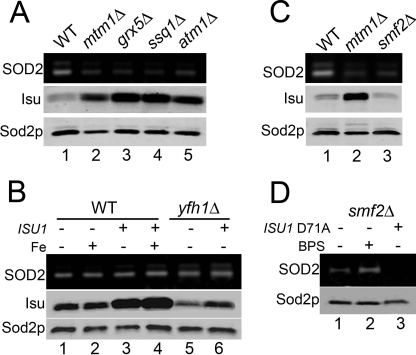
Clues to understanding the origin of SOD2-reactive iron may come from an analysis of the iron-sulfur pathway. The yeast grx5 and ssq1 mutations that promote iron inactivation of Sod2p block both mitochondrial and cytosolic iron-sulfur biogenesis (16). A factor needed for cytosolic, but not mitochondrial, iron-sulfur clusters is yeast ATM1, the homologue to mammalian ABCB7 (32, 33). Previously, knockdown of human ABCB7 was associated with a lowering of SOD2 activity (34), and we now provide evidence that this reflects iron inactivation of the enzyme. As seen in Fig. 4A, deletion of S. cerevisiae ATM1 results in a dramatic rise in mitochondrial iron. This iron is reacting with Sod2p as there is a pronounced loss in Sod2p activity (Fig. 4B, lane 2) that was rescued by iron chelation (lane 3).
FIGURE 4.
Effects of disrupting iron-sulfur biogenesis on Sod2p activity. A and B, the designated strains were grown in YPD medium containing where indicated (BPS: +) 80 μm BPS. A, mitochondrial iron levels were measured as in 3A. B, Sod2p activity and protein levels were monitored as in Fig. 1. C and D, cells that were originally maintained on galactose-containing medium were shifted to glucose for 48 h to deplete Isu1p in the ISU knockdown (KD) cultures. C, top, mitochondrial iron levels monitored as in 3A. C, bottom, and D, whole-cell lysates were analyzed where indicated for Isu (combination of yeast Isu1p and Isu2p) protein levels by immunoblot and for Sod2p activity and protein as in Fig. 1. Strains utilized: WT, BY4741; atm1Δ, LJ206 (A and B); WT, BY4741; ISU KD, LJ403 (C and D).
Another component of the iron-sulfur biogenesis pathway is the yeast Isu proteins (also known as IscU in bacteria and ISCU in mammals) onto which labile iron-sulfur scaffolds are built (17–22, 35). A combined deletion in S. cerevisiae ISU1 and ISU2 is lethal (18, 35); hence we expressed ISU1 under the repressible S. cerevisiae GAL1 promoter in a isu1Δ isu2Δ background. Repression of ISU1 resulted in an increase in mitochondrial iron (Fig. 4C) as previously shown with isu deficiency and with loss of mtm1, grx5, and ssq1 (13, 17, 18, 36–38). However, in the case of isu loss, the elevated iron did not impair Sod2p activity; there was no evidence of iron interactions with the enzyme (Fig. 4D, lane 2).
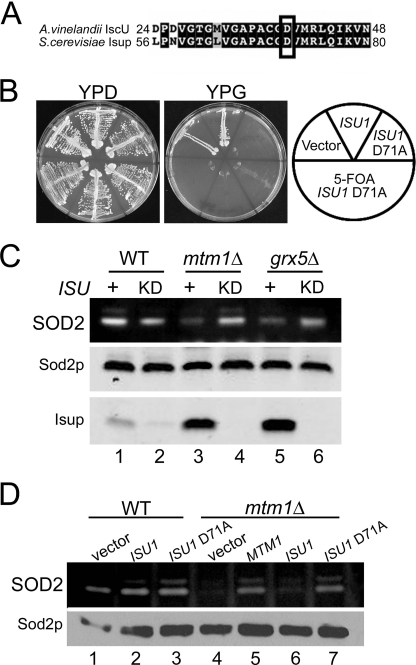
As a second approach to yeast Isu protein inactivation, we sought to obtain a dominant negative Isu1p. Recently, Dean and colleagues (39, 40) created a dominant negative (D39A) IscU from Azotobacter vinelandii that can bind iron and assemble an iron-sulfur cluster but cannot transfer this cluster to recipient iron-sulfur proteins. Iron binding is very stable compared with the labile iron binding of wild type IscU. We created the analogous D71A in S. cerevisiae Isu1p (Fig. 5A). Expression of D71A Isu1p in WT cells rapidly caused the cells to become petite or respiratory deficient, and this defect was not reversed upon subsequent loss of the D71A ISU1 expression plasmid (Fig. 5B), consistent with permanent mitochondrial DNA damage (rho mutations). Rapid loss of mitochondrial DNA is typically seen in yeast cells defective in iron-sulfur biogenesis (17, 23, 36, 41–43). Despite this dominant negative effect of D71A Isu1p on WT mitochondria, there was no loss in Sod2p activity (Fig. 5D, lane 3). Together with the ISU1 down-regulation study of Fig. 4D, these studies show that the iron accumulating in Isu deficient cells does not react with Sod2p. It is not a loss of iron-sulfur biogenesis per se that creates SOD2-reactive iron but rather specific blockages in the pathway (e.g. grx5, ssq1, and atm1 mutants).
FIGURE 5.
Blocking Isu activity in yeast. A, alignment of an N-terminal segment of A. vinelandii IscU versus S. cerevisiae Isu1p. Identical and similar residues are in black and gray; boxed is a conserved aspartate targeted for mutagenesis. B, WT cells transformed with vectors for overproducing either WT Isu1p or D71A Isu1p or with empty vector were tested for growth on fermentable (YPD) and nonfermentable (YPG) carbon sources, the latter being a marker of mitochondrial respiration. Where indicated (+5-fluoroorotic acid), three independent transformants for D71A ISU1 were induced to shed the D71A ISU1 plasmid by growth on 5-fluoroorotic acid prior to growth tests. C and D, whole-cell lysates were analyzed for SOD activity (top) and Sod2p protein levels (bottom) as in Fig. 1. C, cells that were originally maintained on methionine-free medium were shifted to enriched methionine-containing YPD medium for 48 h to deplete Isu1p in the ISU KD cultures. ISU +, cells harboring wild type ISU1 and ISU2 alleles. Isup, combined levels of Isu1p and Isu2p detected by immunoblot using an anti-E. coli IscU antibody. D, WT and mtm1Δ cells were transformed with the indicated plasmids for overexpression of wild type or D71A ISU1 or for MTM1 and were grown in selecting synthetic defined medium. Strains utilized: WT, BY4741 and mtm1Δ, MY019 (A, B, and D); WT, BY4742; ISU KD, LJ436 (C); mtm1Δ, LJ443; mtm1Δ ISU KD, LJ444; grx5Δ, LJ445; grx5Δ ISU KD, LJ446.
Isu as a Potential Source of SOD2-reactive Iron
We reported previously that in mtm1Δ mutants, iron inactivation of Sod2p can be reversed by a deletion in YFH1 encoding iron binding frataxin (7). Because Yfh1p can donate iron to Isu molecules (44), we tested whether loss of the yeast Isu proteins can similarly prevent iron-Sod2p interactions. First, the Isu proteins were depleted by placing the chromosomal ISU1 gene under control of the methionine-repressible MET3 protein in an isu2Δ strain. As seen in Fig. 5C, growth of this strain in enriched (methionine-containing) medium resulted in a major loss of total Isu proteins (lanes 2, 4, and 6; bottom). In the background of a mtm1Δ or grx5Δ strain, such depletion of Isu resulted in restoration of Sod2p activity (Fig. 5C, lanes 4 and 6; see also supplemental Fig. S2). Hence, total loss of Isu proteins can disrupt the trafficking of iron toward Sod2p. To address this further, we exploited the dominant negative D71A Isu1p described above that is expected to bind, but not transfer iron (39, 40). As seen in Fig. 5D, overexpression of D71A ISU1, but not WT ISU1, reversed loss of Sod2p activity in mtm1 mutants (lane 7). The Isu polypeptides are present in this case but are presumably unable to release iron, indicating that iron derived from Isu1p can react with Sod2p.
ISU1 is known to be up-regulated in ssq1Δ (45, 46) and grx5Δ cells (47), and we now report that mtm1Δ strains show a similar increase in Isu protein levels, as do atm1Δ mutants that likewise exhibit iron-inactivation of Sod2p (Fig. 6A and supplemental Fig. S2). In contrast, Isu protein levels do not increase in our yfh1Δ strain that hyperaccumulates a form of iron that does not react with Sod2p (Fig. 6B, lane 5). There appears to be a close correlation between Isu levels and the degree of Sod2p inactivation (supplemental Fig. S2). Nevertheless, elevating Isu protein levels alone is not sufficient to promote iron binding to Sod2p. Overexpression of ISU1 in WT or yfh1Δ cells did not cause Sod2p inactivation, even when cells were treated with iron supplements (Fig. 6B, lanes 3, 4, and 6). Specific disruptions in iron homeostasis are required.
FIGURE 6.
Isu1p levels and iron inactivation of Sod2p. Sod2p activity and protein levels and Isu protein levels were monitored as in Fig. 5. A and C, indicated strains were grown in YPD medium. B and D, strains were grown in selecting synthetic defined medium supplemented where indicated with 2 mm ammonium iron(III) citrate (Fe:+) or 80 μm BPS. As designated (ISU1: + or ISU1 D71A: +), cells were transformed with the pLJ129 or pLJ357 vectors for overexpressing WT or D71A Isu1p. Strains utilized: WT, BY4741; mtm1Δ, MY019; grx5Δ, 2769; ssq1Δ, 5278; atm1Δ, LJ206; yfh1Δ, AN015; smf2Δ, 1878.
We have shown previously that iron can gain access to Sod2p without disruptions in mitochondrial iron if the cells are starved for manganese (7). Yeast smf2Δ mutants accumulate very low manganese but normal iron (7). These cells show a loss in Sod2p activity similar to effects seen in mtm1 mutants (Fig. 6C). However, in this case, Sod2p inactivation is not associated with increased Isu protein levels (Fig. 6C, lane 3). Moreover, Sod2p activity in smf2Δ mutants was not rescued by the dominant negative D71A Isu1p (Fig. 6D, lane 2) even though iron chelation treatments did improve activity (lane 3). Therefore Isu1p is not responsible for the inactivation of Sod2p under conditions of manganese starvation.
DISCUSSION
In eukaryotic mitochondria, iron atoms outnumber manganese by nearly two orders of magnitude (7). Iron is also more widely used as cofactor, including roughly 15 heme and iron-sulfur containing proteins (48) compared with only one or two manganese-containing enzymes. Eukaryotic Sod2p is one of those rare mitochondrial enzymes that has chosen manganese over iron. Yet even so, slippage of the wrong metal can occur, and we have shown that the enzyme switches from the active manganese bound state to the inactive iron-state with specific disruptions in mitochondrial iron homeostasis. The switch in cofactor selection is not caused by a global change in the chemical environment of mitochondrial iron. Rather, a fraction of mitochondrial iron becomes highly reactive with Sod2p, and we have identified the iron-sulfur scaffold protein Isu as one potential source of this reactive iron.
The effects of the mitochondrial environment on Sod2p cofactor selection were reproduced with a heterologous bacterial Mn-SOD expressed in mitochondria. In fact, the impact of the mitochondria on metal selectivity of Mn-SOD was similar to what has previously been reported for this bacterial enzyme expressed in its native host (6, 8). We therefore propose that manganese SOD molecules from bacteria to eukaryotes have evolved to capture manganese based on differential bioavailability of manganese versus iron rather than a specific manganese-metallochaperone.
Although manganese SOD molecules are highly susceptible to changes in metal bioavailability, the same is not true for the highly homologous Fe-SOD from E. coli. When expressed in mitochondria, Fe-SOD exists in the active iron-bound state and is refractory to the changes in manganese and iron homeostasis that dictate metal specificity in Mn-SOD. The affinity constants of Fe-SOD for manganese or iron have not been reported; however, it is possible that the affinity for iron over manganese is sufficiently high to preclude mis-incorporation of the less abundant manganese. Our findings with Fe-SOD have also shed light into the bulk mitochondrial iron that does not typically react with Mn-SOD. What we have denoted previously as the “SOD2-inert iron” (7) may in fact be quite bioavailable but just in a form that cannot compete with manganese for binding to Mn-SOD molecules.
We have identified the iron-sulfur scaffold protein Isu as a potential source for SOD2-reactive iron. Depletion of the yeast Isu polypeptides or expression of a dominant negative D71A Isu1p reversed iron inactivation of Sod2p in mtm1Δ and grx5Δ cells. Based on studies with the highly homologous bacterial IscU proteins, iron binding to wild type Isu proteins is quite labile, facilitating ready transfer of iron to iron-sulfur proteins (21). But in cases where such iron transfer cannot be completed (e.g. yeast grx5, ssq1, and atm1 mutants affecting downstream steps in iron-sulfur biogenesis), the levels of Fe-bound Isu increase in the cell (14). A fraction of this labile iron may then be diverted to Sod2p. In yfh1Δ strains that block an upstream step in iron-sulfur biogenesis, mitochondrial iron should not accumulate on Isu proteins and accordingly, we observed no iron inactivation of Sod2p in our yfh1Δ mutants (7). Although iron binding to wild type Isu is labile, iron binding to Asp71 (or the analogous position in bacterial IscU) mutants is stable (39, 40), explaining the ability of yeast D71A Isu1p to reverse Sod2-inactivation in mtm1Δ mutants.
Although the function of Mtm1p is not yet known, we predict mtm1 mutations affect the iron-sulfur biogenesis pathway. In many regards, mtm1Δ mutations phenocopy grx5 and ssq1 mutants defective in iron-sulfur synthesis including increases in mitochondrial iron (13), induction of the Aft1 iron regulon (7), loss of Sod2p activity (7) and induction of Isu (this study). In our recent studies with a severe knockdown of mtm1 through MET3 transcriptional repression, we observed a loss of iron-sulfur aconitase activity without a loss in mitochondrial DNA.4 Furthermore, recent studies of zebrafish and mammalian orthologues to yeast Mtm1p also implicate a role in mitochondrial iron homeostasis consistent with effects on iron-sulfur biogenesis (49). Based on its ability to phenocopy yeast grx5 and ssq1 mutants, Mtm1p may participate in a downstream component of iron-sulfur biogenesis yet to be determined.
Lastly, it is worth noting that Isu and the iron-sulfur pathway are not the sole source of iron for Sod2p inactivation. In the absence of iron homeostasis defects, a small pool of yeast Sod2p can react with iron, and this pool is expanded when cells are starved for manganese (7). In this case, Isu proteins are not responsible for Sod2p inactivation, and it is likely that other labile iron sources in the mitochondria, e.g. iron-sulfur proteins, may be involved.
Supplementary Material
Acknowledgments
We thank Jerry Kaplan for the Fe-SOD expression plasmid, Larry Vickery for anti-IscU, and Dennis Dean for helpful discussions. XAS spectra measured at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U. S. Dept. of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
This work was supported, in whole or in part, by National Institutes of Health Grants ES-08996 (to V. C. C.) and GM-38047 (awarded to J. E. P.-H.) and the Johns Hopkins University NIEHS Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1 and S2, and Figs. S1 and S2.
L. T. Jensen and V. C. Culotta, unpublished observations.
- SOD
- superoxide dismutase
- XANES
- x-ray absorption near edge structure
- EXAFS
- extended x-ray absorption fine structure
- BPS
- bathophenanthrolinedisulfonate
- WT
- wild type
- KD
- knockdown.
REFERENCES
- 1.Wintjens R., Noël C., May A. C., Gerbod D., Dufernez F., Capron M., Viscogliosi E., Rooman M. (2004) J. Biol. Chem. 279,9248–9254 [DOI] [PubMed] [Google Scholar]
- 2.Carlioz A., Ludwig M. L., Stallings W. C., Fee J. A., Steinman H. M., Touati D. (1988) J. Biol. Chem. 263,1555–1562 [PubMed] [Google Scholar]
- 3.Whittaker J. W. (2003) Biochem. Soc. Trans. 31,1318–1321 [DOI] [PubMed] [Google Scholar]
- 4.Vance C. K., Miller A. F. (1998) J. Am. Chem. Soc. 120,461–467 [Google Scholar]
- 5.Vance C. K., Miller A. F. (2001) Biochemistry 40,13079–13087 [DOI] [PubMed] [Google Scholar]
- 6.Beyer W. F., Jr., Fridovich I. (1991) J. Biol. Chem. 266,303–308 [PubMed] [Google Scholar]
- 7.Yang M., Cobine P. A., Molik S., Naranuntarat A., Lill R., Winge D. R., Culotta V. C. (2006) EMBO J. 25,1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Privalle C. T., Fridovich I. (1992) J. Biol. Chem. 267,9140–9145 [PubMed] [Google Scholar]
- 9.Weisiger R. A., Fridovich I. (1973) J. Biol. Chem. 248,4793–4796 [PubMed] [Google Scholar]
- 10.Lebovitz R. M., Zhang H., Vogel H., Cartwright J., Jr., Dionne L., Lu N., Huang S., Matzuk M. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93,9782–9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duttaroy A., Paul A., Kundu M., Belton A. (2003) Genetics 165,2295–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby K., Hu J., Hilliker A. J., Phillips J. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,16162–16167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luk E., Carroll M., Baker M., Culotta V. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,10353–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) EMBO J. 22,4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouault T. A., Tong W. H. (2005) Nat. Rev. Mol. Cell Biol. 6,345–351 [DOI] [PubMed] [Google Scholar]
- 16.Lill R., Mühlenhoff U. (2005) Trends Biochem. Sci. 30,133–141 [DOI] [PubMed] [Google Scholar]
- 17.Gerber J., Neumann K., Prohl C., Mühlenhoff U., Lill R. (2004) Mol. Cell. Biol. 24,4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garland S. A., Hoff K., Vickery L. E., Culotta V. C. (1999) J. Mol. Biol. 294,897–907 [DOI] [PubMed] [Google Scholar]
- 19.Tong W. H., Rouault T. A. (2006) Cell Metab. 3,199–210 [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Cash V. L., Flint D. H., Dean D. R. (1998) J. Biol. Chem. 273,13264–13272 [DOI] [PubMed] [Google Scholar]
- 21.Agar J. N., Krebs C., Frazzon J., Huynh B. H., Dean D. R., Johnson M. K. (2000) Biochemistry 39,7856–7862 [DOI] [PubMed] [Google Scholar]
- 22.Agar J., Zhneg L., Cash V., Dean D., Johnson M. (2000) J. Am. Chem. Soc. 122,2136–2137 [Google Scholar]
- 23.Jensen L. T., Culotta V. C. (2000) Mol. Cell. Biol. 20,3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikorski R. S., Hieter P. (1989) Genetics 122,19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balzan R., Agius D. R., Bannister W. H. (1999) Biochem. Biophys. Res. Comm 256,63–67 [DOI] [PubMed] [Google Scholar]
- 26.Shaw G. C., Cope J. J., Li L., Corson K., Hersey C., Ackermann G. E., Gwynn B., Lambert A. J., Wingert R. A., Traver D., Trede N. S., Barut B. A., Zhou Y., Minet E., Donovan A., Brownlie A., Balzan R., Weiss M. J., Peters L. L., Kaplan J., Zon L. I., Paw B. H. (2006) Nature 440,96–100 [DOI] [PubMed] [Google Scholar]
- 27.Luk E. E., Culotta V. C. (2001) J. Biol. Chem. 276,47556–47562 [DOI] [PubMed] [Google Scholar]
- 28.Popescu B. F., Pickering I. J., George G. N., Nichol H. (2007) J. Inorg. Biochem. 101,957–966 [DOI] [PubMed] [Google Scholar]
- 29.Laliberté J., Whitson L. J., Beaudoin J., Holloway S. P., Hart P. J., Labbé S. (2004) J. Biol. Chem. 279,28744–28755 [DOI] [PubMed] [Google Scholar]
- 30.Tobin D. A., Pickett J. S., Hartman H. L., Fierke C. A., Penner-Hahn J. E. (2003) J. Am. Chem. Soc. 125,9962–9969 [DOI] [PubMed] [Google Scholar]
- 31.Weng T. C., Waldo G. S., Penner-Hahn J. E. (2005) J. Synchrotron. Radiat. 12,506–510 [DOI] [PubMed] [Google Scholar]
- 32.Kispal G., Csere P., Guiard B., Lill R. (1997) FEBS Lett. 418,346–350 [DOI] [PubMed] [Google Scholar]
- 33.Bekri S., Kispal G., Lange H., Fitzsimons E., Tolmie J., Lill R., Bishop D. F. (2000) Blood 96,3256–3264 [PubMed] [Google Scholar]
- 34.Cavadini P., Biasiotto G., Poli M., Levi S., Verardi R., Zanella I., Derosas M., Ingrassia R., Corrado M., Arosio P. (2007) Blood 109,3552–3559 [DOI] [PubMed] [Google Scholar]
- 35.Schilke B., Voisine C., Beinert H., Craig E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Manzaneque M. T., Tamarit J., Bellí G., Ros J., Herrero E. (2002) Mol. Biol. Cell 13,1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen L. T., Sanchez R. J., Srinivasan C., Valentine J. S., Culotta V. C. (2004) J. Biol. Chem. 279,29938–29943 [DOI] [PubMed] [Google Scholar]
- 38.Lesuisse E., Knight S. A., Courel M., Santos R., Camadro J. M., Dancis A. (2005) Genetics 169,107–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raulfs E. C., O'Carroll I. P., Dos Santos P. C., Unciuleac M. C., Dean D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,8591–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unciuleac M. C., Chandramouli K., Naik S., Mayer S., Huynh B. H., Johnson M. K., Dean D. R. (2007) Biochemistry 46,6812–6821 [DOI] [PubMed] [Google Scholar]
- 41.Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18,3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mühlenhoff U., Richhardt N., Ristow M., Kispal G., Lill R. (2002) Hum. Mol. Genet. 11,2025–2036 [DOI] [PubMed] [Google Scholar]
- 43.Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., Kaplan J. (1997) Science 276,1709–1712 [DOI] [PubMed] [Google Scholar]
- 44.Gerber J., Mühlenhoff U., Lill R. (2003) EMBO Rep 4,906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knieszner H., Schilke B., Dutkiewicz R., D'Silva P., Cheng S., Ohlson M., Craig E. A., Marszalek J. (2005) J. Biol. Chem. 280,28966–28972 [DOI] [PubMed] [Google Scholar]
- 46.Andrew A. J., Song J. Y., Schilke B., Craig E. A. (2008) Mol. Biol. Cell 19,5259–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellí G., Molina M. M., García-Martínez J., Pérez-Ortín J. E., Herrero E. (2004) J. Biol. Chem. 279,12386–12395 [DOI] [PubMed] [Google Scholar]
- 48.Hudder B. N., Morales J. G., Stubna A., Münck E., Hendrich M. P., Lindahl P. A. (2007) J. Biol. Inorg. Chem. 12,1029–1053 [DOI] [PubMed] [Google Scholar]
- 49.Nilsson R. H., Schultz I. J., Pierce E. L., Soltis K. A., Naranuntarat A., Ward D. M., Baughman J. M., Paradkar P. N., Kingsley P. D., Culotta V. C., Kaplan J., Palis J., Paw B. H., Mootha V. K. (2009) Cell Metab., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.