Abstract
Corynebacterium diphtheriae is a Gram-positive, non-spore forming, non-motile, pleomorphic rod belonging to the genus Corynebacterium and the actinomycete group of organisms. The organism produces a potent bacteriophage-encoded protein exotoxin, diphtheria toxin (DT), which causes the symptoms of diphtheria. This potentially fatal infectious disease is controlled in many developed countries by an effective immunisation programme. However, the disease has made a dramatic return in recent years, in particular within the Eastern European region. The largest, and still on-going, outbreak since the advent of mass immunisation started within Russia and the newly independent states of the former Soviet Union in the 1990s. We have sequenced the genome of a UK clinical isolate (biotype gravis strain NCTC13129), representative of the clone responsible for this outbreak. The genome consists of a single circular chromosome of 2 488 635 bp, with no plasmids. It provides evidence that recent acquisition of pathogenicity factors goes beyond the toxin itself, and includes iron-uptake systems, adhesins and fimbrial proteins. This is in contrast to Corynebacterium’s nearest sequenced pathogenic relative, Mycobacterium tuberculosis, where there is little evidence of recent horizontal DNA acquisition. The genome itself shows an unusually extreme large-scale compositional bias, being noticeably higher in G+C near the origin than at the terminus.
INTRODUCTION
Corynebacterium diphtheriae was shown to be the cause of the acute, communicable disease diphtheria after being isolated from diphtheritic pseudomembranes in the late 19th century; shortly after this, the diphtheria toxin (DT) was purified by filtration from C.diphtheriae cultures. Following this discovery, antitoxin prepared from an experimental animal was successfully used by von Behring to treat a case of diphtheria, leading to the introduction of antitoxin therapy. His contribution was acknowledged with the award of the Nobel Prize in Physiology and Medicine in 1901 (1). Despite this history, surprisingly little is known about the biology of this microorganism, although there have been extensive studies published on its clinical pathology and epidemiology (reviewed in 1).
After infection (by direct contact, sneezing or coughing), C.diphtheriae can colonise the skin and/or the upper respiratory tract where it releases DT, causing the symptoms of the disease. The toxin can also be absorbed by the circulatory system and distributed to distant organs, such as the heart (myocardium) or peripheral nervous system. In respiratory diphtheria the disease develops in the posterior structures of the mouth and the proximal pharynx, producing a membrane on one or both tonsils. The microorganism then multiplies on the surface of this membrane, resulting in the formation of the pseudomembrane, which is initially white and becomes grey later on in the infection. The coating of the trachea by the pseudomembrane can reduce the air flow and may eventually result in complete blockage, causing suffocation and death (2).
The lack of molecular investigation of this organism means that there is a limited knowledge of the factors involved in the colonisation of mucosal sites or indeed any other virulence factors that might be associated with invasion, carriage or proliferation. The main reason for this has been the poorly developed genetic systems for C.diphtheriae, making it difficult to identify and characterise these factors.
In contrast to the lack of general knowledge of the organism, the toxin itself, its transcriptional activation, and mechanisms of action have been very well studied. It is known to be encoded on a mobile temperate bacteriophage (corynephage) (3) and is exported via the general secretory pathway. On encountering a host cell it translocates into the cytoplasm and inhibits cellular protein synthesis by ADP ribosylation of elongation factor 2 (1).
Until very recently (4) there has also been a lack of a good animal model for respiratory diphtheria, due to the fact that mice and rats are naturally resistant to DT. Bitransgenic mice have been developed whose cells efficiently express the DT receptor, making them sensitive to the toxin and therefore the first animal model in which this disease and newly developed antidotes can be thoroughly studied.
This project was initiated to generate the genomic sequence and analysis of a pathogenic Corynebacterium in order to help develop and make the most of these new genetic systems so as to gain a better understanding of the biology and virulence of this microorganism. Diphtheria cases are still being reported from every single region of the World Health Organisation and new epidemics occur regularly, as for example in South East Asia and South America. The disease is also still endemic in many parts of the world, which has severe implications for the developed countries that have successful immunisation programmes.
In this work we describe the complete sequence and analysis of C.diphtheriae biotype gravis strain NCTC13129, a clinical toxigenic isolate from the current Eastern European outbreak.
MATERIALS AND METHODS
Bacterial strain
Corynebacterium diphtheriae biotype gravis, NCTC13129 was isolated in 1997 from the pharyngeal membrane of a 72-year-old female with clinical diphtheria who had returned to the UK from a Baltic cruise (5). The organism was cultured onto Columbia blood agar (Oxoid, Basingstoke, UK) and characterised by standard microbiological methods (6). Chromosomal DNA was extracted directly from the plate culture using previously described methods (7).
Sequencing
The genome sequence was obtained from 60 750 end sequences (giving 9.2× coverage) derived from two pUC18 genomic shotgun libraries (with insert sizes of 1.4–2 and 2–4 kb) using dye terminator chemistry on ABI377 automated sequencers. End sequences from a large insert BAC library (pBACe3.6; 1.1× clone coverage, 6–9 kb insert size) were used as a scaffold. All identified repeats were bridged by read-pairs or end-sequenced PCR products. The sequence was assembled, finished and annotated as described previously (8).
Annotation and analysis
The final chromosome sequence was searched with Orpheus (9) and Glimmer2 (10) in order to identify possible coding sequences (CDSs) and the results were curated manually. Predicted proteins were searched against the public databases using FASTA (11) and BLASTP (12), and protein domains were identified using Pfam (13) and Prosite (14). The results of all searches were collated using Artemis (15) to facilitate annotation. Orthologous proteins between C.diphtheriae and M.tuberculosis (16) were identified as reciprocal best matches using FASTA with subsequent manual curation. Pseudogenes had one or more mutations that would prevent full translation; each of the inactivating mutations was subsequently checked against the original sequencing data.
Database submission
The sequence and annotation of the genome have been submitted to the DDBJ/EMBL/GenBank databases with the accession no. BX248353.
RESULTS AND DISCUSSION
General features and metabolic analysis
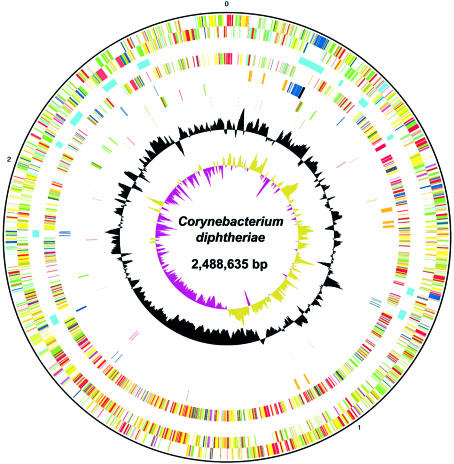
The general features of the C.diphtheriae genome are shown in Figure 1 and Table 1. Metabolic analysis revealed a complete set of enzymes for the glycolysis, gluconeogenesis and pentose-phosphate pathways. The citrate cycle (TCA cycle) appears to be complete except for the conversion between succinate and succinyl-CoA. The usual bacterial enzyme catalysing this step, succinyl-CoA synthetase [encoded by sucC and sucD, which are both present in Corynebacterium efficiens (17)], is absent; instead, C.diphtheriae may utilise the product of DIP1902, a homologue of the Clostridium kluyveri cat1 gene, which has been shown to act as a succinyl-CoA:coenzyme A transferase (18). As expected, both aerobic and anaerobic respiration genes are present. All the de novo amino acid biosynthesis pathways are present, as is the purine nucleotide biosynthetic pathway. Conversely, the pyrimidine pathway seems to lack the final cytidine triphosphate synthetase (PyrG), which is present in M.tuberculosis, Corynebacterium glutamicum (19,20) and C.efficiens, although the pathway leading to the biosynthesis of thymidine nucleotide seems complete. Pantothenate, CoA and biotin production pathways are complete, although that for folic acid is apparently not.
Figure 1.
Circular representation of the C.diphtheriae NCTC13129 chromosome. From the outer to the inner circle: Circle 1, DNA bases (counting clockwise); circles 2 and 3, all genes (forward and reverse strands); circle 4, PAIs; circle 5, genes with orthologues in M.tuberculosis; circle 6, metal-ion transport systems (orange), phage-related genes (navy blue), DT (black); circle 7, putative sortases (red), putative sortase substrates (green); circle 8, repX (brown), IS element pairs (purple); circle 9, G+C content (plotted using a 10 kb window); circle 10, GC skew [(G–C) / (G+C)] (plotted using a 10 kb window). Colour coding for circles 2, 3 and 5: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage and IS elements; grey, miscellaneous.
Table 1. General features of the C.diphtheriae NCTC13129 genome.
| Size (bp) | 2 488 635 |
|---|---|
| G+C content (%) | 53.48 |
| CDSs | 2320 |
| of which pseudogenes | 45 |
| Coding density (%) | 87.9 |
| Average gene length (bp) | 962 |
| Ribosomal RNAs | 5× (16S-23S-5S) |
| Transfer RNAs | 54 |
Chromosomal structure and variation of the G+C content
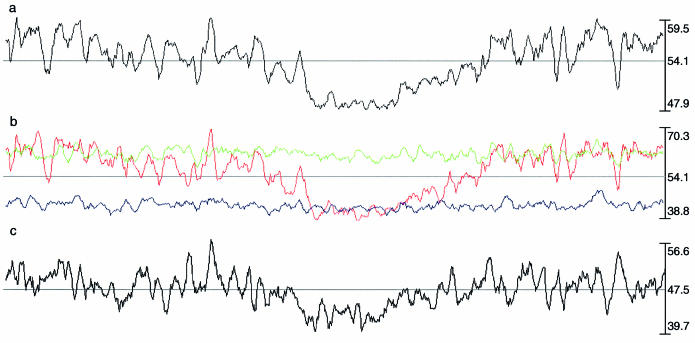
Corynebacteria are characterised by their high G+C content as well as irregular cell morphology; however, within the genus Corynebacterium the G+C content is broad (51–70%) and this reflects its genetic diversity. The almost universal bacterial bias towards guanine (G) on the leading strand of the bidirectional replication fork (21) is conserved, allowing the designation of an origin of replication near base 1, and a terminus around base 1 249 000. However, a highly unusual feature of the C.diphtheriae genome is that the G+C content itself is not constant across the genome (Fig. 2a). Strikingly, there is a region of ∼740 kb (approximate bases 981 700– 1 720 500), which encompasses the terminus of replication, that has a significantly lower G+C content (49.99% overall, reaching a trough of 48.08%) than the remainder of the genome (54.96%). This region does not have clearly defined boundaries; indeed there seems to be a gradual transition from a higher G+C content in the region near the origin to a lower content in the region around the terminus. This change in G+C content is not due to recent acquisition of genes, or to a bias in the position of certain types or classes of gene, as a comparison with the genome of M.tuberculosis (16), a distantly related actinomycete, demonstrates that orthologous genes between the two genomes are spread across both sections of the genome (Fig. 1 and Supplementary Material Fig. 1). This is also evident in closer comparisons; the entire length of the C.diphtheriae genome is co-linear with the backbone of C.glutamicum (19,20) and that of C.efficiens (17), both non-pathogenic bacteria that show no such large genomic changes in their G+C content (Fig. 3). Further investigation of the C.diphtheriae genome indicated that almost all of the variation is due to changes in the third codon position within CDSs (Fig. 2b), and within non-protein-coding regions (Fig. 2c). A recent analysis (22) has shown that many bacterial genomes are structured in this way, with a lower G+C content near the terminus. In most cases, however, this was only detectable by measuring cumulative changes in third-position GC content in all genes across the genome; clearly the bias in C.diphtheriae is considerably stronger than these other genomes. It was hypothesised that this change could be due to structural constraints around the terminus, or to differential mutational pressures around the terminus leading to an increase in GC to AT changes. This would suggest that there is some temporal or physical compartmentalisation of the genome at some stage; the most obvious candidate being chromosomal replication [it has been shown in Escherichia coli that the pre- and post- replication sections of the chromosome occupy different areas of the cell (23)]. It is intriguing in this context that this extreme bias is only apparent in C.diphtheriae, not C.glutamicum or C.efficiens; this may reflect different environmental mutational pressures for the pathogenic versus the environmental species.
Figure 2.
G+C content of the C.diphtheriae genome. (a) Total G+C content (using 20 kb window). (b) Frame-specific G+C content of CDSs (using 20 kb window): green, frame 1; blue, frame 2; red, frame 3. (c) G+C content of non-protein-coding regions (using 2 kb window). The predicted origin of replication is situated on the left-hand side of the figures.
Figure 3.
Linear genomic comparison of C.diphtheriae (top) with C.efficiens (bottom). The coloured ticks represent the genes in the six reading frames; those in C.diphtheriae are colour coded as for Figure 1. The red lines in between the genomes represent DNA:DNA similarities (BLASTN matches) between the two DNA sequences. The plots above and below the genomes represent G+C content plotted over a 50 kb window.
The occurrence of Rag (RGNAGGGS) motifs within the C.diphtheriae genome agrees with studies performed by Lobry and Louran (24). These motifs correspond to a family of G-rich octamers whose skew strongly shifts near the origin and the terminus of replication, and this is maintained in the C.diphtheriae genome even in the low G+C region surrounding the terminus of replication. The point at which the skew shifts near the terminus is marked by dif, a site devoted to chromosome dimer resolution. It has been proposed that these polarised Rag motifs are involved in facilitating the attachment of the septum-anchored protein FtsK to the chromosome, so preventing the capture of this region by the septum and facilitating dimer resolution.
Pathogenicity islands (PAIs)
Local anomalies in nucleotide composition, such as G+C content, GC skew [(G–C) / (G+C)] and/or dinucleotide frequency of the DNA can potentially be indicative of recent acquisition of DNA. In the C.diphtheriae NCTC13129 genome we have identified 13 regions (including the corynephage) matching some or all of these criteria, many of which are flanked by tRNAs (Fig. 1 and Table 2). Subsequent comparisons with C.glutamicum and C.efficiens have shown that none of these regions are present in the genomes of these two environmental strains. It is also likely that there are other, smaller, regions that may have been horizontally acquired. Many genes that could contribute to the pathogenicity of C.diphtheriae are found within these putative islands. These PAIs encode the vast majority of the fimbrial and fimbria-related genes, as well as iron-uptake systems, a potential siderophore biosynthesis system and a lantibiotic biosynthesis system. This is reminiscent of the situation in the Enterobacteriaceae, where alternative (often mobile) pathogenicity determinants, allowing different host interactions and pathogenic lifestyles, are superimposed on a stable backbone encoding core functions (25), and similar to that of another Gram-positive pathogen, Staphylococcus aureus, in which some pathogenic determinants such as staphylococcal superantigen determinants are carried on staphylococcal PAIs (SaPIs) (26). However, this is in strong contrast to the situation described in the closest sequenced pathogenic relative of C.diphtheriae, M.tuberculosis, where pathogenicity appears to be a function of diverse factors encoded throughout the genome, and PAIs seem to be absent (16).
Table 2. Regions with anomalous G+C content, GC skew or dinucleotide frequency.
| Genes | G+C (%) | Length (bp) | Flanked by | Putative function |
|---|---|---|---|---|
| DIP0180–DIP0222 | 52.22 | 36 566 | 2× tRNA | Corynephage and DT gene |
| DIP0223–DIP0244 | 56.92 | 17 535 | tRNA | Fimbriae-related, putative sortase-like proteins, IS element |
| DIP0282–DIP0287 | 55.53 | 6671 | tRNA | Putative iron transport system |
| DIP0334–DIP0357 | 51.24 | 18 355 | tRNA | Phage- and plasmid-related genes, DNA repair protein, secreted polysaccharide degradation enzyme |
| DIP0438–DIP0445 | 52.22 | 8767 | Ribosomal proteins | Metal transport, surface-anchored proteins |
| DIP0752–DIP0766 | 50.90 | 15 057 | rRNA | Possible lantibiotic biosynthesis |
| DIP0795–DIP0820 | 59.69 | 19 019 | tRNA | Phage-related proteins, DNA methylase |
| DIP1645–DIP1663 | 50.40 | 20 228 | – | Secreted Ala-rich protein |
| DIP1817–DIP1837 | 55.15 | 14 638 | tRNA | Potential prophage |
| DIP2010–DIP2015 | 49.37 | 11 082 | – | Fimbriae related |
| DIP2066–DIP2093 | 52.31 | 30 065 | – | Fimbriae-associated protein, putative Sdr family, adhesion-related, transposases, IS elements |
| DIP2148–DIP2168 | 58.19 | 30 000 | – | Phage-related proteins and IS element, transposases, putative siderophore biosynthesis and export system, putative metal-ion uptake system |
| DIP2208–DIP2234 | 50.70 | 25 947 | – | Phage-related proteins, fimbrial-related and surface-anchored proteins, sortase homologues |
Diphtheria toxin and iron acquisition
DT is one of the most widely studied bacterial toxins (reviewed in 1). In NCTC13129 the tox gene (DIP0222) encoding DT is situated within the right-hand end of an integrated corynephage (bases 154 153–190 718), just inside the att site, within a discrete region of low C+G content (42.54%) (Fig. 4). This arrangement, suggesting recent acquisition of the tox gene by the phage, is similar to that in several pathogenicity determinant-encoding phages in Streptococcus pyogenes (27). DtxR is an iron-dependent negative-regulatory protein in C.diphtheriae that has been shown, under high iron conditions, to transcriptionally repress the tox gene, the corynebacterial siderophore and some other components of the high-affinity iron-uptake system (28). Iron limitation is a common mechanism by which hosts can suppress bacterial growth, and thus low iron is a common environmental cue for pathogenic bacteria, to which the expression of DT has been coupled. Pathogenic bacteria need specialised mechanisms for acquiring iron, often by the manufacture of secreted high-affinity iron sequestration molecules termed siderophores. Only one ferrisiderophore receptor has been described before in C.diphtheriae, Irp6A. It is situated upstream of a putative iron-uptake system (DIP0109–DIP0111) under the control of a DtxR recognised promoter (29). Siderophores are often manufactured by complex polyketide or non-ribosomal peptide synthases, and two large candidate genes for siderophore biosynthesis are DIP2160 (7.9 kb), a predicted modular polyketide synthase with similarity to the Streptomyces verticillus bleomycin biosynthesis polyketide synthase BlmVIII (30), and DIP2161 (5.2 kb), a predicted non-ribosomal peptide synthase with similarities to the Pseudomonas aeruginosa pyochelin synthetase PchF (31), which are situated together in a potential PAI (Table 2). Downstream of these genes are a pair of ABC transporters with similarity to the Yersinia pestis ATP-binding protein YbtP required for iron transport, itself situated just downstream of the Yersiniabactin biosynthesis cluster (32). In all, seven putative iron-uptake systems have been found in the C.diphtheriae genome, two of which have been previously described: the siderophore receptor IrpA6 and the hemin utilisation gene cluster hmu (Supplementary Material Table 1). Of these seven systems, only two are present in C.glutamicum (hmu and that encoded by DIP1059–DIP1063) and none in C.efficiens.
Figure 4.
Linear representation of the inserted corynephage (CDSs from DIP0180 to tox-DIP0222), showing the DNA (black central line) and flanking tRNAs (blue), the phage-related CDSs (pink), CDSs with no significant database matches (light green), putative membrane-associated proteins (dark green), a pseudogene (brown) and the toxin gene (tox, black). The top graph represents the CG content in this region (calculated with a window size of 500 bp), highlighting the low-G+C region encoding tox on the right-hand side.
Fimbriae
Fimbriae (or pili) in C.diphtheriae have been previously described (33) although not molecularly characterised. The only actinomycetes in which fimbriae have been fully characterised are Actinomyces naeslundii and Actinomyces viscosus (34). These are the dominant commensal Actinomyces spp. on dental and mucosal surfaces of numerous animal hosts, although some have been implicated in infection. They present type 1 and type 2 fimbriae that bind to a number of host proteins. These fimbrial systems are completely unlike any described in Gram-negative systems, but their components instead show similarity to sortases and sortase-processed proteins. Sortases are membrane-bound transpeptidases that covalently link surface proteins to the cell wall peptidoglycan. This is achieved through recognition of one of the conserved motifs LP/AXTG or NPQTG just upstream of a C-terminal hydrophobic signal peptide sequence and subsequent cleavage at, and linkage of, the threonine via an amide bond to an amino group of the peptidoglycan. Such systems are often used by Gram-positive pathogens to attach host-interacting proteins to the bacterial surface (35). The mechanisms of production and polymerisation of the Actinomyces fimbriae are unknown, but the obvious possibility is that the sortase-like proteins are involved in anchoring or even polymerising the fimbrial subunits. The C.diphtheriae genome contains six genes encoding putative sortases: DIP0233, DIP0236, DIP2012, DIP2224, DIP2225 and DIP2272 (Supplementary Material Table 2). The last of these appears to be part of the backbone of the chromosome, as it is present in both C.glutamicum and C.efficiens, and hence may be the ‘housekeeping’ sortase, while the other five (which are more closely related to each other than to DIP2272) are located in potential PAIs and are not present in the two non-pathogenic corynebacterial genomes. It seems possible, therefore, that C.diphtheriae has recently acquired a sortase-related fimbrial system similar to that in Actinomyces that would aid the bacteria in its early stages of invasion and adherence to the host cell surfaces. A total of 18 CDSs were found with correctly situated potential sortase anchor sites; many of these are associated with fimbrial genes, and are similar to fimbria-related proteins from Actinomyces (Supplementary Material Table 3). Other significant sortase-anchored proteins include DIP2093, which shows weak similarity to members of the Ser–Asp repeat (Sdr) family of adhesins from staphylococci (36).
The cell surface
In addition to such apparently recently acquired elements, some other genes important for the pathogenic lifestyle of C.diphtheriae are found in what could be considered to be core regions. For example, the cell wall of actinomycetes is considered to be an important pathogenicity factor. The cell walls of C.diphtheriae and M.tuberculosis share several common features; both contain an arabinogalactan polymer that anchors an outer lipid-rich domain to the murein sacculus of the cell. The detailed structure of the corynebacterial arabinogalactan, however, remains to be defined. Although corynomycolic acids are significantly smaller than their mycobacterial counterparts, their basic construction is similar. Like the mycolic acids, they are alpha-alkyl, beta-hydroxy fatty acids produced via a Claisen-like condensation of two fatty acyl chains (37). Fatty acid synthesis in plants and mammals occurs via a multifunctional polypeptide (FAS-I type system encoded by fas) that carries all of the necessary enzymatic functions. In most bacteria, however, de novo synthesis occurs via a dissociated multi-enzyme FAS-II system. In mycobacteria de novo fatty acid synthesis is carried out through FAS-I, with further extensions performed by FAS-II leading to the long-chain mycolic acids (37). Consistent with the fact that direct condensation of FAS-I products should be sufficient for the synthesis of corynomycolates, our analysis of the C.diphtheriae genome has revealed that only a fas homologue is present. Moreover, this suggests that FAS-I may be a common means of de novo fatty acid synthesis in actinomycetes. However, FAS-II systems capable of de novo synthesis do occur in Streptomyces (38).
The esterification of mycolates to the arabinogalactan is catalysed by a family of mycolyltransferases known as the antigen 85 complex (FbpA,B,C1,C2) (39). A similar function has been ascribed to the product of csp1 in C.glutamicum (40). In both M.tuberculosis and C.diphtheriae the genes encoding these enzymes are situated in a highly conserved region of the genome that carries several cell wall-related functions. In mycobacteria and C.diphtheriae two ‘mycolyltransferase’ genes lie downstream of the galactosyltransferase implicated in cell wall galactan polymerisation. In M.tuberculosis fbpA is followed by a homologue, fbpC1, which lacks the catalytic triad found in other members of the antigen 85 complex; sequence alignments suggest that both C.diphtheriae enzymes (encoded by csp1 and DIP2194) possess this catalytic triad, which is necessary for corynomycolyltransferase activity.
In mycobacteria the complete embCAB cluster is required to produce the arabinan domain of arabinogalactan, and these three membrane proteins have been implicated as arabinosyltransferases. Only one putative arabinosyltransferase gene, emb (DIP0159), is apparent in the C.diphtheriae genome. In M.tuberculosis, arabinogalactan is attached to peptidoglycan via a rhamnose-N-acetylglucosamine disaccharide linker unit. All the necessary enzymes appear to be conserved in C.diphtheriae to form this linker. The genes required for the synthesis of the activated sugar donor dTDP-rhamnose (rmlABCD) in M.tuberculosis have recently been characterised (41). In C.diphtheriae NCTC13129 the orthologues of rmlCD appear to be fused (DIP0361) to form a bifunctional protein. The orthologue of rfbE, which encodes the putative ligase implicated in attachment of arabinogalactan to the peptidoglycan, is also located close to emb in a similar genetic context to that in M.tuberculosis. The major difference in the organisation of the genes involved in mycolylarabinogalactan synthesis in these bacteria is that emb and rfbE are located 468.8 kb distant from the glfT homologue in C.diphtheriae rather than in a single cluster as in M.tuberculosis.
It is known that some strains of C.diphtheriae exhibit sialidase (neuraminidase) and trans-sialidase activity (42). As these activities have been linked to virulence in several other microbial pathogens, and may enhance fimbrial-mediated adhesion in actinomycetes by unmasking receptors on mammalian cells (43), they represent potential virulence factors in C.diphtheriae. Furthermore, sialidases and trans-sialidases have proven attractive drug and vaccine targets in some pathogens. The NCTC13129 genome encodes two putative sialidases. DIP0330 is encoded within a four-gene insertion relative to the C.glutamicum genome but appears to lack a signal peptide or trans-membrane domain, and DIP0543 is encoded by a single-gene insertion and possesses a signal peptide, a coiled-coil domain and a C-terminal trans- membrane domain.
The recent diphtheria epidemics have emphasised that continuous expansion in the depth of knowledge of basic biological and genetic mechanisms, which could affect the organism’s adaptability and pathogenicity, will remain as one of our most powerful tools in the fight against diphtheria. The data from the genome sequence has allowed us to characterise a number of putative virulence factors, such as adhesins or fimbrial-related proteins, which could be used as targets for diagnostic reagents, antimicrobials and as potential vaccine candidates against invasive diphtheria. Unlike its closest sequenced pathogenic relative, M.tuberculosis (16), C.diphtheriae appears to have recently acquired many genes necessary for survival, attachment and virulence in the host. This difference may be a reflection of the different environments of the two organisms; M.tuberculosis is a predominantly intracellular pathogen and thus has less opportunity for genetic exchange than does the extracellular C.diphtheriae.
SUPPLEMENTARY MATERIAL
Supplementary Material, including a linear gene map and functional classification of identified genes is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to David Hopwood for his critical reading of the manuscript. We would like to acknowledge the support of the Wellcome Trust Sanger Institute core sequencing and informatics groups. This work was supported by the Wellcome Trust through its Beowulf Genomics initiative.
DDBJ/EMBL/GenBank accession no. BX248353
REFERENCES
- 1.Holmes R.K. (2000) Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis., 181 (Suppl. 1), S156–S167. [DOI] [PubMed] [Google Scholar]
- 2.Hadfield T.L., McEvoy,P., Polotsky,Y., Tzinserling,V.A. and Yakovlev,A.A. (2000) The pathology of diphtheria. J. Infect. Dis., 181 (Suppl. 1), S116–S120. [DOI] [PubMed] [Google Scholar]
- 3.Leong D. and Murphy,J.R. (1985) Characterization of the diphtheria tox transcript in Corynebacterium diphtheriae and Escherichia coli. J. Bacteriol., 163, 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha J.H., Chang,M.Y., Richardson,J.A. and Eidels,L. (2003) Transgenic mice expressing the diphtheria toxin receptor are sensitive to the toxin. Mol. Microbiol., 49, 235–240. [DOI] [PubMed] [Google Scholar]
- 5.Public Health Laboratory Service (1997) Diptheria acquired during a cruise in the Baltic Sea. Commun. Dis. Rep. Weekly, 7, 137–138. [PubMed] [Google Scholar]
- 6.Efstratiou A. and George,R.C. (1999) Laboratory guidelines for the diagnosis of infections caused by Corynebacterium diphtheriae and C. ulcerans. World Health Organization. Commun. Dis. Public Health, 2, 250–257. [PubMed] [Google Scholar]
- 7.De Zoysa A., Efstratiou,A., George,R.C., Jahkola,M., Vuopio-Varkila,J., Deshevoi,S., Tseneva,G. and Rikushin,Y. (1995) Molecular epidemiology of Corynebacterium diphtheriae from northwestern Russia and surrounding countries studied by using ribotyping and pulsed-field gel electrophoresis. J. Clin. Microbiol., 33, 1080–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhill J., Achtman,M., James,K.D., Bentley,S.D., Churcher,C., Klee,S.R., Morelli,G., Basham,D., Brown,D., Chillingworth,T. et al. (2000) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature, 404, 502–506. [DOI] [PubMed] [Google Scholar]
- 9.Frishman D., Mironov,A., Mewes,H.W. and Gelfand,M. (1998) Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res., 26, 2941–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bairoch A. (1991) PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res., 19 (Suppl.), 2241–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 16.Cole S.T., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S.V., Eiglmeier,K., Gas,S., Barry,C.E. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 17.Nishio Y., Nakamura,Y., Kawarabayasi,Y., Usuda,Y., Kimura,E., Sugimoto,S., Matsui,K., Yamagishi,A., Kikuchi,H., Ikeo,K. et al. (2003) Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res., 13, 1572–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohling B. and Gottschalk,G. (1996) Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J. Bacteriol., 178, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda M. and Nakagawa,S. (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol., 62, 99–109. [DOI] [PubMed] [Google Scholar]
- 20.Kalinowski J., Bathe,B., Bartels,D., Bischoff,N., Bott,M., Burkovski,A., Dusch,N., Eggeling,L., Eikmanns,B.J., Gaigalat,L. et al. (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol., 104, 5–25. [DOI] [PubMed] [Google Scholar]
- 21.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 22.Daubin V. and Perriere,G. (2003) G+C3 structuring along the genome: a common feature in prokaryotes. Mol. Biol. Evol., 20, 471–483. [DOI] [PubMed] [Google Scholar]
- 23.Sherratt D.J., Lau,I.F. and Barre,F.X. (2001) Chromosome segregation. Curr. Opin. Microbiol., 4, 653–659. [DOI] [PubMed] [Google Scholar]
- 24.Lobry J.R. and Louarn,J.M. (2003) Polarisation of prokaryotic chromosomes. Curr. Opin. Microbiol., 6, 101–108. [DOI] [PubMed] [Google Scholar]
- 25.Hentschel U. and Hacker,J. (2001) Pathogenicity islands: the tip of the iceberg. Microbes Infect., 3, 545–548. [DOI] [PubMed] [Google Scholar]
- 26.Novick R.P., Schlievert,P. and Ruzin,A. (2001) Pathogenicity and resistance islands of staphylococci. Microbes Infect., 3, 585–594. [DOI] [PubMed] [Google Scholar]
- 27.Ferretti J.J., McShan,W.M., Ajdic,D., Savic,D.J., Savic,G., Lyon,K., Primeaux,C., Sezate,S., Suvorov,A.N., Kenton,S. et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J.H., Wang,T., Ault,K., Liu,J., Schmitt,M.P. and Holmes,R.K. (1997) Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun., 65, 4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Y., Lee,J.H. and Holmes,R.K. (2002) Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol., 184, 4846–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du L., Sanchez,C., Chen,M., Edwards,D.J. and Shen,B. (2000) The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol., 7, 623–642. [DOI] [PubMed] [Google Scholar]
- 31.Quadri L.E., Keating,T.A., Patel,H.M. and Walsh,C.T. (1999) Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE and PchF. Biochemistry, 38, 14941–14954. [DOI] [PubMed] [Google Scholar]
- 32.Fetherston J.D., Bertolino,V.J. and Perry,R.D. (1999) YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol., 32, 289–299. [DOI] [PubMed] [Google Scholar]
- 33.Yanagawa R. and Honda,E. (1976) Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect. Immun., 13, 1293–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T., Khah,M.K., Slavnic,S., Johansson,I. and Stromberg,N. (2001) Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect. Immun., 69, 7224–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallen M.J., Lam,A.C., Antonio,M. and Dunbar,K. (2001) An embarrassment of sortases—a richness of substrates? Trends Microbiol., 9, 97–102. [DOI] [PubMed] [Google Scholar]
- 36.McCrea K.W., Hartford,O., Davis,S., Eidhin,D.N., Lina,G., Speziale,P., Foster,T.J. and Hook,M. (2000) The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology, 146, 1535–1546. [DOI] [PubMed] [Google Scholar]
- 37.Kremer L., Baulard,A.R. and Besra,G.S. (2000) Genetics of mycolic acid biosynthesis. In Hatfull,G.F. and Jacobs,W.R.,Jr (eds), Molecular Genetics of Mycobacteria. ASM Press, Washington, DC, pp. 173–190. [Google Scholar]
- 38.Revill W.P., Bibb,M.J., Scheu,A.K., Kieser,H.J. and Hopwood,D.A. (2001) Beta-ketoacyl acyl carrier protein synthase III (FabH) is essential for fatty acid biosynthesis in Streptomyces coelicolor A3(2). J. Bacteriol., 183, 3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kremer L., Maughan,W.N., Wilson,R.A., Dover,L.G. and Besra,G.S. (2002) The M. tuberculosis antigen 85 complex and mycolyltransferase activity. Lett. Appl. Microbiol., 34, 233–237. [DOI] [PubMed] [Google Scholar]
- 40.Puech V., Bayan,N., Salim,K., Leblon,G. and Daffe,M. (2000) Characterization of the in vivo acceptors of the mycoloyl residues transferred by the corynebacterial PS1 and the related mycobacterial antigens 85. Mol. Microbiol., 35, 1026–1041. [DOI] [PubMed] [Google Scholar]
- 41.Ma Y., Stern,R.J., Scherman,M.S., Vissa,V.D., Yan,W., Jones,V.C., Zhang,F., Franzblau,S.G., Lewis,W.H. and McNeil,M.R. (2001) Drug targeting Mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob. Agents Chemother., 45, 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattos-Guaraldi A.L., Duarte Formiga,L.C. and Pereira,G.A. (2000) Cell surface components and adhesion in Corynebacterium diphtheriae. Microbes Infect., 2, 1507–1512. [DOI] [PubMed] [Google Scholar]
- 43.Yeung M.K. (1999) Molecular and genetic analyses of Actinomyces spp. Crit. Rev. Oral Biol. Med., 10, 120–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.