Abstract
As a stable analog for ADP-sensitive phosphorylated intermediate of sarcoplasmic reticulum Ca2+-ATPase E1PCa2·Mg, a complex of E1Ca2·BeFx, was successfully developed by addition of beryllium fluoride and Mg2+ to the Ca2+-bound state, E1Ca2. In E1Ca2·BeFx, most probably E1Ca2·BeF3−, two Ca2+ are occluded at high affinity transport sites, its formation required Mg2+ binding at the catalytic site, and ADP decomposed it to E1Ca2, as in E1PCa2·Mg. Organization of cytoplasmic domains in E1Ca2·BeFx was revealed to be intermediate between those in E1Ca2·AlF4− ADP (transition state of E1PCa2 formation) and E2·BeF3−·(ADP-insensitive phosphorylated intermediate E2P·Mg). Trinitrophenyl-AMP (TNP-AMP) formed a very fluorescent (superfluorescent) complex with E1Ca2·BeFx in contrast to no superfluorescence of TNP-AMP bound to E1Ca2·AlFx. E1Ca2·BeFx with bound TNP-AMP slowly decayed to E1Ca2, being distinct from the superfluorescent complex of TNP-AMP with E2·BeF3−, which was stable. Tryptophan fluorescence revealed that the transmembrane structure of E1Ca2·BeFx mimics E1PCa2·Mg, and between those of E1Ca2·AlF4−·ADP and E2·BeF3−. E1Ca2·BeFx at low 50–100 μm Ca2+ was converted slowly to E2·BeF3− releasing Ca2+, mimicking E1PCa2·Mg → E2P·Mg + 2Ca2+. Ca2+ replacement of Mg2+ at the catalytic site at approximately millimolar high Ca2+ decomposed E1Ca2·BeFx to E1Ca2. Notably, E1Ca2·BeFx was perfectly stabilized for at least 12 days by 0.7 mm lumenal Ca2+ with 15 mm Mg2+. Also, stable E1Ca2·BeFx was produced from E2·BeF3− at 0.7 mm lumenal Ca2+ by binding two Ca2+ to lumenally oriented low affinity transport sites, as mimicking the reverse conversion E2P· Mg + 2Ca2+ → E1PCa2·Mg.
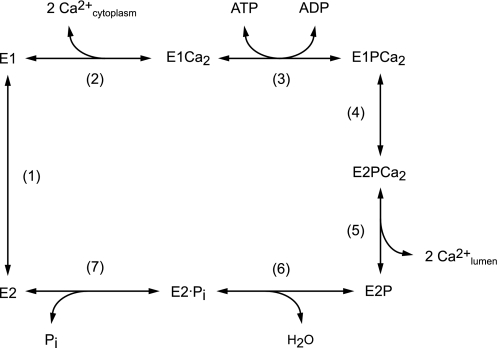
Sarcoplasmic reticulum Ca2+-ATPase (SERCA1a),2 a representative member of the P-type ion transporting ATPases, catalyze Ca2+ transport coupled with ATP hydrolysis (Fig. 1) (1–9). The enzyme forms phosphorylated intermediates from ATP or Pi in the presence of Mg2+ (10–13). In the transport cycle, the enzyme is first activated by cooperative binding of two Ca2+ ions at high affinity transport sites (E2 to E1Ca2, steps 1–2) (14) and autophosphorylated at Asp351 with MgATP to form the ADP-sensitive phosphoenzyme (E1P, step 3), which reacts with ADP to regenerate ATP in the reverse reaction. Upon this E1P formation, the two bound Ca2+ are occluded in the transport sites (E1PCa2). Subsequent isomeric transition to the ADP-insensitive form (E2PCa2), i.e. loss of ADP sensitivity at the catalytic site, results in rearrangement of the Ca2+ binding sites to deocclude Ca2+, reduce the affinity, and open the lumenal gate, thus releasing Ca2+ into the lumen (E2P, steps 4–5). Finally Asp351-acylphosphate in E2P is hydrolyzed to form the Ca2+-unbound inactive E2 state (steps 6 and 7). Mg2+ bound at the catalytic site is required as a physiological catalytic cofactor in phosphorylation and dephosphorylation and thus for the transport cycle. The cycle is totally reversible, e.g. E2P can be formed from Pi in the presence of Mg2+ and absence of Ca2+, and subsequent Ca2+ binding at lumenally oriented low affinity transport sites of E2P reverses the Ca2+-releasing step and produces E1PCa2, which is then decomposed to E1Ca2 by ADP.
FIGURE 1.
Ca2+ transport cycle of Ca2+-ATPase.
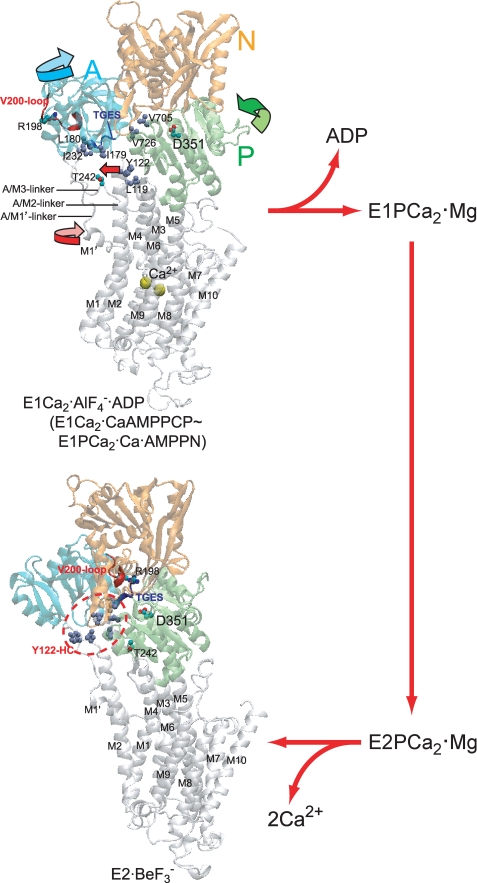
Various intermediate structural states in the transport cycle were fixed as their structural analogs produced by appropriate ligands such as AMP-PCP (non-hydrolyzable ATP analog) or metal fluoride compounds (phosphate analogs), and their crystal structures were solved at the atomic level (15–22). The three cytoplasmic domains, N, P, and A, largely move and change their organization state during the transport cycle, and the changes are coupled with changes in the transport sites. Most remarkably, in the change from E1Ca2·AlF4−·ADP (the transition state for E1PCa2 formation, E1PCa2·ADP·Mg‡) to E2·BeF3− (the ground state E2P·Mg) (23–25), the A domain largely rotates by more than 90° approximately parallel to the membrane plane and associates with the P domain, thereby destroying the Ca2+ binding sites, and opening the lumenal gate, thus releasing Ca2+ into the lumen (see Fig. 2). E1PCa2·Ca·AMP-PN formed by CaAMP-PNP without Mg2+ is nearly the same as E1Ca2·AlF4−·ADP and E1Ca2·CaAMP-PCP in their crystal structures (17, 18, 22).
FIGURE 2.
Structure of SERCA1a and its change during processing of phosphorylated intermediate. E1Ca2·AlF4−·ADP (the transition state analog for phosphorylation E1PCa2·ADP·Mg‡) and E2·BeF3− (the ground state E2P analog (25)) were obtained from the Protein Data Bank (PDB accession code 1T5T (17) and 2ZBE (21), respectively). Cytoplasmic domains N (nucleotide binding), P (phosphorylation), and A (actuator), and 10 transmembrane helices (M1–M10) are indicated. The arrows on the domains, M1′ and M2 (Tyr122) in E1Ca2·AlF4−·ADP, indicate their approximate motions predicted for E1PCa2·ADP·Mg‡ → E2P·Mg. The phosphorylation site Asp351, TGES184 of the A domain, Arg198 (tryptic T2 site) on the Val200 loop (DPR198AV200NQD) of the A domain, and Thr242 (proteinase K site) on the A/M3-linker are shown. Seven hydrophobic residues gather in the E2P state to form the Tyr122-hydrophobic cluster (Y122-HC); Tyr122/Leu119 on the top part of M2, Ile179/Leu180/Ile232 of the A domain, and Val705/Val726 of the P domain. The overall structure of E1Ca2·AlF4−·ADP is virtually the same as those of E1Ca2·CaAMP-PCP and E1PCa2·Ca·AMP-PN (17, 18, 22).
Despite these atomic structures, yet unsolved is the structure of E1PCa2·Mg, the genuine physiological intermediate E1PCa2 with bound Mg2+ at the catalytic site without the nucleotide. Its stable structural analog has yet to be developed. E1PCa2·Mg is the major intermediate accumulating almost exclusively at steady state under physiological conditions. Its rate-limiting isomerization results in Ca2+ deocclusion/release producing E2P·Mg as a key event for Ca2+ transport. In E1Ca2·CaAMP-PCP, E1Ca2·AlF4−·ADP, and E1PCa2·Ca·AMP-PN, the N and P domains are cross-linked and strongly stabilized by the bound nucleotide and/or Ca2+ at the catalytic site, thus they are crystallized (17, 18, 22). Kinetically, E1PCa2·Ca formed with CaATP is markedly stabilized due to Ca2+ binding at the catalytic Mg2+ site, and its isomerization to E2P is strongly retarded in contrast to E1PCa2·Mg (26, 27). Thus, the bound Ca2+ at the catalytic Mg2+ site likely produces a significantly different structural state from that with bound Mg2+.
Therefore, it is now essential to develop a genuine E1PCa2·Mg analog without bound nucleotide and thereby gain further insight into the structural mechanism in the Ca2+ transport process. It is also crucial to further clarify the structural importance of Mg2+ as the physiological catalytic cation. In this study, we successfully developed the complex E1Ca2·BeFx, most probably E1Ca2·BeF3−, as the E1PCa2·Mg analog by adding beryllium fluoride (BeFx) to the E1Ca2 state without any nucleotides. For its formation, Mg2+ binding at the catalytic site was required and Ca2+ substitution for Mg2+ was absolutely unfavorable, revealing a likely structural reason for its preference as the physiological cofactor. In E1Ca2·BeF3−, two Ca2+ ions bound at the high affinity transport sites are occluded. It was also produced from E2·BeF3− by lumenal Ca2+ binding at the lumenally oriented low affinity transport sites, mimicking E2P·Mg + 2Ca2+ → E1PCa2·Mg. All properties of the newly developed E1Ca2·BeF3− fulfilled the requirements as the E1PCa2·Mg analog, and hence we were able to uncover the hitherto unknown nature of E1PCa2·Mg as well as structural events occurring in the phosphorylation and isomerization processes. Also, we successfully found the conditions that perfectly stabilize the E1Ca2·BeF3− complex.
EXPERIMENTAL PROCEDURES
Preparation of SR Vesicles and Treatment with BeFx, AlFx, and AlFx·ADP
SR vesicles were prepared from rabbit skeletal muscle as described (28). The content of the phosphorylation site in the vesicles determined according to Barrabin et al. (29) was 4.49 ± 0.22 nmol/mg of vesicle protein (n = 5). The Ca2+-dependent ATPase activity determined at 25 °C in a mixture containing 5 μg/ml microsomal protein, 1 mm ATP, 1.7 μm A23187, 7 mm MgCl2, 0.1 m KCl, 50 mm MOPS/Tris (pH 7.0), and 0.6 mm CaCl2 with 0.5 mm EGTA (or 2 mm EGTA without added CaCl2) was 1.87 ± 0.14 μmol/min/mg of vesicle protein (n = 3). The Ca2+-ATPase was purified from the vesicles by deoxycholate as described (30, 31). The E1Ca2 state ATPase was incubated with fluoride compounds, 2 mm potassium fluoride and 100 μm BeSO4 or AlCl3, at 25 °C for 30 min in the presence of 0.1 mm Ca2+, 15 mm MgCl2, 0.1 m KCl, 30 mm MOPS/Tris buffer (pH 7.0), unless stated otherwise. E1Ca2· AlF4−·ADP was formed by including 50 μm ADP in the above AlFx incubation mixture as described (32). E2·BeF3−, E2·AlF4−, and E2·MgF42− were produced as described (23–25).
Determination of EP
EP formation was performed with 3 μm [γ-32P]ATP in 100 (or 50) μm Ca2+ at 0 °C for 3 s, and terminated by trichloroacetic acid containing carrier Pi. The amount of EP formed was determined as described previously (28). The background level determined with excess 5 mm EGTA was less than 0.5% of the phosphorylation sites.
Ca2+ Binding and Occlusion
45Ca2+ binding and occlusion at the transport sites was determined at 25 °C with 2 ml of the SR vesicle mixture (0.2 mg/ml protein) with a 0.45-μm nitrocellulose membrane filter (Millipore) as described (31). In some cases, the vesicles on the filter were washed for 10 s by perfusion with 2 ml of a washing solution containing 5 mm EGTA. The amount of Ca2+ bound at the transport sites was obtained by subtracting the nonspecific Ca2+ binding level determined as described in the figure legends.
Proteolysis
SR vesicles (0.45 mg/ml protein) were treated at 25 °C with trypsin (0.3 mg/ml) or proteinase K (0.1 mg/ml) as described (23, 24) and as noted in the figure legends. The samples were subjected to Laemmli SDS-PAGE (33) and densitometric analyses of the Coomassie Brilliant Blue R-250-stained gels (23, 24).
Fluorescence Measurements
The TNP-AMP fluorescence and intrinsic tryptophan fluorescence of Ca2+-ATPase (0.06 mg/ml protein) were measured on a RF-5300PC spectrofluorophotometer (Shimadzu, Kyoto, Japan) with excitation and emission wavelengths 408 and 540 nm for TNP-AMP (with band widths 5 and 10 nm), and 290 and 338.4 nm for tryptophan (with bandwidth 1.5 and 5 nm), unless otherwise described (28).
Miscellaneous
Trypsin (l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated) and proteinase K were obtained from Sigma. TNP-AMP was synthesized according to Hiratsuka (34). Protein concentrations were determined by the method of Lowry et al. (35) with bovine serum albumin as a standard. Free Ca2+ concentrations were calculated by the Calcon program. Data were analyzed by nonlinear regression using the program Origin (Microcal Software, Inc., Northampton, MA). Three-dimensional models of the enzyme were reproduced by the program VMD (36).
RESULTS
Formation of E1PCa2 Analogs by Fluoride Compounds
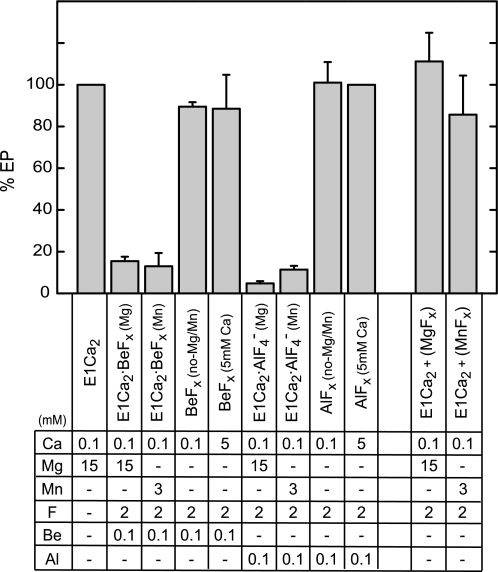
The Ca2+-ATPase of SR vesicles in 100 μm Ca2+ and 15 mm Mg2+ was incubated with beryllium fluoride (BeFx) or aluminum fluoride (AlFx) for 30 min at 25 °C. The ability of EP formation from ATP was almost completely lost (actually within 1 min) (Fig. 3), showing the stable complex formation with BeFx and AlFx. No inhibition occurred with Mg2+ and fluoride without beryllium and aluminum (E1Ca2+(MgFx)). Thus, MgFx (MgF42−) was not able to produce a complex with E1Ca2, in contrast to E2·MgF42− formation from E2, the E2·Pi product analog in E2P hydrolysis (19, 25). This finding agrees with the in-line phosphorylation of E1Ca2 to E1PCa2 (37), in which there is no state with non-covalently bound Pi.
FIGURE 3.
Inhibition of EP formation by binding of BeFx and AlFx to E1Ca2. The E1Ca2 state ATPase of SR vesicles in 0.1 or 5 mm free Ca2+ was treated for 30 min at 25 °C with fluoride compounds in the presence of the indicated concentrations of the ligands. Subsequently, the samples were cooled to 0 °C and phosphorylated for 3 s by 3 μm [γ-32P]ATP. The amount of EP formed was shown as the percentage of the control value obtained without F−, Be2+, and Al3+. The EP formation was not inhibited when the incubations were performed with Be2+ or Al3+ but without F−. The error bars show the S. D. of five independent experiments.
The binding of Mg2+ at the catalytic site as a physiological cation is nevertheless required for EP formation. Actually in Fig. 3, Mg2+ was required for complex formation with BeFx in 100 μm Ca2+. The apparent Mg2+ affinity (K0.5 of 5 mm, supplemental Fig. S1) was consistent with that of the catalytic site in phosphorylation from ATP or Pi (e.g. Refs. 11, 12, and 38–41). The BeFx-induced inhibition also occurred with Mn2+ with apparent affinity (K0.5 of 0.6 mm) significantly higher than that of Mg2+, as found in E1PCa2 formation and ATP hydrolysis with Mn2+ (40, 42).
When Ca2+ over millimolar amounts was added in place of Mg2+, the EP formation was not inhibited by BeFx (BeFx(5 mm Ca)). Therefore, Ca2+ substitution probably at the catalytic Mg2+ site abolished complex formation with BeFx. Although CaATP as a substrate and Ca2+ bound at the catalytic Mg2+ site are able to function for E1PCa2 formation (26, 27), Ca2+ bound at the catalytic site likely produces different structure from the Mg2+ bound structure (as in fact found, see below). The complex formation of E1Ca2 by AlFx in 100 μm Ca2+ also required Mg2+ or Mn2+ with somewhat higher apparent affinities than those for BeFx-induced complex formation, and was abolished by Ca2+ binding at the catalytic site.
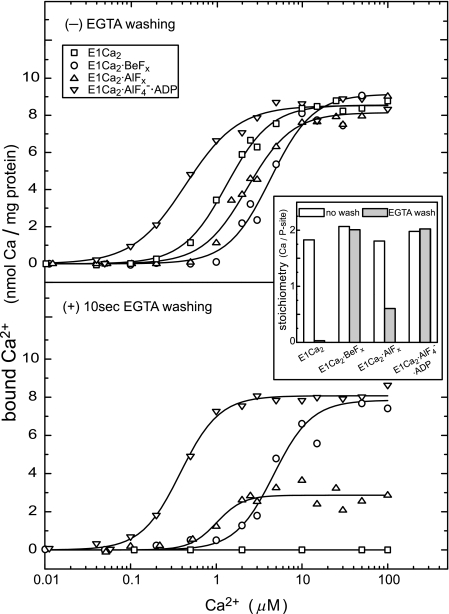
Ca2+ Binding and Occlusion at Transport Sites
In Fig. 4, Ca2+ binding and occlusion in formation of the Ca2+-ATPase complexes with fluoride compounds were determined in 15 mm Mg2+ with and without a 10-s EGTA filter washing. In all the cases without washing, Ca2+ was bound to the Ca2+-ATPase with high affinities; K0.5 at sub-micromolar to millimolar ranges, Hill coefficient ∼2, and maximum levels of 9–10 nmol/mg of protein, i.e. the stoichiometry of two Ca2+ per phosphorylation site (inset at 50 μm Ca2+). Therefore, E1Ca2·BeFx and E1Ca2·AlF4−·ADP/E1Ca2·AlFx were produced by cooperative binding of two Ca2+ ions at high affinity transport sites. This finding agrees with the property of the sites for Ca2+ binding and the resulting enzyme activation for phosphorylation by ATP as nicely demonstrated for the first time by Inesi et al. (14). Upon the EGTA washing of E1Ca2 that is complexed with BeFx, the two bound Ca2+ were not removed, and therefore occluded in the complex as “E1Ca2·BeFx.” The two Ca2+ are occluded also in E1Ca2·AlF4−·ADP and less strongly in E1Ca2·AlFx.
FIGURE 4.
Ca2+ binding and occlusion at transport sites. The E1Ca2 state ATPase of SR vesicles in various concentrations of 45Ca2+ and 15 mm MgCl2 was incubated with BeFx, AlFx, and AlFx·ADP or without these compounds (E1Ca2) for 30 min at 25 °C. The amounts of bound (upper panel) and occluded (lower panel) 45Ca2+ were determined without and with the perfusion of the membrane filter with a 5 mm EGTA-containing washing solution (without CaCl2 and fluoride compounds otherwise as the above incubation solution). The nonspecific Ca2+ binding was determined by including 10 μm thapsigargin before the addition of fluoride compounds, and subtracted. When ADP was used for E1Ca2·AlF4−·ADP, 5 μm A23187 was included to avoid Ca2+ accumulation in the vesicles by ATP produced from ADP due to adenylate kinase in the vesicles. In the inset, the stoichiometries of bound Ca2+ (open bars) and occluded Ca2+ (closed bars) to the phosphorylation site (P-site) were determined at saturating 50 μm 45Ca2+. Solid lines in the upper panel show the least squares fit to the Hill equation. K0.5 of Ca2+ and Hill coefficients obtained were 1.3 μm and 1.7 (E1Ca2), 4.3 μm and 1.7 (E1Ca2·BeFx), 2.3 μm and 1.7 (E1Ca2·AlFx), and 0.4 μm and 1.4 (E1Ca2·AlF4−·ADP). In the lower panel, the values for E1Ca2·BeFx and E1Ca2·AlF4−·ADP are essentially not altered by EGTA washing (4.7 μm and 1.9, and 0.4 μm and 1.9, respectively).
Note that the Ca2+ affinity became 2–3-fold lower for BeFx and AlFx. This may be because the Ca2+-free E2 state produces E2·BeF3− and E2·AlF4− (25), and therefore competes with Ca2+ binding for formation of E1Ca2·BeFx and E1Ca2·AlFx. On the other hand, the observed ∼3-fold Ca2+-affinity increase in formation of E1Ca2·AlF4−·ADP is probably brought about by the fact that ADP together with AlFx strongly stabilizes the cross-linked N-P domains (17, 18), which is unfavorable for formation of the Ca2+-free E2 and E2·AlF4−, because for these structures the A domain should rotate into the opened space between the N and P domains and associate with them (19, 23, 24).
Cytoplasmic Structure in E1Ca2·BeFx Is Intermediate between Those in E1Ca2·AlF4−·ADP and E2·BeF3−
Proteolytic analysis was made to reveal the organization state of the cytoplasmic domains in the newly developed E1PCa2·Mg analog E1Ca2·BeFx, and to compare with E1Ca2·AlF4−·ADP/E1Ca2·AlFx and E2·BeF3− (E2P·Mg) (see the typical cleavage in supplemental Fig. S2). The initial rate of the “A1” appearance upon cleavage at the T2 site (Arg198 on the Val200 loop of the A domain) in E1Ca2·BeFx was substantially slower than the rapid cleavage of E1Ca2·AlF4−·ADP and E1Ca2·AlFx as well as E1Ca2 (Table 1). The slowed T2 cleavage was also observed when E1Ca2·BeFx was formed with 3 mm Mn2+ in place of 15 mm Mg2+ (data not shown). Also important was the slow but definitely occurring T2 cleavage in E1Ca2·BeFx, in sharp contrast to its complete resistance in E2·BeF3−. Therefore A-P domain organization at the Val200 loop in E1Ca2·BeFx is intermediate between those in E1Ca2·AlF4−·ADP/E1Ca2·AlFx and E2·BeF3−.
TABLE 1.
Summary of fluorescence changes and proteolysis rates
Maximal TNP-AMP fluorescence intensity at saturating 4 μm TNP-AMP is given as % value of that of E2·BeF3− without A23187. Tryptophan fluorescence change upon complex formation with the ligand from E1Ca2 (or from other state when indicated in parentheses) is shown as % value of the intensity of E1Ca2 (see also supplemental Fig. S3E). The cleavage rate at the T2 site (Arg198) with trypsin and the digestion rate of the 110-kDa ATPase chain with proteinase K were obtained by the detailed time course analysis in the initial 1 (T2) and 30 min (proteinase K), and shown as % values of those determined with E1Ca2 in 0.1 mm Ca2+. Some of these experiments were done at 0.05 mm Ca2+ instead of 0.1 mm, but the results were virtually the same and therefore are represented with 0.1 mm Ca2+ for simplicity. It should also be mentioned that the digestion rates in E1Ca2 were not altered by 5 or 0.7 mm Ca2+ or by A23187 (being 97–101% of the rates of E1Ca2 in 0.1 mm Ca2+ without A23187), and that the ligand-free E2 state was also rapidly digested by trypsin and proteinase K, and TG binding to E2 and A23187 did not alter essentially the rapid cleavage rates (Refs. 23–25). The other Ca2+-ATPase complexes were produced under the same buffer conditions as those for E1Ca2·BeFx formation, otherwise as follows and noted below: E1Ca2·MgAMP-PCP by 5 mm MgAMP-PCP; E1Ca2·CaAMP-PCP by 5 mm CaAMP-PCP; E1PCa2·Ca·AMP-PN by 5 mm CaAMP-PNP; E1PCa2·Ca by 5 mm CaATP. E1Ca2·BeFx + 5 mm Ca2+, the E1Ca2·BeFx complex was formed in 15 mm Mg2+ and 100 μm Ca2+ and then incubated with the subsequently added 5 mm Ca2+ for 3 h; E1Ca2 + 5 mm Ca2+ + BeFx, the E1Ca2 state ATPase was incubated with BeFx for 10 min in the presence of 5 mm Ca2+ without Mg2+.
| ATPase state (→ consequent state, if altered) | Ca2+ (mm)/Mg2+ (mm) | Relative TNP-AMP fluorescence intensity, see Fig. 5A and supplemental Fig. S5 | Change in tryptophan fluorescence from E1Ca2 (or from another state), supplemental Fig. S3 ↑ increase, ↓ decrease | Relative digestion rate |
|
|---|---|---|---|---|---|
| Trypsin (T2), supplemental Fig. S2 | Proteinase K, supplemental Fig. S2 | ||||
| mm | % | % of E1Ca2 level | % | ||
| E1Ca2 | 0.1/15 | 7 | (3.27 ↑ from E2) | 100 | 100 |
| E1Ca2·MgAMP-PCP | 0.1/15 | 0 | 64 | 25 | |
| E1Ca2·CaAMP-PCP | 5/0 | 0.80 ↑ | 61 | 9 | |
| E1Ca2·AlF4−·ADP | 0.1/15 | 0.84 ↑ | 65 | 3 | |
| E1PCa2·Ca·AMP-PN | 5/0 | 0.77 ↑ | 65 | 4 | |
| E1Ca2·AlFx | 0.1/15 | 7 | 0 | 70 | 10 |
| E1PCa2·Ca | 5/0 | 0.89 ↓ | 80 | 6 | |
| E1PCa2·Mg | 0.1/15 | 1.18 ↓ | |||
| E1Ca2·BeFx | 0.1/15 | 75 | 1.27 ↓ | 35 | 13 |
| E1Ca2·BeFx + 5 mm Ca2+ (→ partially E1Ca2) | 0.1/15 + 5 mm Ca2+ | 30 | 70 | 32 | |
| E1Ca2 + 5 mm Ca2+ + BeFx | 5/0 | 16 | 0 | 116 | 90 |
| E2·BeF3− | 0/15 | 100 | (0.66 ↑ from E2) | 0 | 0 |
| E1Ca2·BeFx + TG (→ E2·BeF3−(TG)) | 0.1/15 | 64 | (5.35 ↓ from E1Ca2·BeFx) | 0 | 0 |
| E2·BeF3− + TG (→ E2·BeF3−(TG)) | 0/15 | 65 | (4.62 ↓ from E2·BeF3−) | 0 | 0 |
| E1Ca2·BeFx + A23187 (→ E2·BeF3−) | 0.1/15 | 100 | 10 | 0 | |
| E2·BeF3− + A23187 | 0/15 | 100 | 0 | 0 | |
| E1PCa2·Mg (0.7 mm Ca2+) | 0.7/15 | 1.19 ↓ | |||
| E1Ca2·BeFx (0.7 mm Ca2+) | 0.7/15 | 75 | 1.27 ↓ | 39 | 15 |
| E1Ca2·BeFx (0.7 mm Ca2+ + A23187) | 0.7/15 | 75 | 32 | 8 | |
| E2·BeF3− + 0.7 mm Ca2+ | 0/15 + 0.7 mm Ca2+ | 100 | (0 from E2·BeF3−) | 0 | 2 |
| E2·BeF3− + 0.7 mm Ca2+ + A23187 (→ E1Ca2·BeF3−) | 0/15 + 0.7 mm Ca2+ | 75 | 38 | 16 | |
All complexes were almost completely resistant to proteinase K at the major site of Thr242 on the A/M3-linker that produces the “p83” fragment. Therefore, in E1Ca2·BeFx, the A domain is rotated perpendicular to the membrane plane from its position in E1Ca2 thereby causing the A/M3-linker strain, as in E1Ca2·CaAMP-PCP, E1Ca2·AlF4−·ADP (18, 19, 24), and E1Ca2·AlFx.
These analyses revealed that in the change E1Ca2·AlF4−· ADP → E1Ca2·BeFx (i.e. upon the ADP release from the transition state), the A domain moves, i.e. probably rotates to some extent parallel to the membrane plane likely due to the A/M3-linker strain, and thereby Arg198 on the Val200 loop comes close to the P domain. In the subsequent change, E1Ca2·BeFx → E2·BeF3−, the A domain rotates further (by the A/M3-linker strain as predicted to be motive force (18, 19, 43, 44)) and produces its tight association with the P domain at the Val200 loop, mimicking E1PCa2·Mg → E2P·Mg + 2Ca2+.
Ca2+ Ligation at the Catalytic Mg2+ Site
The proteolysis further revealed that E1Ca2·BeFx was not produced from E1Ca2 in 5 mm Ca2+ without Mg2+ (E1Ca2 + 5 mm Ca2+ + BeFx in Table 1), and that E1Ca2·BeFx produced in 15 mm Mg2+ and 50–100 μm Ca2+ was decomposed to the E1Ca2 by 5 mm Ca2+ (E1Ca2·BeFx + 5 mm Ca2+), as shown by the rapid cleavage rates at the T2 and proteinase K sites. In E1PCa2·Ca and E1Ca2· CaAMP-PCP formed in 5 mm Ca2+ without Mg2+ (Table 1),3 the T2 site was also rapidly cleaved, in contrast to its substantially slowed cleavage in E1Ca2·BeFx formed with Mg2+. Thus, for organization of the cytoplasmic domains at the T2 site (Arg198), E1Ca2·CaAMP-PCP and E1PCa2·Ca are very similar to E1Ca2·AlF4−·ADP, but differ from E1Ca2·BeFx. The close similarity between E1Ca2·CaAMP-PCP and E1Ca2·AlF4−·ADP is in agreement with their nearly same atomic structures and previous observations (17, 18, 45). Also notably, structure E1PCa2·Ca·AMP-PN formed by CaAMP-PNP in 10 mm Ca2+ without Mg2+ (22) is almost identical with those of E1Ca2·CaAMP-PCP and E1Ca2·AlF4−·ADP (see also Table 1).
In E1Ca2·CaAMP-PCP and E1PCa2·Ca·AMP-PN, the N-P domain cross-linked state is stabilized by Ca2+ bound at catalytic Mg2+ site I (Asp351/Thr353/Asp703 and the phosphate) and by the nucleotide to be nearly identical to the state stabilized with AlF4− plus ADP in E1Ca2·AlF4−·ADP (17, 18, 22). The results on E1PCa2·Ca further indicated that such an N-P domain closed state is stabilized solely by site I Ca2+ ligation without the nucleotide. The stabilization of this state in E1PCa2·Ca is consistent with its markedly retarded isomerization to E2P (27), because isomerization requires the A domain rotation into the space between the N and P domains. In E1Ca2·BeFx formed with Mg2+ at the catalytic site (site I), such a Ca2+ ligation effect is obviously not present. Therefore, the N and P domains are probably more easily separated from each other, and the A domain can rotate into the space between the N and P domains to some extent thus resulting in partial T2 resistance (but not yet as completely as in E2·BeF3−). As the cause of the Ca2+-induced E1Ca2·BeFx to E1Ca2 decomposition, Ca2+ replacement of Mg2+ at site I altered the domain organization state and made the BeFx ligation unfavorable (see “Discussion”).
TNP-AMP Superfluorescence
TNP-AMP binds to the nucleotide binding site with an extremely high affinity (46–48), and in the E2P ground state and its analog E2·BeF3−, the bound TNP-AMP develops its extremely high fluorescence “superfluorescence” (25), which reflects a strongly hydrophobic atmosphere around Asp351 (49, 50). On the other hand, it has been controversial whether E1PCa2 develops TNP-AMP superfluorescence, mostly because its tight binding to the nucleotide binding site prevents phosphorylation to form E1PCa2·Mg, so the TNP-AMP·E1PCa2·Mg complex is not formed in significant amounts. Nakamoto and Inesi (47), nevertheless, predicted the development of superfluorescence in E1PCa2.
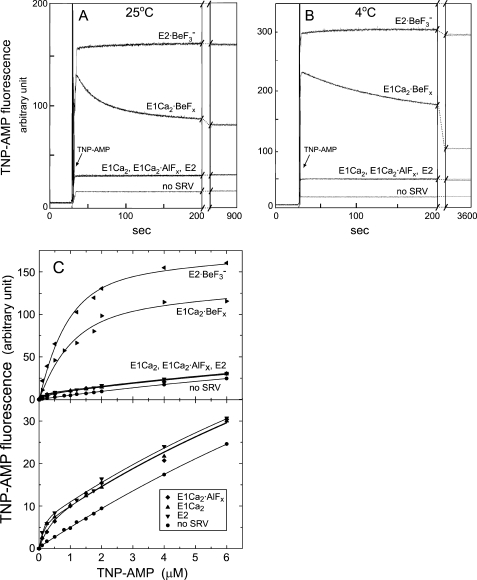
Here, with E1Ca2·BeFx as the E1PCa2·Mg analog, we examined the superfluorescence without ATP. In Fig. 5, A and B, we first formed E1Ca2·BeFx in 15 mm Mg2+ and 50 μm Ca2+, then TNP-AMP was added to give a saturating level of 4 μm. E1Ca2·BeFx rapidly developed superfluorescence, and then the fluorescence decreased slowly (much more slowly and extensively at 4 °C).4 The proteolysis after the loss of superfluorescence revealed that E1Ca2·BeFx was decomposed to E1Ca2 (data not shown). Thus, E1Ca2·BeFx develops superfluorescence, and the TNP-AMP binding per se causes its decomposition to E1Ca2, in sharp contrast to the completely stable E2·BeF3− even after TNP-AMP binding. The maximum superfluorescence level of E1Ca2·BeFx was slightly lower than that of E2·BeF3− (Fig. 5, A-C), which is the same as that of E2P·Mg formed from Pi (25). The results clearly revealed that the atmosphere around Asp351 in E1Ca2·BeFx is strongly hydrophobic, similar to E2·BeF3−, although the cytoplasmic domain organization in E1Ca2·BeFx distinctly differs from and did not yet reach the most compactly organized state in E2·BeF3−.
FIGURE 5.
TNP-AMP superfluorescence of E1Ca2·BeFx and E2·BeF3−. E1Ca2·BeFx and E1Ca2·AlFx were produced in 50 μm Ca2+ and 15 mm MgCl2, then at 25 (A) and 4 °C (B), a small volume of TNP-AMP was added to give saturating 4 μm, and the TNP-AMP fluorescence was followed. The fluorescence of E2·BeF3− produced with BeFx in the absence of Ca2+, the E1Ca2 and E2 states without the fluoride compounds, and without SR vesicles (no SRV) were also followed. C, the TNP-AMP fluorescence intensities were measured at various concentrations of TNP-AMP at 25 °C, otherwise as described for A. For E1Ca2·BeFx, the maximum level of transient superfluorescence was determined by extrapolating its decrease to the time of TNP-AMP addition. In the lower panel in C, the low fluorescence was replotted on the expanded scale. The maximum fluorescence intensities at saturating 4 μm TNP-AMP were obtained by subtracting the background level without TNP-AMP and the level of 4 μm TNP-AMP without the SR vesicles, and given as the relative values in Table 1.
E1Ca2·AlFx as well as E2 and E1Ca2 did not develop superfluorescence despite high affinity TNP-AMP binding. Therefore the catalytic site in E1Ca2·AlFx is hydrophilic and differs critically from the strongly hydrophobic site in E1Ca2·BeFx. Note also that E2·AlF4− (E2-P‡) and E2·MgF42− (E2·Pi) do not develop TNP-AMP superfluorescence (25). The superfluorescence therefore develops solely with Ca2+-ATPase complexed with BeFx; E1Ca2·BeFx and E2·BeF3−.
The superfluorescence development of E1Ca2·BeFx in the presence of 15 mm Mg2+ and 50 μm Ca2+ was rapidly diminished with increasing Ca2+ over millimolar concentrations of Ca2+ (see Fig. 8 and Table 1). Also, inclusion of 5 mm Ca2+ without Mg2+ in the E1Ca2·BeFx formation mixture abolished the superfluorescence (Table 1). The results agree with the above findings that E1Ca2·BeFx is not produced from and decomposed to E1Ca2 by Ca2+ ligation at the catalytic Mg2+ site (site I).
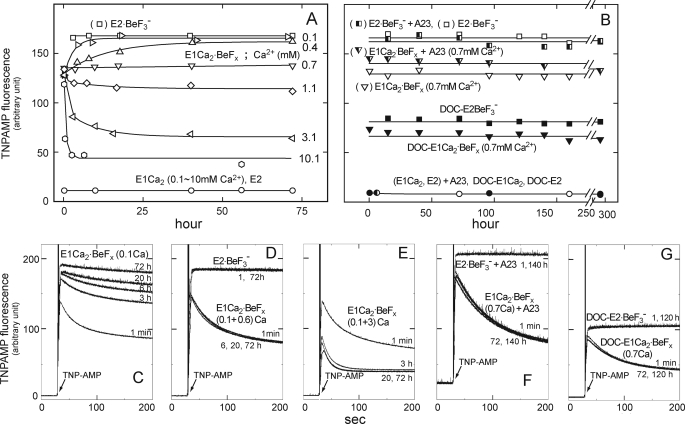
FIGURE 8.
Stability of E1Ca2·BeFx in various Ca2+ concentrations. A, E1Ca2·BeFx was first produced in SR vesicles in 0.1 mm Ca2+ and 15 mm MgCl2 with BeFx. Subsequently, Ca2+ was changed to 0.1 (unchanged), 0.4, 0.7, 1.1, 3.1, and 10.1 mm, and the incubations continued at 25 °C for 72 h. At the indicated periods, the superfluorescence with 4 μm TNP-AMP was examined at 25 °C, and the maximum levels obtained as described in the legend to Fig. 6 are shown. E2·BeF3− formed without Ca2+, E1Ca2 in 0.1–10 mm Ca2+, and E2 without Ca2+ were also incubated. B, in the presence of 0.7 mm Ca2+, E1Ca2 in SR vesicles was first incubated with and without 1.2 μm A23187 (A23), then BeFx was added (E1Ca2·BeFx). E1Ca2·BeFx (DOC-E1Ca2·BeFx) was also produced from E1Ca2 in 0.7 mm Ca2+ of the Ca2+-ATPase purified and delipidated from SR vesicles by deoxycholate (DOC) treatment (30). The incubation was continued for 12 days, otherwise as indicated in A. E2·BeF3− (E2·BeF3−, DOC-E2·BeF3−) without Ca2+ and E1Ca2 in 0.7 mm Ca2+ (E1Ca2, DOC-E1Ca2) and E2 without Ca2+ (E2, DOC-E2) were also incubated. C–G, the fluorescence traces upon TNP-AMP addition were shown for the representative samples with incubation periods and Ca2+ concentration (mm). Note that at 0.7 mm Ca2+, both with and without A23187, the development of the E1Ca2·BeFx characteristic transient superfluorescence remained perfectly the same for 12 days. By contrast, the transient superfluorescence was converted to the stable and higher superfluorescence characteristic of E2·BeF3− at 0.1 and 0.4 mm Ca2+, and it was markedly reduced by 3 and 10 mm Ca2+ due to decomposition to E1Ca2.
Transmembrane Domain Structure
The 12 tryptophan residues among 13 in the Ca2+-ATPase are located at the transmembrane region. The tryptophan fluorescence changes in fact reflect the transmembrane domain structural changes, i.e. rearrangements of the transmembrane helices upon Ca2+ binding to the high affinity transport sites and during the transport cycle (28, 51, 52) as found originally by Dupont and Leigh (53). As summarized in Table 1 with typical fluorescence traces in supplemental Fig. S3, the fluorescence changes were determined at 4 °C upon formation of the E1PCa2 analogs by the addition of fluoride compounds to E1Ca2 in 15 mm Mg2+ and 100 μm Ca2+. E1Ca2·BeFx formation decreased fluorescence by 1.3% very similar to the decrease in E1PCa2·Mg formation from E1Ca2 by MgATP, i.e. in E1Ca2·MgATP → E1PCa2·Mg (52). In contrast, E1Ca2· AlFx formation did not cause any change. The E1Ca2·AlF4−·ADP formation increased the fluorescence by 0.8%. (F− alone and ADP alone did not cause any change, except the dilution (F−) and absorption of excitation light (ADP).) Thus the transmembrane structure of E1Ca2·BeFx mimics that of E1PCa2· Mg, but those of E1Ca2·AlF4−·ADP and E1Ca2·AlFx differ substantially although the Ca2+ ions are occluded at the transport sites (or less strongly in E1Ca2·AlFx, Fig. 4). This observation is consistent with the finding in proteolysis and TNP-AMP superfluorescence that organization of the cytoplasmic domains and structure at the catalytic site in E1Ca2·BeFx substantially differ from those in E1Ca2·AlF4−·ADP and E1Ca2·AlFx. It is concluded that the transmembrane structure with the occluded Ca2+ adopts not simply one state, but changes with the change in the cytoplasmic region during phosphoryl transfer and ADP release (see the diagram of tryptophan fluorescence change in supplemental Fig. S3E (with Ref. 54)).
Upon formation of E1Ca2· CaAMP-PCP and E1PCa2·Ca·AMP-PN by CaAMP-PCP and CaAMP-PNP, respectively, the fluorescence increased by 0.8% equal to that upon E1Ca2·AlF4−·ADP formation (Table 1), in agreement with their essentially identical structures with occluded Ca2+ (17, 18, 22, 45). By contrast, the fluorescence did not change upon formation of the E1Ca2·MgAMP-PCP, which is the Ca2+-unoccluded state (28, 45), and in rapid equilibrium with E1Ca2.
Upon the exclusive accumulation of E1PCa2·Ca by CaATP without Mg2+, tryptophan fluorescence decreased by 0.9%, being slightly less than that by formation of E1PCa2·Mg and E1Ca2·BeFx (Table 1). Thus in the overall structure, E1PCa2·Ca may be between E1Ca2·CaAMP-PCP and E1PCa2·Mg (E1Ca2·BeFx), and closer to the latter state. Although Ca2+ ligation at catalytic Mg2+ site I in E1PCa2·Ca favors the N-P domain closed state, similar to E1Ca2·CaAMP-PCP, the absence of the N-P domain cross-linking nucleotide in E1PCa2·Ca likely altered the overall structure slightly.
Upon formation of E2·BeF3− from E2 by BeFx and Mg2+ without Ca2+, the fluorescence increased by 0.7%, mimicking the change upon E2P·Mg formation from E2 with Pi and Mg2+, and reflecting the opening of the lumenal gate from the closed state (25). As a consequence, the fluorescence of E1Ca2·BeFx was definitely higher by ∼1.3% than that of E2·BeF3−, showing their distinct difference in the transmembrane structure. In agreement, the previous kinetic analysis have shown (28) that tryptophan fluorescence decreases by ∼1% in the isomerization/Ca2+ release, E1PCa2·Mg → E2P·Mg + 2Ca2+, reflecting the transmembrane structural change from the Ca2+-occluded state to the Ca2+-released and lumenally opened state.
Upon the addition of thapsigargin (TG) to E1Ca2·BeFx and E2·BeF3−, tryptophan fluorescence decreased rapidly by 5.4 and 4.6%, respectively, and reached the level of E2·BeF3− with bound TG (E2·BeF3−(TG), see Table 1). TNP-AMP superfluorescence (supplemental Fig. S5, A and B) and proteolysis (Table 1) also demonstrated that E1Ca2·BeFx was converted by TG to E2·BeF3−(TG). Importantly, as described under supplemental Fig. S4, two Ca2+ occluded in E1Ca2·BeFx are most likely released into the lumen by the TG-induced structural perturbation and trapped in the lumen by the bound TG, as TG fixes the lumenal gate in the closed state and suppresses Ca2+ leakage (16, 55).
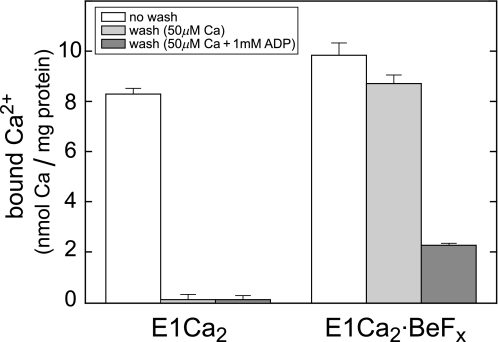
E1Ca3·BeFx Is ADP-sensitive
In Fig. 6, two 45Ca2+ occluded in E1Ca2·BeFx were rapidly removed by washing with 1 mm ADP, whereas the occluded 45Ca2+ remained completely without ADP. Thus ADP caused the loss of Ca2+ occlusion. In agreement, ADP binding to E1Ca2·BeFx increased tryptophan fluorescence to the E1Ca2 level, and resulted in the tryptic T2 site cleavage as E1Ca2 with bound ADP (data not shown). By contrast, ADP binding to E2·BeF3− did not alter its structure (data not shown). The ADP-induced decomposition of E1Ca2·BeFx to E1Ca2 was also demonstrated with the ADP-induced loss of TNP-AMP superfluorescence, in contrast to normal superfluorescence development in E2·BeF3− after ADP incubation (data not shown). Thus E1Ca2·BeFx is ADP-sensitive as E1PCa2·Mg, and E2·BeF3− is ADP-insensitive as E2P·Mg.
FIGURE 6.
ADP causes the loss of Ca2+ occlusion in E1Ca2·BeFx. E1Ca2·BeFx or E1Ca2 were produced in 50 μm 45Ca2+ and 15 mm Mg2+ as described in the legend to Fig. 4, then subjected to membrane filtration without and with perfusion for 20 s by the same buffer containing 50 μm non-radioactive Ca2+ (in place of 45Ca2+) with and without 1 mm ADP. The amounts of 45Ca2+ specifically bound and occluded in the Ca2+-ATPase were determined by subtracting the nonspecific background level (2.03 ± 0.04 nmol/mg) obtained with the above perfusion without ADP of the E1Ca2 state. The error bars show the S. D. of three independent experiments.
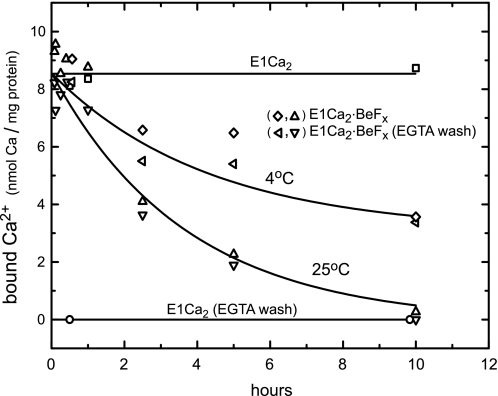
Conversion of E1Ca2·BeFx to E2·BeF3− at 50 μm Ca2+
In Fig. 7, E1Ca2·BeFx was first formed in SR vesicles with BeFx at 25 °C in 50 μm Ca2+ and 15 mm Mg2+, then further incubated at 25 and 4 °C in the presence of these ligands. The amount of bound and occluded Ca2+ was lost slowly (t½ = ∼2 h at 25 °C and ∼7 h at 4 °C). TNP-AMP superfluorescence (Fig. 8) and tryptic and proteinase K proteolyses (data not shown) revealed that E1Ca2·BeFx turned to E2·BeF3− with Ca2+ loss. Thus E1Ca2· BeFx proceeded its spontaneous slow conversion to E2·BeF3−, as the autoisomerization of E1PCa2·Mg to E2P·Mg. The Ca2+ ions released into the lumen may leak out during such long periods. In E1Ca2·AlFx and E1Ca2·AlF4−·ADP, the amount of bound (occluded) Ca2+ was not decreased during the above 10-h incubation at 25 °C (data not shown). The proteolysis showed that these complexes were not converted to the Ca2+-released forms, E2·AlF4− (E2·AlFx) with and without ADP (data not shown). The results indicate that the product state E1PCa2·Mg in the phosphoryl transfer acquires the structure ready for autoisomerization to E2P·Mg releasing Ca2+, whereas the transition state structure is not yet fully prepared for autoisomerization to the Ca2+-released E2P form. Interestingly, as described in supplemental Figs. S4 and S5 (with Refs. 56 and 57), the conversion E1Ca2·BeFx → E2·BeF3− was markedly accelerated by the transmembrane structural perturbation with hydrophobic reagents such as A23187, lasalocid, and C12E8, as well as TG. In contrast, E1Ca2·AlFx and E1Ca2·AlF4−·ADP were resistant against these reagents.
FIGURE 7.
Conversion of E1Ca2·BeFx to E2·BeF3− with Ca2+ release. The complex E1Ca2·BeFx was produced at 25 °C in 50 μm 45Ca2+ and 15 mm MgCl2 as described in the legend to Fig. 4. The complex was then further incubated in the presence of these ligands at 25 and 4 °C for various periods, and the bound 45Ca2+ was determined with and without EGTA washing as described in the legend to Fig. 4A. In the control, the bound 45Ca2+ in E1Ca2 without BeFx was determined.
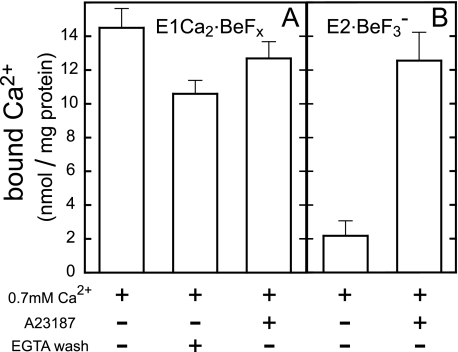
E1Ca2·BeFx Is Perfectly Stabilized at 0.7 mm Ca2+
As found here, Ca2+ binding at high affinity transport sites in E1Ca2 is obligatorily required for E1Ca2·BeFx formation, whereas millimolar high Ca2+ (Ca2+ ligation at the catalytic Mg2+ site I) decomposes this complex to E1Ca2. Furthermore, E1Ca2·BeFx at 50 μm Ca2+ is spontaneously and slowly converted to E2·BeFx releasing Ca2+, and the conversion is markedly accelerated by transmembrane perturbation with hydrophobic reagents such as C12E8 and A23187 (see supplemental materials). The results showed that E1Ca2·BeFx as the E1PCa2·Mg analog possesses the structure prepared for its isomerization to E2·BeF3− with Ca2+ release as E1PCa2·Mg → E2P·Mg + 2Ca2+. On the other hand, it is essential for crystallographic studies to find conditions to perfectly stabilize the E1Ca2·BeFx complex. In Fig. 8A, E1Ca2·BeFx was first formed in 0.1 mm Ca2+ and 15 mm Mg2+, then further incubated with various concentrations of Ca2+ with and without A23187. The structural state was monitored by TNP-AMP superfluorescence (Fig. 8), proteolysis, and tryptophan fluorescence (see Table 1 for representative data). Then we successfully found that Ca2+ at a very narrow concentration range, 0.7 mm, perfectly stabilizes E1Ca2·BeFx and maintains this complex for at least 12 days at 25 °C (and 4 °C) even in the presence of A23187. The 45Ca2+ binding measurements on E1Ca2·BeFx in 0.7 mm 45Ca2+ demonstrated that two Ca2+ ions are bound and occluded in this complex (Fig. 9A).
FIGURE 9.
Two Ca2+ are bound in E1Ca2·BeFx formed from E1Ca2 and from E2·BeF3− at 0.7 mm Ca2+. A, SR vesicles in 0.7 mm 45Ca2+ were incubated with BeFx in the absence or presence of 5 μm A23187, otherwise as described in the legend to Fig. 4. The amount of bound Ca2+ was obtained with subtraction of the background level (10.1 ± 0.5 nmol/mg (n = 6)) determined by EGTA washing the vesicles incubated without BeFx and A23187. The occluded Ca2+ was determined by EGTA washing in the absence of A23187 and by subtraction of the background level. (Here the determination of occluded Ca2+ in A23187 by EGTA washing was not feasible, because in the absence of (or even in 0.1 mm) Ca2+, A23187 converts E1Ca2·BeFx very rapidly to E2·BeF3− releasing Ca2+ (supplemental Figs. 4 and 5). B, E2·BeF3− was first produced in SR vesicles without A23187 and Ca2+. Subsequently, the samples were diluted 10 times with the buffer containing 45CaCl2 and BeFx with and without 5 μm A23187, to give 0.7 mm 45Ca2+ and the same buffer conditions as in A. At 15 s after dilution, the amount of bound 45Ca2+ was determined without EGTA washing and by subtracting the nonspecific Ca2+ binding (1.0 ± 0.1 nmol/mg (n = 6)) determined by EGTA washing the sample incubated without BeFx and A23187.
The perfectly stable E1Ca2·BeFx was produced from E1Ca2 even in the presence of A23187 if 0.7 mm Ca2+ was included before BeFx addition (Fig. 8, B and F, and Table 1). Also, E1Ca2·BeFx was successfully produced with the Ca2+-ATPase purified from SR vesicles by delipidation with deoxycholate (30); in this case again, by including 0.7 mm Ca2+ before BeFx addition. E1Ca2·BeFx thus produced with the purified and delipidated Ca2+-ATPase was perfectly stable at least for 12 days at 4 and 25 °C in 0.7 mm Ca2+ (Fig. 8, B and G, at 25 °C).
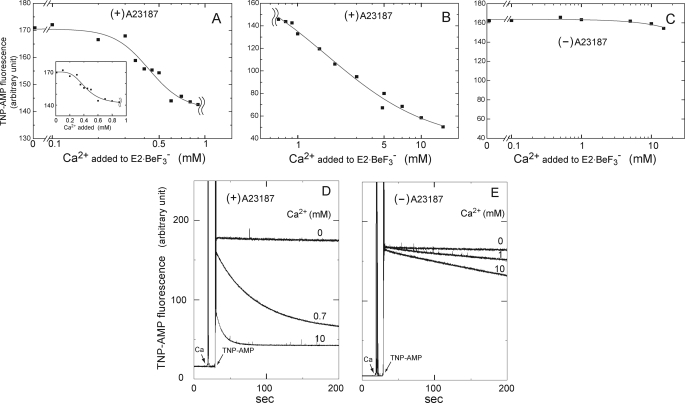
E1Ca2·BeF3− Is Produced from E2·BeF3− by Lumenal Ca2+ Binding
We successfully found also that E1Ca2·BeFx (E1Ca2·BeF3−) can be produced from E2·BeF3− by lumenal Ca2+ binding, as mimicking the lumenal Ca2+-induced reverse transition, E2P·Mg + 2Ca2+ → E1PCa2·Mg. In Fig. 10, we added various concentrations of Ca2+ to E2·BeF3− formed in SR vesicles in 15 mm Mg2+ without Ca2+ in the presence and absence of A23187, then at 10 s after Ca2+ addition the structural state was examined by TNP-AMP superfluorescence. With increasing Ca2+ to 0.7 mm in the presence of A23187, the stable superfluorescence of E2·BeF3− was converted to the transient and slightly lower superfluorescence characteristic of E1Ca2·BeFx with K0.5 of 0.4 mm Ca2+ and a Hill coefficient of 4 (Fig. 10, A and D). A further Ca2+ increase in the millimolar range caused the marked loss of superfluorescence with K0.5 of 1.7 mm and a Hill coefficient of 1 (Fig. 10, B and D). The proteolysis also clearly showed that E2·BeF3− was converted to E1Ca2·BeFx by 0.7 mm Ca2+ in A23187 (Table 1), and this complex was further decomposed to E1Ca2 by 10 mm Ca2+ (data not shown). In Fig. 9B, two 45Ca2+ were shown to be bound producing E1Ca2·BeFx, when 0.7 mm 45Ca2+ was added to E2·BeF3− in the presence of A23187. In contrast, in the absence of A23187, E2·BeF3− was neither converted to E1Ca2·BeFx nor decomposed to E1Ca2 even at 10 mm Ca2+ (Figs. 9B and 10, C and E, and Table 1 (proteolysis at 0.7 mm Ca2+)).
FIGURE 10.
Formation of E1Ca2·BeF3− from E2·BeF3− by lumenal Ca2+ binding. E2·BeF3− was first produced in SR vesicles with BeFx in the presence and absence of 1.2 μm A23187 at 25 °C in 0.5 mm EGTA, 15 mm MgCl2, 0.1 m KCl, and 30 mm MOPS/Tris (pH 7.0). Then a small volume of CaCl2 was added to give various Ca2+ concentrations as indicated. At 10 s after this Ca2+ addition, TNP-AMP fluorescence was monitored with 4 μm TNP-AMP. In A and B, the maximum intensity in the presence of A23187 was plotted in semi-log scale at 0–0.9 mm Ca2+ (A) and 0.7–15 mm Ca2+ (B). Note also the different scales in the ordinates. The inset in A is the linear plot at 0–0.9 mm Ca2+ to clearly show the saturation of the first phase of the Ca2+-dependent change, i.e. the formation of E1Ca2·BeF3− from E2·BeF3−. In C, the maximum superfluorescence intensity in the absence of A23187 was plotted at 0–15 mm Ca2+. In D and E, the traces of superfluorescence development upon TNP-AMP addition in the presence of A23187 (D) or its absence (E) were shown at the representative Ca2+ concentrations (0, 0.7 or 1.0, and 10 mm). Solid lines in A and B show the least squares fit to the Hill equation. Apparent Ca2+ affinity (K0.5) and Hill coefficients at 0–0.9 mm Ca2+ are 0.43 mm and 3.8 with the intensity decrease from 170 to 142, and those at Ca2+ over 0.7 mm are 1.7 mm and 1.0 with a further intensity decrease to 36.
The results demonstrated that E1Ca2·BeFx, most probably E1Ca2·BeF3−, was produced from E2·BeF3− by the lumenal Ca2+ binding at the lumenally oriented low affinity transport sites, and further that Ca2+ substitution of Mg2+ at the catalytic site in E1Ca2·BeF3− produced from E2·BeF3− caused its decomposition to E1Ca2, therefore as the change E2·BeF3− + 2Ca2+ → E1Ca2·BeF3− → E1Ca2. Note that Mg2+ bound at the catalytic site in E2P·Mg is occluded, whereas it is not and therefore is exchangeable in E1PCa2·Mg (42). Thus, these two distinctly different states of the ligated Mg2+ at the catalytic site (site I) in E2P·Mg and E1PCa2·Mg are obviously mimicked here by the respective analogs E2·BeF3− and E1Ca2·BeF3−. The perfect stabilization of E1Ca2·BeF3− achieved by 0.7 mm Ca2+ (Fig. 10) obviously involves lumenal Ca2+ binding and prevention of the Ca2+ release into the lumen. The stabilization by 0.7 mm Ca2+ in the absence of A23187 is probably due to Ca2+ moved passively into the vesicle lumen during the long incubation periods. All these findings show that the forward and reverse transition, E1PCa2·Mg ↔ E2P·Mg + 2Ca2+, is mimicked by the forward and reverse conversion, E1Ca2·BeF3− ↔ E2·BeF3− + 2Ca2+.
It is of interest to note the Hill coefficient of 4 in the lumenal Ca2+-induced reverse conversion, E2·BeF3− + 2Ca2+ → E1Ca2·BeF3− at 0.1–0.7 mm Ca2+ in Fig. 10A. This might be indicative of the existence of lumenal Ca2+ access sites in addition to transport sites and their possible cooperative involvement in lumenal Ca2+ access to the transport sites. In fact, two such sites besides the two transport sites have been suggested by the kinetics and protein-chemical study on the lumenal loops (58, 59).
DISCUSSION
Formation of E1Ca2·BeF3−
As a structural analog of the physiological intermediate E1PCa2·Mg, the E1Ca2·BeFx complex was successfully produced by BeFx binding to the E1Ca2 state Ca2+-ATPase and from E2·BeF3− by lumenal Ca2+ binding to the lumenally oriented low affinity transport sites. All the revealed properties of E1Ca2·BeFx met the requirements for the E1PCa2·Mg analog; i.e. two Ca2+ occluded at the transport sites, Mg2+ bound (but not occluded) at the catalytic site, the ADP-released but still ADP-sensitive state, and its isomerization to the ADP-insensitive Ca2+-released state E2P·Mg (E2·BeF3−) and reversal by lumenal Ca2+ binding, E1PCa2·Mg ↔ E2P·Mg + 2Ca2+.
Furthermore, the coordination chemistry of beryllium fluoride, actually BeF3−, fulfills the requirement of E1Ca2·BeFx as the E1PCa2·Mg analog. In chemistry, beryllium fluoride compounds are known to adopt tetrahedral geometry with the Be-F 1.55-Å bond length, thereby making them strictly isomorphous to the tetrahedral phosphate group (60). Moreover, because of the high charge density due to the small size, beryllium is able to coordinate the aspartate-oxygen in addition to F−. The -O-BeF3− thus produced with Asp351-oxygen in fact possesses the tetrahedral geometry superimposable with the covalently bound phosphate at the aspartate, as actually seen in E2·BeF3−, the E2P·Mg ground-state analog (21, 22, 25). MgF42− also possesses the tetrahedral geometry, but magnesium is not able to be coordinated directly with the Asp351-oxygen, as seen in E2·MgF42−, the E2·Pi analog. AlF4− in E1Ca2·AlF4−·ADP and E2·AlF4− (17, 18, 20) (or AlF3 in some cases in the haloacid dehalogenase superfamily (61)) possesses planar geometry, in which Asp351-oxygen and ADP β-phosphate or the hydrolytic water (E2·AlF4−) coordinate the aluminum at apical positions producing the bipyramidal structure superimposable to the penta-coordinated phosphorus in the transition state of in-line phosphoryl transfer E1PCa2·ADP·Mg‡ and acylphosphate hydrolysis E2-P·Mg‡. Thus all chemical properties of the Pi analogs agree with the conclusion that E1Ca2·BeFx is the analog for E1PCa2·Mg, and BeFx is most probably BeF3−, i.e. E1Ca2·BeF3−.
Here note that the replacement of phosphate with BeF3− produces stabilization of the E1PCa2·Mg structure with the same geometry of BeF3− as phosphate, and therefore probably with the same binding residues for them within the catalytic site. The E1Ca2·BeF3− stability is likely brought about by the specific chemical nature of fluoride. Namely, it possesses a significantly higher electronegativity than oxygen (actually the highest among all atoms) and a small size, therefore producing stronger BeF3− binding in the catalytic site and fixing the intermediate structure.
Structure of E1Ca2·BeF3−
Then with the newly developed E1Ca2·BeF3−, we explored its structural properties and uncovered the hitherto unknown nature of the physiological intermediate E1PCa2·Mg and structural changes during the phosphoryl transfer/ADP release and subsequent EP isomerization/Ca2+ release. The observed proteinase K resistance of Thr242 on the A/M3-linker revealed that, in E1PCa2·Mg (E1Ca2·BeF3−) the A domain is already rotated perpendicular to the membrane plane from the position in E1Ca2, thereby bringing up its junction with the A/M3-linker and imposing a strain on this linker, similarly to E1Ca2·AlF4−·ADP and E1Ca2·CaAMP-PCP (17, 18). As described for the E1Ca2·CaAMP-PCP structure (18), the A/M3-linker strain, i.e. the A domain perpendicular rotation is brought about by bending the P domain due to binding of the phosphate moiety and Ca2+ at the catalytic site (Mg2+-site I, Asp351/Thr353/Asp703) on the P domain (see Figs. 4 and 5 in Ref. 18).5 Our results revealed that such a strained state is achieved even without the N-P domain cross-linking nucleotide but solely with BeF3− and Mg2+ binding at the catalytic site, and therefore remains in E1PCa2·Mg after ADP release.
The strain of the A/M3-linker thus imposed has been predicted with the atomic structure (18, 19) to function as a motive force for the A domain rotation parallel to the membrane in the E1P to E2P isomerization. The partial resistance at T2 site Arg198 in E1Ca2·BeF3− (as compared with the rapid cleavage in E1Ca2·CaAMP-PCP/E1Ca2·AlF4−·ADP and E1Ca2·AlFx) further indicated that in E1PCa2·Mg, the A domain is already likely rotated parallel to the membrane to some extent from the position in E1Ca2·MgATP*/E1PCa2·ADP·Mg‡. Thus, the A/M3-linker strain is likely functioning for this partial A domain rotation during the phosphoryl transfer/ADP release to produce E1PCa2·Mg, and further for the large and complete rotation to achieve the tight A-P domain association at Arg198 on the Val200 loop in the Ca2+-released state E2P·Mg (E2·BeF3−). The A-P domain interaction at the Val200 loop is actually critical for formation of the proper Ca2+-released structure, E2P·Mg and its analog E2·BeF3− (25, 62, 63).
Here, it is of interest to note that residues Asp351, Thr353, and Asp703 ligating Mg2+ and phosphate will come more proximate to each other during E1PCa2·ADP·Mg‡ → E1PCa2·Mg + ADP (E1Ca2·AlF4−·ADP/E1Ca2·AlFx → E1Ca2·BeFx). As a consequence, a further P domain bending and more strain for the A/M3-linker will likely be induced by this coordination-chemical change, thereby contributing to inducing the A domain rotations during E1PCa2·Mg formation and subsequent isomerization to E2PCa2·Mg (besides the release of the N-P domain cross-linking nucleotide ADP). In any case, our results show that E1PCa2·Mg (E1Ca2·BeF3−) as the product of the phosphorylation reaction acquires the structure ready for isomerization and Ca2+ deocclusion/release (i.e. ready for the large A domain rotation to produce E2P·Mg (E2·BeF3−)), whereas the transition state structure in the phosphorylation (E1Ca2·AlF4−·ADP and E1Ca2·AlFx) is not yet fully prepared. Note again that the E1PCa2·Mg structure before such motions for its isomerization to E2P·Mg is stabilized with replacement of phosphate with BeF3− in E1Ca2·BeF3−.
Important also, we found that the Ca2+-occluded transmembrane structure adopts not simply one state, but will proceed through changes during the phosphoryl transfer and ADP release to form E1PCa2·Mg (see supplemental Fig. S3E). The structural change is probably coupled with the above described motions of the P and A domains (more bending and rotation) during this process. In the subsequent Ca2+ deocclusion/release in E1PCa2·Mg → E2P·Mg + 2Ca2+, the transmembrane structure changes further (52), which was also clearly mimicked here in the change E1Ca2·BeF3− → E2·BeF3−. Thus, the structures of the transmembrane domain as well as the cytoplasmic domains in E1Ca2·BeF3− (E1PCa2·Mg) are intermediate between those of E1Ca2·AlF4−·ADP (E1PCa2·ADP·Mg‡) and E2·BeF3− (E2P·Mg).
Mg2+ as Physiological Catalytic Cation
Important questions regarding the E1Ca2·BeF3− structure are why Ca2+ coordination at the catalytic Mg2+ site (site I, Asp351/Thr353/Asp703) is absolutely unfavorable for E1Ca2·BeF3− formation, and why the Mg2+-coordinated structure E1Ca2·BeF3− differs from Ca2+-coordinated E1PCa2·Ca, E1Ca2·CaAMP-PCP, and E1PCa2·Ca·AMP-PN structures as well as from E1Ca2· AlF4−·ADP/E1Ca2·AlFx, especially in A domain positioning. These questions may be relevant to the questions of why forward isomerization of E1PCa2·Ca to E2P is markedly retarded in contrast to E1PCa2·Mg and thus why Mg2+ is preferred as the catalytic cation. In stringent coordination chemistry, the coordination distance of Mg2+ is shorter than Ca2+, typically by 0.2 Å (e.g. 2.1 versus 2.3 Å (64, 65)). As a consequence, in the case of E1Ca2·CaAMP-PCP, the distance between the γ-phosphate and Asp351-oxygen becomes 3.24 Å, being greater by 0.3 Å than that predicted in E1Ca2·MgAMP-PCP. Therefore MgAMP-PCP (MgATP) binding would result in steric clash, and E1Ca2·CaAMP-PCP is more stable than E1Ca2·MgAMP-PCP, and therefore has less tendency to decompose to E1Ca2 (also in the forward direction to the EP formation and its decay in the case of E1Ca2·CaATP) (45). In E1Ca2·BeF3− formed here with Mg2+, the direct coordination between Asp351 and the beryllium and their proximate positioning would probably favor the closely positioned ligand residues (Thr353/Asp703/Asp351) for BeF3− and Mg2+ but not for Ca2+. Therefore Ca2+ substitution of Mg2+ probably disrupted the precise geometry and decomposed the E1Ca2·BeF3− complex. Also, a possible difference in the coordination number might be involved; Mg2+ prefers definitely six, whereas Ca2+ can accommodate seven or eight ligands (65–69).
Furthermore, the difference in A domain positioning between the Mg2+-coordinated state E1Ca2·BeF3− and the Ca2+-coordinated states may be reasonably understood by the consequence of the stringent coordination chemistry. Namely, because the shorter coordination distance of Mg2+, P domain bending, and the resulting A domain rotation perpendicular to the membrane will be greater in the Mg2+-coordinated state. Therefore the strain of the A/M3-linker and A domain rotation parallel to the membrane will be more in the Mg2+ state E1Ca2·BeFx. In this context, it is also reasonable that E1PCa2·Mg is more rapidly isomerized to E2P with less energy barrier for the large A domain rotation, in contrast to the retarded isomerization in E1PCa2·Ca that is stabilized by the likely conformational inadequacy. Here note that the cause of the E1PCa2·Ca stabilization is obviously different from that of E1PCa2·Mg stabilization produced by replacement of phosphate with BeF3− (see the above discussion for E1Ca2·BeF3− stabilization).
Previously it was documented (45, 64, 70) that destabilization of the non-covalent complex E1Ca2·MgATP by Mg2+ (as found with MgAMP-PCP versus CaAMP-PCP) together with stabilization of the transition state by Mg2+ (as found with E1Ca2·AlF4−·ADP bound Mg2+ at both sites I and II) leads to a decrease of the activation energy and a rapid phosphoryl transfer. As another critical reason for Mg2+ preference for catalysis, we predict here by exploring the property of the E1Ca2·BeFx that the Mg2+ bound at the catalytic site produces the proper E1PCa2 structure, which is ready for rapid transition to E2P in this rate-limiting process of the transport cycle.
Hydrophobic Catalytic Site in E1Ca2·BeF3−
The microenvironment around Asp351 in E1PCa2·Mg was further predicted by TNP-AMP superfluorescence in E1Ca2·BeF3− to be strongly hydrophobic and thus a closed state, and this will become even more in the change E1PCa2·Mg → E2P·Mg + 2Ca2+ (E1Ca2·BeF3− → E2·BeF3−). The observed distinct difference between E1Ca2·BeF3− and E2·BeF3− (transient versus stable and slightly higher superfluorescence) is probably ascribed to the distinct difference in the organization state of cytoplasmic domains. The superfluorescence, nevertheless, developed solely in the Ca2+-ATPase complexed with beryllium fluoride, E1Ca2·BeF3− and E2·BeF3− (E2P·Mg), but no development in E1Ca2·AlFx and in E2·AlF4− and E2·MgF42− (25). Therefore, the hydrophobic closed catalytic site is accomplished by the direct coordination and close proximity of the beryllium with Asp351-oxygen and by the specific coordination of the tetrahedral -O-BeF3−, i.e. Asp351-acylphosphate within the catalytic site. This is obviously not the case in AlF4− (the penta-coordinated phosphorus of the transition states) and MgF42− (non-covalently bound Pi), thus in these states, the catalytic site is more accessible to nonspecific water molecules.
Whether E1PCa2·Mg develops the superfluorescence had been controversial. In addition to the obvious problem of TNP-AMP competition against ATP for phosphorylation, the observed TNP-AMP-induced decomposition of E1Ca2·BeF3− further revealed that the E1PCa2·Mg structure may be similarly disrupted rapidly by TNP-AMP binding, therefore making it virtually impossible to examine the superfluorescence development in E1PCa2·Mg. The TNP-AMP-induced E1Ca2·BeF3− decomposition might have occurred by means of a similar structural change as the ADP-induced one, i.e. disruption of the cytoplasmic domain organization and possible BeF3− release. The most important conclusion here is that the hydrophobic and closed property of the phosphorylated catalytic site both in E1PCa2·Mg and E2P·Mg may be requisite to avoid a possible attack of nonspecific water molecules on the Asp351-acylphosphate thus accomplishing Ca2+ release into the lumen and energy coupling.
Formation of E1Ca2·BeF3− from E2·BeF3− and Perfect Stabilization of E1Ca2·BeF3−
E1Ca2·BeF3− was produced also from E2·BeF3− by binding two lumenal Ca2+ to the lumenally oriented low affinity transport sites at 0.7 mm Ca2+ and 15 mm Mg2+, as mimicking the reverse transition E2P·Mg + 2Ca2+ → E1PCa2·Mg. At the critical concentration of 0.7 mm Ca2+ in 15 mm Mg2+, E1Ca2·BeF3− is perfectly stabilized without decomposition to E1Ca2 or conversion to E2·BeF3−. The perfect E1Ca2·BeF3− stabilization is obviously achieved by preventing Ca2+ release into the lumen and by avoiding the absolutely unfavorable Ca2+ substitution of Mg2+ in site I at the most appropriately balanced concentrations of Ca2+ and Mg2+. As noted in the last paragraph under “Results,” stabilization of E1Ca2·BeF3− might possibly involve lumenal Ca2+ access at the putative lumenal gating sites besides the transport sites. If this is the case, the gate-opening and Ca2+ release into the lumen takes place when the lumenal Ca2+ is low enough to avoid the possible lumenal Ca2+ access to the gate.
Integrated Picture of EP Processing
Recently, we successfully identified and trapped for the first time the intermediate state E2PCa2·Mg, ADP-insensitive EP with two Ca2+ occluded at transport sites, by elongating the A/M1′-linker (71), and revealed that the proper length of this linker is critical for inducing structural changes for Ca2+ deocclusion and release from E2PCa2·Mg. This dependence on the length of the linker is probably because the length controls the extent of strain between the A domain and M1′, which causes motions of the cytoplasmic A and P domains thereby transmitting the structural signal to the transmembrane transport sites. In trapped E2PCa2·Mg, the A domain is already largely rotated, and A-P domain associations at Val200 and TGES184 loops are already produced, although the interaction network is not produced properly at the Tyr122-hydrophobic cluster (71), which is critical for Ca2+ deocclusion/release and E2P hydrolysis (72–74). In the Ca2+-released E2P·Mg, this cluster is formed from seven residues of the A (Ile179/Leu180/Ile232) and P (Val705/Val726) domains and the top part of M2 (Leu119/Tyr122) (see Fig. 2).
The results indicated that the successive structural changes take place as follows: in E1PCa2·Mg → E2PCa2·Mg, the A domain rotates largely (further from the position in E1PCa2·Mg) into the space between the N and P domains and docks onto the Asp351-acylphosphate of the P domain, thereby causing loss of ADP sensitivity and also the strain of the A/M1′-linker (because the A domain is brought above the P domain). The strain thus imposed will cause inclinations of the A and P domains and the connected M2 and M4/M5 thereby rearranging the helices to destroy Ca2+ sites and open the lumenal gate thus to release Ca2+. Upon these motions, the Tyr122-hydrophobic cluster is produced from the inclined A and P domains and M2. Hence, interactions at this cluster and at the Val200 loop stabilize the Ca2+-released structure E2P·Mg, and also produce the catalytic site for the acylphosphate hydrolysis to occur after Ca2+ release, ensuring energy coupling (63, 72–74). Atomic level structural studies of E1Ca2·BeF3− as E1PCa2·Mg and the trapped intermediate state E2PCa2·Mg will contribute to further understanding of EP processing, Ca2+ handling, and E2P hydrolysis.
Supplementary Material
Acknowledgment
We thank Dr. Chikashi Toyoshima, University of Tokyo, for helpful discussions.
This work was supported by a grant-in-aid for scientific research (B) (to H. S.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The proteolytic analysis of E1PCa2·Ca was made possible by its markedly retarded decay due to Ca2+ ligation at the catalytic site (27) and feedback inhibition by the high lumenal Ca2+. Analysis of E1PCa2·Mg formed from MgATP was not feasible because of its very rapid turnover, thus of a very rapid ATP exhaustion.
The fluorescence level after the decrease of the transient superfluorescence of E1Ca2·BeFx was somewhat higher than the non-superfluorescent low level of TNP-AMP bound to E1Ca2 especially at 25 °C. We obtained the results indicating that a small fraction of E2·BeF3− was produced even in the presence of 50 μm Ca2+ (more at 25 than at 4 °C) after the TNP-AMP-induced E1Ca2·BeFx decomposition to E1Ca2, and therefore remained somewhat superfluorescence (data not shown).
As depicted in Figs. 4 and 5 by Toyoshima et al. (18) for the change E1Ca2 → E1Ca2·CaAMP-PCP, the top part of the first half of the P domain (Pβ1–Pβ4) moves together as a result of γ-phosphate and Ca2+ (Mg2+) binding, because Thr353 just above Pβ1 coordinates to both ligands. Furthermore, Pβ5 twists upon binding of Ca2+ (Mg2+) because of the coordination by Asp703, which causes Pα5–Pα7 tilting. Thus the P domain is bent. This bending causes the perpendicular A domain rotation because the P7 helix moves upwards and tilts so that Gly156–Lys158 on the A domain is brought up as they are in contact with Ala725–Val726 on top of the P7 helix.
- SERCA1a
- adult fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase
- SR
- sarcoplasmic reticulum
- EP
- phosphoenzyme
- E1P
- ADP-sensitive phosphoenzyme
- E2P
- ADP-insensitive phosphoenzyme
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- TG
- thapsigargin
- TNP-AMP
- 2′(3′)-O-(trinitrophenyl)-AMP
- C12E8
- octaethylene glycol monododecyl ether
- N
- nucleotide binding
- P
- phosphorylation
- A
- actuator
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- AMP-PCP
- adenosine 5′-(β,γ-methylene)triphosphate
- AMP-PN
- adenosine 5′-diphosphoramidate.
REFERENCES
- 1.Hasselbach W., Makinose M. (1961) Biochem. Z. 333, 518–528 [PubMed] [Google Scholar]
- 2.Ebashi S., Lipmann F. (1962) J. Cell Biol. 14, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inesi G., Sumbilla C., Kirtley M. E. (1990) Physiol. Rev. 70, 749–760 [DOI] [PubMed] [Google Scholar]
- 4.Møller J. V., Juul B., le Maire M. (1996) Biochim. Biophys. Acta 1286, 1–51 [DOI] [PubMed] [Google Scholar]
- 5.MacLennan D. H., Rice W. J., Green N. M. (1997) J. Biol. Chem. 272, 28815–28818 [DOI] [PubMed] [Google Scholar]
- 6.McIntosh D. B. (1998) Adv. Mol. Cell. Biol. 23A, 33–99 [Google Scholar]
- 7.Toyoshima C., Inesi G. (2004) Annu. Rev. Biochem. 73, 269–292 [DOI] [PubMed] [Google Scholar]
- 8.Toyoshima C. (2008) Arch. Biochem. Biophys. 476, 3–11 [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima C. (2009) Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto T., Tonomura Y. (1968) J. Biochem. 64, 137–145 [DOI] [PubMed] [Google Scholar]
- 11.de Meis L., Masuda H. (1974) Biochemistry 13, 2057–2062 [DOI] [PubMed] [Google Scholar]
- 12.Masuda H., de Meis L. (1973) Biochemistry 12, 4581–4585 [DOI] [PubMed] [Google Scholar]
- 13.Kanazawa T., Boyer P. D. (1973) J. Biol. Chem. 248, 3163–3172 [DOI] [PubMed] [Google Scholar]
- 14.Inesi G., Kurzmack M., Coan C., Lewis D. E. (1980) J. Biol. Chem. 255, 3025–3031 [PubMed] [Google Scholar]
- 15.Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C., Nomura H. (2002) Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 17.Sørensen T. L., Møller J. V., Nissen P. (2004) Science 304, 1672–1675 [DOI] [PubMed] [Google Scholar]
- 18.Toyoshima C., Mizutani T. (2004) Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 19.Toyoshima C., Nomura H., Tsuda T. (2004) Nature 432, 361–368 [DOI] [PubMed] [Google Scholar]
- 20.Olesen C., Sørensen T. L., Nielsen R. C., Møller J. V., Nissen P. (2004) Science 306, 2251–2255 [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima C., Norimatsu Y., Iwasawa S., Tsuda T., Ogawa H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19831–19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 23.Danko S., Daiho T., Yamasaki K., Kamidochi M., Suzuki H., Toyoshima C. (2001) FEBS Lett. 489, 277–282 [DOI] [PubMed] [Google Scholar]
- 24.Danko S., Yamasaki K., Daiho T., Suzuki H., Toyoshima C. (2001) FEBS Lett. 505, 129–135 [DOI] [PubMed] [Google Scholar]
- 25.Danko S., Yamasaki K., Daiho T., Suzuki H. (2004) J. Biol. Chem. 279, 14991–14998 [DOI] [PubMed] [Google Scholar]
- 26.Shigekawa M., Wakabayashi S., Nakamura H. (1983) J. Biol. Chem. 258, 8698–8707 [PubMed] [Google Scholar]
- 27.Wakabayashi S., Shigekawa M. (1987) J. Biol. Chem. 262, 11524–11531 [PubMed] [Google Scholar]
- 28.Nakamura S., Suzuki H., Kanazawa T. (1994) J. Biol. Chem. 269, 16015–16019 [PubMed] [Google Scholar]
- 29.Barrabin H., Scofano H. M., Inesi G. (1984) Biochemistry 23, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 30.Meissner G., Fleischer S. (1974) Methods Enzymol. 32, 475–481 [DOI] [PubMed] [Google Scholar]
- 31.Kubota T., Daiho T., Kanazawa T. (1993) Biochim. Biophys. Acta 1163, 131–143 [DOI] [PubMed] [Google Scholar]
- 32.Troullier A., Girardet J. L., Dupont Y. (1992) J. Biol. Chem. 267, 22821–22829 [PubMed] [Google Scholar]
- 33.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34.Hiratsuka T. (1982) Biochim. Biophys. Acta 719, 509–517 [DOI] [PubMed] [Google Scholar]
- 35.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 36.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 37.Webb M. R., Trentham D. R. (1981) J. Biol. Chem. 256, 4884–4887 [PubMed] [Google Scholar]
- 38.Hasselbach W., Fassold E., Migala A., Rauch B. (1981) Fed. Proc. 40, 2657–2661 [PubMed] [Google Scholar]
- 39.González D. A., Ostuni M. A., Lacapère J. J., Alonso G. L. (2006) Biophys. Chem. 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 40.Yamada S., Ikemoto N. (1980) J. Biol. Chem. 255, 3108–3119 [PubMed] [Google Scholar]
- 41.Kanazawa T. (1975) J. Biol. Chem. 250, 113–119 [PubMed] [Google Scholar]
- 42.Ogurusu T., Wakabayashi S., Shigekawa M. (1991) J. Biochem. 109, 472–476 [DOI] [PubMed] [Google Scholar]
- 43.Möller J. V., Lenoir G., Marchand C., Montigny C., le Maire M., Toyoshima C., Juul B. S., Champeil P. (2002) J. Biol. Chem. 277, 38647–38659 [DOI] [PubMed] [Google Scholar]
- 44.Holdensen A. N., Andersen J. P. (2009) J. Biol. Chem. 284, 12258–12265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picard M., Toyoshima C., Champeil P. (2005) J. Biol. Chem. 280, 18745–18754 [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T., Inesi G. (1982) J. Biol. Chem. 257, 11510–11516 [PubMed] [Google Scholar]
- 47.Nakamoto R. K., Inesi G. (1984) J. Biol. Chem. 259, 2961–2970 [PubMed] [Google Scholar]
- 48.Dupont Y., Chapron Y., Pougeois R. (1982) Biochem. Biophys. Res. Commun. 106, 1272–1279 [DOI] [PubMed] [Google Scholar]
- 49.de Meis L., Martins O. B., Alves E. W. (1980) Biochemistry 19, 4252–4261 [DOI] [PubMed] [Google Scholar]
- 50.Dupont Y., Pougeois R. (1983) FEBS Lett. 156, 93–98 [DOI] [PubMed] [Google Scholar]
- 51.Champeil P., Le Maire M., Moller J. V., Riollet S., Guillain F., Green N. M. (1986) FEBS Lett. 206, 93–98 [DOI] [PubMed] [Google Scholar]
- 52.Suzuki H., Kanazawa T. (1995) J. Biol. Chem. 270, 3089–3093 [DOI] [PubMed] [Google Scholar]
- 53.Dupont Y., Leigh J. B. (1978) Nature 273, 396–398 [DOI] [PubMed] [Google Scholar]
- 54.Obara M., Suzuki H., Kanazawa T. (1988) J. Biol. Chem. 263, 3690–3697 [PubMed] [Google Scholar]
- 55.Inesi G., Lewis D., Toyoshima C., Hirata A., de Meis L. (2008) J. Biol. Chem. 283, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 56.Murphy A. J., Coll R. J. (1993) J. Biol. Chem. 268, 23307–23310 [PubMed] [Google Scholar]
- 57.Lakowitz J. R. (1983) Principles of Fluorescence Spectroscopy, Plenum Press, New York [Google Scholar]
- 58.Myung J., Jencks W. P. (1994) Biochemistry 33, 8775–8785 [DOI] [PubMed] [Google Scholar]
- 59.Webb R. J., Khan Y. M., East J. M., Lee A. G. (2000) J. Biol. Chem. 275, 977–982 [DOI] [PubMed] [Google Scholar]
- 60.Petsko G. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 538–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W., Cho H. S., Kim R., Jancarik J., Yokota H., Nguyen H. H., Grigoriev I. V., Wemmer D. E., Kim S. H. (2002) J. Mol. Biol. 319, 421–431 [DOI] [PubMed] [Google Scholar]
- 62.Daiho T., Suzuki H., Yamasaki K., Saino T., Kanazawa T. (1999) FEBS Lett. 444, 54–58 [DOI] [PubMed] [Google Scholar]
- 63.Kato S., Kamidochi M., Daiho T., Yamasaki K., Gouli W., Suzuki H. (2003) J. Biol. Chem. 278, 9624–9629 [DOI] [PubMed] [Google Scholar]
- 64.Picard M., Jensen A. M., Sørensen T. L., Champeil P., Møller J. V., Nissen P. (2007) J. Mol. Biol. 368, 1–7 [DOI] [PubMed] [Google Scholar]
- 65.Peeraer Y., Rabijns A., Collet J. F., Van Schaftingen E., De Ranter C. (2004) Eur. J. Biochem. 271, 3421–3427 [DOI] [PubMed] [Google Scholar]
- 66.Stokes D. L., Green N. M. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 445–468 [DOI] [PubMed] [Google Scholar]
- 67.Yang W., Lee H. W., Hellinga H., Yang J. J. (2002) Proteins Struct. Funct. Genet. 47, 344–356 [DOI] [PubMed] [Google Scholar]
- 68.Shannon R. D. (1976) Acta Crystallogr. Sect. A 32, 751–767 [Google Scholar]
- 69.Falke J. J., Drake S. K., Hazard A. L., Peersen O. B. (1994) Q. Rev. Biophys. 27, 219–290 [DOI] [PubMed] [Google Scholar]
- 70.Suzuki H., Nakamura S., Kanazawa T. (1994) Biochemistry 33, 8240–8246 [DOI] [PubMed] [Google Scholar]
- 71.Daiho T., Yamasaki K., Danko S., Suzuki H. (2007) J. Biol. Chem. 282, 34429–34447 [DOI] [PubMed] [Google Scholar]
- 72.Yamasaki K., Daiho T., Danko S., Suzuki H. (2004) J. Biol. Chem. 279, 2202–2210 [DOI] [PubMed] [Google Scholar]
- 73.Wang G., Yamasaki K., Daiho T., Suzuki H. (2005) J. Biol. Chem. 280, 26508–26516 [DOI] [PubMed] [Google Scholar]
- 74.Yamasaki K., Wang G., Daiho T., Danko S., Suzuki H. (2008) J. Biol. Chem. 283, 29144–29155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.