Abstract
Laminins are large heterotrimeric glycoproteins with many essential functions in basement membrane assembly and function. Cell adhesion to laminins is mediated by a tandem of five laminin G-like (LG) domains at the C terminus of the α chain. Integrin binding requires an intact LG1-3 region, as well as contributions from the coiled coil formed by the α, β, and γ chains. We have determined the crystal structure at 2.8-Å resolution of the LG1-3 region of the laminin α2 chain (α2LG1-3). The three LG domains adopt typical β-sandwich folds, with canonical calcium binding sites in LG1 and LG2. LG2 and LG3 interact through a substantial interface, but LG1 is completely dissociated from the LG2-3 pair. We suggest that the missing γ chain tail may be required to stabilize the interaction between LG1 and LG2-3 in the biologically active conformation. A global analysis of N-linked glycosylation sites shows that the β-sandwich faces of LG1 are free of carbohydrate modifications in all five laminin α chains, suggesting that these surfaces may harbor the integrin binding site. The α2LG1-3 structure provides the first atomic view of the integrin binding region of laminins.
The laminins constitute a major class of cell-adhesive glycoproteins that are intimately involved in basement membrane assembly and function. Their essential roles in embryo development and tissue function have been demonstrated by numerous genetic studies and the analysis of severe human diseases resulting from mutations in laminin genes (1–4). All laminins are heterotrimers composed of three different gene products, termed α, β, and γ chains. At present, 16 mouse and human laminins are known, assembled from five α, three β, and three γ chains. The different laminins have characteristic expression patterns and functions in the embryo and adult animal (1). Laminins are cross-shaped molecules: the three short arms are composed of one chain each, while the long arm is a coiled coil of all three chains, terminating in a tandem of five laminin G-like (LG)2 domains, LG1-5, contributed by the α chain (2). Basement membrane assembly requires polymerization via the short arms and cell attachment via the LG1-5 region (5, 6).
Cell adhesion to laminins is mediated by multiple receptors: integrins bind to the LG1-3 region, whereas α-dystroglycan, heparan sulfate proteoglycans, and sulfated glycolipids bind predominantly to sites in the LG4-5 pair (7). Integrins are heterodimers with a large extracellular domain consisting of one α and one β chain, which both span the cell membrane and engage in transmembrane signaling (8). Of the 24 mouse and human integrins, the major laminin binding integrins are α3β1, α6β1, α7β1, and α6β4, which have distinct affinities for the different laminin isoforms (9). Although some studies have reported integrin binding or integrin-mediated cell adhesion to isolated LG domains or tandems (10–12), there is strong evidence to suggest that the coiled coil region and an intact γ chain tail are required for full integrin binding to the laminin LG1-3 region (13–18). Compared with integrin binding to collagen and fibronectin, which is understood in atomic detail (19, 20), the laminin-integrin interaction remains poorly characterized in structural terms. We previously determined crystal structures of the LG4-5 region of the laminin α1 and α2 chains and defined their receptor binding sites (21–23). Here, we report the crystal structure of the remainder of the laminin α2 receptor binding region, LG1-3.
EXPERIMENTAL PROCEDURES
Expression Construct
DNA coding for residues 2136–2746 (QANSI … MVHGP) of the mouse laminin α2 chain was obtained by PCR amplification from a cDNA kindly provided by Takako Sasaki. The sequence corresponds to NCBI entry NP_032507, but is missing residues 2476–2479 corresponding to exon 53 of the mouse LAMA2 gene (24). The PCR primers added EcoRI and NheI sites at the 5′-end and NotI and BamHI sites at the 3′-end. The PCR product was cloned into pBluescript II KS+ using EcoRI and BamHI. Using strand overlap extension PCR, the codons for residues 2566–2578 (TLTPPRRKRRQTT) were replaced by a GGA triplet coding for glycine. The sequence-verified insert was cloned into a modified pCEP-Pu vector (25) using NheI and NotI. After cleavage of the BM-40 sequence signal, a vector-derived APLA sequence remains at the N terminus of the secreted recombinant α2LG1-3 protein, and a AAAHHHHHH sequence is added at the C terminus.
Protein Production
The laminin α2LG1-3 protein was purified from the conditioned medium of episomally transfected 293-EBNA cells. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) (Invitrogen), transfected using FuGENE reagent (Roche Applied Science), and selected with 1 μg/ml puromycin (Sigma). Confluent cells were maintained for 12 h in DMEM with 10% FCS supplemented with 5 μm kifunensine (Industrial Research Ltd), washed twice with phosphate-buffered saline, and incubated with serum-free DMEM supplemented with 5 μm kifunensine for 3 days. The conditioned medium was loaded onto a 5-ml HisTrap column (GE Healthcare) and the α2LG1-3 protein eluted with 0.5 m imidazole in Tris-buffered saline buffer pH 7.4 supplemented with 5 mm calcium (TBS-Ca). The protein was further purified by size exclusion chromatography on a Superdex 200 HR10/30 column (GE Healthcare) run at 0.5 ml/min in TBS-Ca buffer. To remove the N-linked glycan, the α2LG1-3 protein was incubated with endoglycosidase H (expressed in Escherichia coli using a plasmid kindly provided by Dan Leahy) overnight at room temperature in an enzyme:protein ratio of 1:10. The deglycosylated protein was purified from the reaction mixture by a second size exclusion chromatography step in TBS-Ca buffer and concentrated to 7 mg/ml for crystallization.
Crystallization and Diffraction Data Collection
Initial crystallization screening was carried out in 100-nl sitting drops using a Mosquito Nanodrop robot (TTP Labtech). Large, needle-shaped crystals were grown in 2-μl hanging drops using 0.1 m bis-Tris propane pH 6.5–7.0, 0.2 m sodium citrate, 18–22% PEG3350 as precipitant and flash-frozen to 100 K in precipitant solution supplemented with 20% glycerol. The crystals were found to belong to space group P6x with unit cell dimensions a = b = 138.83 Å, c = 73.89 Å; there is one α2LG1-3 molecule in the asymmetric unit, resulting in a solvent content of ∼60%. Diffraction data to 2.8-Å resolution were collected from a weakly diffracting single crystal at 100 K on beamline 10.1 at the SRS Daresbury (λ = 1.045 Å). The diffraction data were processed with MOSFLM and programs of the CCP4 suite (26). Data processing statistics are summarized in Table 1.
TABLE 1.
Crystallographic statistics
| Data collection and reduction | |
| Space group | P65 |
| Unit cell dimensions (Å) | a = b = 138.83, c = 73.89 |
| Resolution range (Å)a | 20.0 (2.95) - 2.80 |
| Unique reflections | 19248 |
| Multiplicity | 3.7 (3.3) |
| Completeness (%) | 95.5 (89.1) |
| Mean I/σ(I) | 11.6 (3.8) |
| Rmerge | 0.099 (0.380) |
| Refinement | |
| Reflections (working set/test set) | 17351/1894 |
| Protein atoms | 3937 |
| Sugar atoms | 64 (4 N-acetylglucosamines) |
| Solvent atoms | 60 H2O, 3 Ca2+ |
| Rcryst/Rfree | 0.213/0.266 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.4 |
| B-factors (Å2) | 1.5 |
| Ramachandran plot (%)b | 82.1/16.6/1.3/0.0 |
a Numbers in parentheses refer to data in the highest resolution shell.
b Residues in most favored, additionally allowed, generously allowed, and disallowed regions (54).
Structure Solution and Refinement
The α2LG1-3 structure was solved in space group P65 by molecular replacement with PHASER (27, 28). Using a partial poly-Ala/Ser model of laminin α2LG4 (22), domains LG1 and LG2 of α2LG1-3 could be located (translation Z-score of 10.3; Rfree = 0.520). All attempts to locate LG3 at this stage proved unsuccessful. After cautious rebuilding with O (29) and refinement with CNS (30), the improved LG1-2 model gave a translation Z-score of 31.4 in PHASER, and LG3 could be located in a partial translation function using a partial poly-Ala/Ser model of laminin α2LG5 (31) (translation Z-score of 13.8, Rfree = 0.476). The α2LG1-3 model was completed in many rounds of rebuilding and refinement. The final model (Rfree = 0.266) encompasses residues 2142–2707, but the following residues are missing due to lack of electron density: 2146–2149 and 2310–2313 in LG1; 2325–2332 in the LG1-2 linker; 2564–2578, 2598–2601, and 2664–2672 in LG3. Refinement statistics are summarized in Table 1. The figures were made with PYMOL. The coordinates of the α2LG1-3 structure have been deposited in the Protein Data Bank (entry code: 2wjs).
RESULTS
Crystal Structure Determination
The laminin α2LG1-3 region contains a basic sequence in LG3, 2571-RRKRR-2575, that is proteolytically cleaved during protein expression in human embryonic kidney 293 cells (24). Our initial efforts in crystallizing α2LG1-3 focused on a triple mutant, R2571A/K2573A/R2575A, which had been shown to be resistant to proteolytic processing (32). However, we found that these three mutations did not completely suppress proteolysis. The RRKRR sequence is located in a long loop that is only present in LG3, but not in any of the other four LG domains of laminin α chains (31). We therefore replaced the entire predicted loop sequence, TLTPPRRKRRQTT, with a single glycine residue. This deletion mutant, hereafter simply termed α2LG1-3, was highly expressed by the 293 cells with no evidence of proteolytic processing. After extensive screening, the best crystals of this construct did not diffract beyond 4-Å resolution. The α2LG1-3 region contains six potential N-linked glycosylation sites, and we suspected that the large amount of glycan was contributing to the poor order of our crystals. We therefore expressed the α2LG1-3 protein in the presence of the α-mannosidase inhibitor kifunensine and removed the resulting immature glycan with endoglycosidase H (33). The endoglycosidase H-treated protein could be crystallized in a new crystal form that diffracted to 2.8-Å resolution. The α2LG1-3 structure was solved by molecular replacement and refined to a free R-factor of 0.266 (Table 1).
Crystal Structure of Laminin α2LG1-3
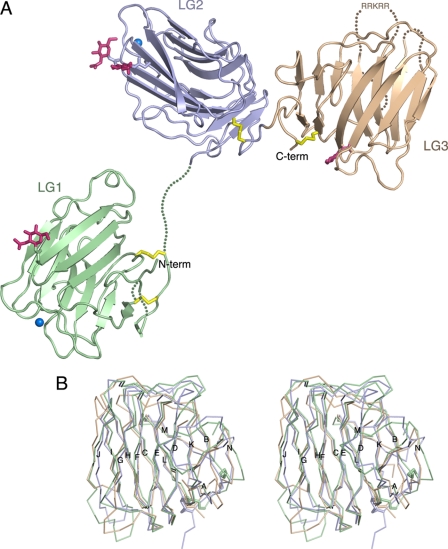
The overall structure of laminin α2LG1-3 in our crystals is shown in Fig. 1A. While there is a substantial interface between domains LG2 and LG3 (see below), LG1 surprisingly makes no contact with either LG2 or LG3, and seven residues in the LG1-2 linker have no electron density. SDS-PAGE analysis of dissolved crystals confirmed that the crystallized protein is intact and not proteolysed at the LG1-2 junction (data not shown). The gap between LG1 and LG2 measures 22.5 Å, a distance that can be spanned by the seven disordered residues in the LG1-2 linker. The unique molecule shown in Fig. 1A is the only one with a physically possible distance between LG1 and LG2; in the crystal lattice, all alternative combinations of LG2–3 with symmetry-related LG1 domains are physically impossible. The open conformation of α2LG1-3 is stabilized by a multitude of crystal packing interactions involving LG1. The dissociation of LG1 from the remainder of the molecule is likely to be a consequence of the missing coiled coil and β and γ chain tails (see “Discussion”).
FIGURE 1.
Crystal structure of laminin α2LG1-3. A, schematic representation of the structure. LG1, LG2, and LG3 are in green, blue, and brown, respectively. Disulfide bridges are in yellow, N-linked carbohydrate moieties are in magenta, and calcium ions are shown as sky blue spheres. Disordered regions of the polypeptide chain are indicated by dotted lines. The location of the RRKRR motif in LG3 (see text) is indicated. B, stereoview of a superposition of Cα traces of LG1 (green), LG2 (blue), and LG3 (brown). Disulfide bridges are shown as thick sticks. The 14 canonical β-strands of laminin LG domains (31) have been labeled sequentially, A–N.
The three LG domains of laminin α2LG1-3 adopt the canonical β-sandwich fold of the LG domain superfamily, including the conserved disulfide bridge near the domain C terminus (7, 34) (Fig. 1B). An additional disulfide bridge (between Cys-2150 and Cys-2321) links the N and C termini of LG1. The A-B and L-N loops are disordered in LG1, but the two disulfide bridges in this region have strong electron density and unambiguously anchor the domain termini. The coiled coil of laminin α chains is predicted to extend right up to β-strand A (35), which has weak density in our structure. LG2 and the linker to LG3 could be traced without interruption. In LG3 three spatially adjacent loops are disordered: D-E (the site of the engineered 12-residue deletion), F-G, and K-L. Four N-acetylglucosamine moieties remaining after endoglycosidase H digestion are included in the model (Fig. 1A). They are attached, respectively, to Asn-2236 (LG1), Asn-2356, and Asn-2431 (LG2), and Asn-2554 (LG3). Patchy electron density at Asn-2644 (LG3) suggests that this residue is also modified. The only potential glycosylation site that does not appear to be modified is Asn-2547 (LG3). Strong spherical electron densities in LG1 and LG2 indicated the presence of two metal ions, which were modeled as calcium (present at 2.5 mm concentration in the crystallization mixture). The calcium ion in LG1 is bound to the side chains of Asp-2189 and Asp-2268 and the carbonyl oxygens of residues 2206 and 2266, with an average metal-ligand density of 2.6 Å. The calcium ion in LG2 is bound to the side chains of Asp-2383 and Asp-2457 and the carbonyl oxygens of residues 2400 and 2455, with an average metal-ligand density of 2.5 Å. The calcium binding sites of α2LG1-3 are in equivalent positions to the functionally important calcium binding sites of laminin α1LG4-5 (21) and α2LG4-5 (22, 31), agrin (36), and neurexins (37–40). At the calcium binding site in LG2, there is additional strong electron density 2.7 Å from the carboxylate group of Asp-2383; this density was modeled satisfactorily as another calcium ion, but it could well be derived from some other, unidentified, buffer component.
Comparison of the three LG domains of laminin α2LG1-3 emphasizes the conserved β-sandwich core of the three domains and shows that LG3 diverges most from the common template of laminin LG domains (Fig. 1B and data not shown). The segments that make up the top rim of the LG domain, especially the B-C and J-K linkers, follow a unique path in LG3, and the long D-E loop that is deleted in our construct would project from this region. LG3 also has an unusually short I-J hairpin. The divergent structure of LG3 explains why this domain was difficult to locate in molecular replacement searches.
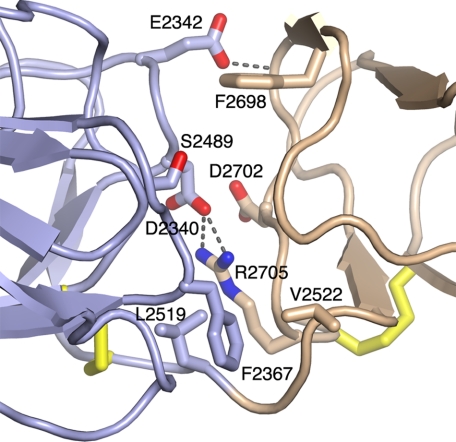
The LG2–3 interface in the laminin α2LG1-3 structure buries as much as 990 Å2 of solvent-accessible surface and thus may be stable under physiological conditions. Prominent features of the interface are: an apolar cluster near the LG2–3 linker, consisting of Phe-2367 (LG2), Leu-2519 (LG2), and Val-2522 (LG3); a salt bridge between Asp-2340 (LG2) and Arg-2705 (LG3); and a hydrogen bond between Glu-2342 (LG2) and the main chain of LG3 (Fig. 2). The relative orientation of LG domains in the α2LG2-3 pair differs substantially from that previously observed in α2LG4-5 (22). When the two pairs are superimposed on their respective N-terminal LG domains (LG2 and LG4), a rotation by 58° degrees is required to bring their C-terminal LG domains (LG3 and LG5) into superposition (not shown).
FIGURE 2.
Structure of the LG2-3 interface. LG2 is in blue and LG3 in brown. Selected residues are shown in atomic detail and have been labeled. Disulfide bridges are in yellow. Dashed lines indicate hydrogen bonds. Glu-2342 accepts a hydrogen bond from a main chain amide nitrogen in LG3. The view direction is similar to Fig. 1A.
Interaction with the γ Chain Tail
Recent studies have shown that a glutamic acid at the C terminus of the laminin γ chain is essential for integrin binding, most likely by promoting an active LG1-3 conformation (15, 18, 41). We used isothermal titration calorimetry to determine whether the crystallized α2LG1-3 protein interacts with a synthetic peptide spanning the last nine residues of the mouse laminin γ1 chain, but no binding could be observed up to a peptide concentration of 70 μm (data not shown). Thus, it appears that the γ chain tail interacts with the α chain only when these two elements are joined in an intact laminin heterotrimer.
Location of Functional Sites
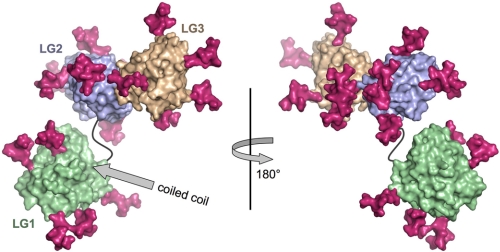
The available data suggest that both the coiled coil and LG1-3 region of laminins are required for integrin binding, but the precise location of the integrin binding site is unknown (13–18). Because bulky carbohydrate modifications often shield much of the surface of extracellular proteins, their absence from certain regions may be indicative of functionally important protein-protein interaction sites (42, 43). We reasoned that the general location of the integrin binding site is likely to be conserved in all laminin α chains and investigated whether there are any large surface regions in LG1-3 that are not obstructed by carbohydrate modifications. This analysis revealed that the overall distribution of putative N-linked carbohydrates in the LG1-3 region is markedly uneven: considering all five mouse and human laminin α chains, there are a total of five sites in LG1, ten in LG2, and another ten in LG3 (Fig. 3 and data not shown). Strikingly, both faces of the LG1 β-sandwich are free of carbohydrate in all laminin α chains, whereas LG2 and LG3 are modified on every face in at least one α chain (Fig. 4). Based on this observation, we speculate that LG1 may provide an essential protein surface for integrin recognition in all laminin heterotrimers.
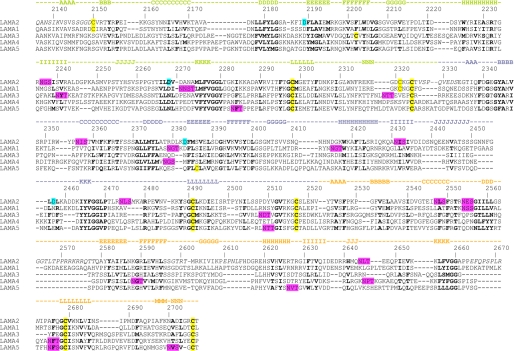
FIGURE 3.
Sequence alignment of the LG1-3 regions of mouse laminin α chains. The sequences used were: laminin α1 (SwissProt P19137), laminin α2 (NCBI NP_032507), laminin α3 (SwissProt Q61789), laminin α4 (SwissProt P97927), laminin α5 (NCBI NP_001074640). The sequence numbering and secondary structure elements of α2LG1-3 are indicated above the alignment. LG1, LG2, and LG3 are distinguished by green, blue, and brown coloring, respectively. Regions that are missing from the α2LG1-3 structure due to alternative splicing, deletion mutagenesis or crystal disorder are in italics. Residues in bold are structurally important in α2LG1-3 and conserved in all laminin α chains. Cysteines, calcium ligands, and putative N-linked glycosylation sites are highlighted in yellow, cyan, and magenta, respectively.
FIGURE 4.
Location of N-linked glycosylation sites in the LG1-3 structure. Shown are two surface representations of the α2LG1-3 structure (LG1, green; LG2, blue; LG3, brown) related by a rotation of 180° about a vertical axis. The LG1-2 linker is shown as a black line. The predicted attachment site in LG1 of the coiled coil region is indicated by a gray arrow. Every N-linked glycosylation site present in one or more of the five mouse or human laminin α chains has been mapped onto the α2LG1-3 structure (see text) and marked by a branched core hexasaccharide (magenta). The left view is similar to the one in Fig. 1A.
Functionally important binding sites are often located by analyzing the evolutionary conservation of a protein surface, provided that the mode binding is conserved in all homologues. Unfortunately, the laminin α chains are not well suited for such an approach, because the different α chains are quite divergent (the sequence identity for any pairwise alignment typically is 20–30%) and do not all interact with the same integrin heterodimer (9).
The RRKRR sequence in LG3 of the laminin α2 chain has been shown to be cleaved by a furin-like protease in 293 and Schwann cells (32, 44). Furin processing reduces the affinity of laminin-211 for heparin, but not for α-dystroglycan or integrins, and is required for acetylcholine receptor clustering on myotubes (44). The RRKRR sequence is located between β-strands D and E of α2LG3 (Fig. 1A). Compared with all other laminin LG domains, α2LG3 has an unusually long D-E loop that is predicted to project from the body of the LG3 domain (the D-E loop of α1LG3 is also long, but does not contain any basic residues; Fig. 3). To obtain the α2LG1-3 crystals, the D-E loop in LG3 was shortened by 12 residues. The resulting short-cut, as well as two adjacent loops (F-G and K-L) are disordered in the crystal. Whether this disorder is intrinsic to LG3, due to the D-E loop deletion, or a result of the dissociation of LG1 from LG2-3 remains to be established by further structural studies.
DISCUSSION
The LG1-3 region of laminin α chains is necessary for integrin binding, but it is not sufficient: several studies have shown that the coiled coil formed by the three laminin chains and, in particular, a glutamic acid in the third position from the C terminus of the γ chain are also required (13–17). This situation represents a challenge for the structure determination of an integrin-binding laminin fragment. While heterotrimeric (truncated) laminins have been successfully produced for biochemical analyses (13–16, 44–47), these molecules are either too large or not available in sufficient quantity or purity for crystallization. As an important first step toward a structural understanding of laminin-integrin interactions, we have determined the crystal structure of laminin α2LG1-3.
Electron micrographs of the distal end of the long arm of the cross-shaped laminin molecules reveal LG1-3 as three globules in a triangular arrangement and in close contact with the rod-shaped coiled coil (48). We previously proposed a model for LG1-3, in which the three LG domains are arranged like the leaves of a clover, forming three distinct interfaces between the constituent domains (7). To our surprise, the α2LG1-3 structure reported here has only one such interface, between LG2 and LG3, whereas LG1 is completely dissociated from the LG2-3 pair (Fig. 1A). Although we cannot exclude the possibility that this open conformation is a crystal artifact, it is more likely that it has resulted from the absence of the coiled coil and γ chain tail in the crystallized construct. This interpretation is supported by electron microscopy of a truncated heterotrimeric laminin-332, which shows a closed LG1-3 conformation when the γ2 chain C terminus is intact, and a more open conformation when the last three residues of the γ2 chain (including the critical glutamic acid) are deleted (18). Because peptides spanning the γ1 or γ2 chain tails neither bind integrins nor inhibit integrin binding to laminins, it has been suggested that the critical glutamic acid is not directly ligated by integrins, but plays an indirect role in maintaining an active LG1-3 conformation (15, 18). An attractive scenario is that the γ chain tail is required for LG1 to form a stable contact with LG2-3, which could explain why we see substantial disorder near the domain termini of LG1 in our crystals. We are particularly intrigued by the disorder of a highly conserved IGLWNF motif preceding β-strand N, which is directly adjacent to the N-terminal β-strand A (Figs. 1 and 3). In this regard, it is interesting to note that there is no gap between the predicted coiled coil domain (35) and the N terminus of LG1 in our structure. The C-terminal disulfide bridge between the β and γ chains (35) is thus likely to be near the center of the closed LG1-3 structure, giving the critical glutamic acid in the γ chain potential access to much of the LG1-3 surface.
Assuming that the γ chain tail is not directly involved in integrin binding, which of the three LG domains in LG1-3 harbors the integrin binding site? Two linear peptides derived from the laminin α1LG1-3 region have been shown to promote strong cell adhesion and spreading, which could be inhibited by anti-integrin antibodies (49). The minimal active sequences, SIYITRF for peptide AG-10 and IAFQRN for peptide AG-32, map to β-strands H of LG1 and LG2 (Fig. 3) and are almost completely buried in the structure (the arginine residues also contribute to the LG domain core). We conclude that these sequences are unlikely to be involved in integrin binding. The isolated LG3 domain of the laminin α3 chain, obtained by bacterial expression, has been reported to interact with integrin α3β1 (10, 12). Kim et al. (12) identified a PPFLMLLKGSTR motif as crucial for this interaction; however, our structure-based sequence analysis (Fig. 3) shows that this sequence is not part of the α3LG3 fold, but is located in the LG3-4 linker. Moreover, a laminin α3LG1-3 construct expressed in mammalian cells does not bind to integrin α3β1, but additionally requires the presence of the β3 and γ2 chains (16). The structural requirements for integrin α3β1 binding to laminin-332 thus remain to be clarified.
The most detailed studies of integrin binding have been carried out with laminin-511. Truncation from the α5 chain C terminus shows that deletion of LG4-5 has no effect on integrin binding, but that further deletion of LG3 abrogates integrin binding (14). On the other hand, experiments in vivo with laminin α5/α1 chimeras have shown that α5LG1-2 account for most of the functions of α5LG1-5 in embryonic development (50). This finding is slightly at odds with results obtained in vitro using purified α5/α1 chimeric laminins, which showed that only chimeras containing α5LG1-3 support integrin-mediated cell adhesion; replacement of α5LG3 with its counterpart from the α1 chain abrogated binding (45). The cell adhesion molecule Lutheran/B-CAM also requires an intact α5LG1-3 region for laminin-511 binding and competes with integrins for binding (45, 51). Finally, the epitope of the function-blocking monoclonal antibody 4C7 has been mapped to LG1 of the α5 chain and shown to require an intact LG1-3 structure for 4C7 binding (52). The collective biochemical data thus suggest that the active (closed) conformation of α5LG1-3 depends critically on long-range interactions between all three LG domains. Our unbiased analysis of N-linked glycosylation sites in the LG1-3 region highlights LG1 as a plausible integrin binding domain (Fig. 4). The rationale for this analysis was that the integrin binding site should be free of obstructing carbohydrate modifications in all five laminin α chains. Two other major extracellular matrix molecules, collagen and fibronectin, employ linear sequence motifs to bind integrins (8, 19, 20). Thus, it could be argued that only a small surface area of laminin LG1-3 has to be accessible for integrins to bind. However, we think that the integrin binding surface in laminins is likely to be more extensive, given the complex requirement for both an intact coiled coil and LG1-3 structure.
Calcium ions are intimately involved in the biological functions of the laminin LG4-5 domains (21, 23, 31), the LG3 domains of agrin (36) and perlecan (53), and the LG/LNS domains of neurexins (37–40). Our structure shows that LG1 and LG2 of α2LG1-3 contain similar calcium binding sites, but it is currently unknown whether these sites are involved in any of the biological activities of the laminin LG1-3 region.
In summary, we have determined an open structure of laminin LG1-3. Localized disorder in LG1 suggests that this domain interacts with the LG2-3 pair in the closed, biologically active, conformation of heterotrimeric laminins. A global analysis of glycosylation sites in the laminin LG1-3 region suggests a prominent role for LG1 in integrin binding. These findings will be useful in guiding further investigations into laminin-receptor interactions.
Acknowledgments
We thank Alexander Nyström and Jan Talts (University of Copenhagen, Denmark) for their contribution to the early stages of this project. We thank Takako Sasaki (Universität Erlangen-Nürnberg, Germany) for the laminin α2 cDNA; Dan Leahy (Johns Hopkins University, Baltimore, MD) for the endoglycosidase H expression plasmid; the staff at beamline 10.1 at the SRS Daresbury for help with x-ray data collection; and Peter Brick for critical reading of the manuscript.
This work was supported by a Wellcome Trust Senior Research Fellowship (to E. H.) (Refs. 054334 and 083942).
The atomic coordinates and structure factors (code 2wjs) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LG
- laminin G-like
- R.m.s.
- root mean-squared.
REFERENCES
- 1.Schéele S., Nyström A., Durbeej M., Talts J. F., Ekblom M., Ekblom P. (2007) J. Mol. Med. 85,825–836 [DOI] [PubMed] [Google Scholar]
- 2.Aumailley M., Bruckner-Tuderman L., Carter W. G., Deutzmann R., Edgar D., Ekblom P., Engel J., Engvall E., Hohenester E., Jones J. C., Kleinman H. K., Marinkovich M. P., Martin G. R., Mayer U., Meneguzzi G., Miner J. H., Miyazaki K., Patarroyo M., Paulsson M., Quaranta V., Sanes J. R., Sasaki T., Sekiguchi K., Sorokin L. M., Talts J. F., Tryggvason K., Uitto J., Virtanen I., von der Mark K., Wewer U. M., Yamada Y., Yurchenco P. D. (2005) Matrix Biol. 24,326–332 [DOI] [PubMed] [Google Scholar]
- 3.Colognato H., Yurchenco P. D. (2000) Dev. Dyn. 218,213–234 [DOI] [PubMed] [Google Scholar]
- 4.Miner J. H., Yurchenco P. D. (2004) Annu. Rev. Cell Dev. Biol. 20,255–284 [DOI] [PubMed] [Google Scholar]
- 5.Li S., Liquari P., McKee K. K., Harrison D., Patel R., Lee S., Yurchenco P. D. (2005) J. Cell Biol. 169,179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKee K. K., Harrison D., Capizzi S., Yurchenco P. D. (2007) J. Biol. Chem. 282,21437–21447 [DOI] [PubMed] [Google Scholar]
- 7.Timpl R., Tisi D., Talts J. F., Andac Z., Sasaki T., Hohenester E. (2000) Matrix Biol. 19,309–317 [DOI] [PubMed] [Google Scholar]
- 8.Hynes R. O. (2002) Cell 110,673–687 [DOI] [PubMed] [Google Scholar]
- 9.Nishiuchi R., Takagi J., Hayashi M., Ido H., Yagi Y., Sanzen N., Tsuji T., Yamada M., Sekiguchi K. (2006) Matrix Biol. 25,189–197 [DOI] [PubMed] [Google Scholar]
- 10.Shang M., Koshikawa N., Schenk S., Quaranta V. (2001) J. Biol. Chem. 276,33045–33053 [DOI] [PubMed] [Google Scholar]
- 11.Yu H., Talts J. F. (2003) Biochem. J. 371,289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J. M., Park W. H., Min B. M. (2005) Exp. Cell Res. 304,317–327 [DOI] [PubMed] [Google Scholar]
- 13.Deutzmann R., Aumailley M., Wiedemann H., Pysny W., Timpl R., Edgar D. (1990) Eur. J. Biochem. 191,513–522 [DOI] [PubMed] [Google Scholar]
- 14.Ido H., Harada K., Futaki S., Hayashi Y., Nishiuchi R., Natsuka Y., Li S., Wada Y., Combs A. C., Ervasti J. M., Sekiguchi K. (2004) J. Biol. Chem. 279,10946–10954 [DOI] [PubMed] [Google Scholar]
- 15.Ido H., Nakamura A., Kobayashi R., Ito S., Li S., Futaki S., Sekiguchi K. (2007) J. Biol. Chem. 282,11144–11154 [DOI] [PubMed] [Google Scholar]
- 16.Künneken K., Pohlentz G., Schmidt-Hederich A., Odenthal U., Smyth N., Peter-Katalinic J., Bruckner P., Eble J. A. (2004) J. Biol. Chem. 279,5184–5193 [DOI] [PubMed] [Google Scholar]
- 17.Sung U., O'Rear J. J., Yurchenco P. D. (1993) J. Cell Biol. 123,1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navdaev A., Heitmann V., Desantana, Evangelista K., Mörgelin M., Wegener J., Eble J. A. (2008) Exp. Cell Res. 314,489–497 [DOI] [PubMed] [Google Scholar]
- 19.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Cell 101,47–56 [DOI] [PubMed] [Google Scholar]
- 20.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296,151–155 [DOI] [PubMed] [Google Scholar]
- 21.Harrison D., Hussain S. A., Combs A. C., Ervasti J. M., Yurchenco P. D., Hohenester E. (2007) J. Biol. Chem. 282,11573–11581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tisi D., Talts J. F., Timpl R., Hohenester E. (2000) EMBO J. 19,1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wizemann H., Garbe J. H., Friedrich M. V., Timpl R., Sasaki T., Hohenester E. (2003) J. Mol. Biol. 332,635–642 [DOI] [PubMed] [Google Scholar]
- 24.Talts J. F., Mann K., Yamada Y., Timpl R. (1998) FEBS Lett. 426,71–76 [DOI] [PubMed] [Google Scholar]
- 25.Kohfeldt E., Maurer P., Vannahme C., Timpl R. (1997) FEBS Lett. 414,557–561 [DOI] [PubMed] [Google Scholar]
- 26.CCP4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50,760–763 [DOI] [PubMed] [Google Scholar]
- 27.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61,458–464 [DOI] [PubMed] [Google Scholar]
- 28.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60,432–438 [DOI] [PubMed] [Google Scholar]
- 29.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A. 47,110–119 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54,905–921 [DOI] [PubMed] [Google Scholar]
- 31.Hohenester E., Tisi D., Talts J. F., Timpl R. (1999) Mol. Cell 4,783–792 [DOI] [PubMed] [Google Scholar]
- 32.Talts J. F., Timpl R. (1999) FEBS Lett. 458,319–323 [DOI] [PubMed] [Google Scholar]
- 33.Chang V. T., Crispin M., Aricescu A. R., Harvey D. J., Nettleship J. E., Fennelly J. A., Yu C., Boles K. S., Evans E. J., Stuart D. I., Dwek R. A., Jones E. Y., Owens R. J., Davis S. J. (2007) Structure 15,267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudenko G., Hohenester E., Muller Y. A. (2001) Trends Biochem. Sci. 26,363–368 [DOI] [PubMed] [Google Scholar]
- 35.Beck K., Hunter I., Engel J. (1990) FASEB J. 4,148–160 [DOI] [PubMed] [Google Scholar]
- 36.Stetefeld J., Alexandrescu A. T., Maciejewski M. W., Jenny M., Rathgeb-Szabo K., Schulthess T., Landwehr R., Frank S., Ruegg M. A., Kammerer R. A. (2004) Structure 12,503–515 [DOI] [PubMed] [Google Scholar]
- 37.Arac̨ D., Boucard A. A., Ozkan E., Strop P., Newell E., Südhof T. C., Brunger A. T. (2007) Neuron 56,992–1003 [DOI] [PubMed] [Google Scholar]
- 38.Chen X., Liu H., Shim A. H., Focia P. J., He X. (2008) Nat. Struct. Mol. Biol. 15,50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabrichny I. P., Leone P., Sulzenbacher G., Comoletti D., Miller M. T., Taylor P., Bourne Y., Marchot P. (2007) Neuron 56,979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen K. C., Kuczynska D. A., Wu I. J., Murray B. H., Sheckler L. R., Rudenko G. (2008) Structure 16,422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taniguchi Y., Ido H., Sanzen N., Hayashi M., Sato-Nishiuchi R., Futaki S., Sekiguchi K. (2009) J. Biol. Chem. 284,7820–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Science 291,2370–2376 [DOI] [PubMed] [Google Scholar]
- 43.Barclay A. N. (2003) Semin. Immunol. 15,215–223 [DOI] [PubMed] [Google Scholar]
- 44.Smirnov S. P., McDearmon E. L., Li S., Ervasti J. M., Tryggvason K., Yurchenco P. D. (2002) J. Biol. Chem. 277,18928–18937 [DOI] [PubMed] [Google Scholar]
- 45.Kikkawa Y., Sasaki T., Nguyen M. T., Nomizu M., Mitaka T., Miner J. H. (2007) J. Biol. Chem. 282,14853–14860 [DOI] [PubMed] [Google Scholar]
- 46.Doi M., Thyboll J., Kortesmaa J., Jansson K., Iivanainen A., Parvardeh M., Timpl R., Hedin U., Swedenborg J., Tryggvason K. (2002) J. Biol. Chem. 277,12741–12748 [DOI] [PubMed] [Google Scholar]
- 47.Kortesmaa J., Yurchenco P., Tryggvason K. (2000) J. Biol. Chem. 275,14853–14859 [DOI] [PubMed] [Google Scholar]
- 48.Bruch M., Landwehr R., Engel J. (1989) Eur. J. Biochem. 185,271–279 [DOI] [PubMed] [Google Scholar]
- 49.Nomizu M., Kim W. H., Yamamura K., Utani A., Song S. Y., Otaka A., Roller P. P., Kleinman H. K., Yamada Y. (1995) J. Biol. Chem. 270,20583–20590 [DOI] [PubMed] [Google Scholar]
- 50.Kikkawa Y., Miner J. H. (2006) Dev. Biol. 296,265–277 [DOI] [PubMed] [Google Scholar]
- 51.Kikkawa Y., Moulson C. L., Virtanen I., Miner J. H. (2002) J. Biol. Chem. 277,44864–44869 [DOI] [PubMed] [Google Scholar]
- 52.Ido H., Harada K., Yagi Y., Sekiguchi K. (2006) Matrix Biol. 25,112–117 [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez E. M., Reed C. C., Bix G., Fu J., Zhang Y., Gopalakrishnan B., Greenspan D. S., Iozzo R. V. (2005) J. Biol. Chem. 280,7080–7087 [DOI] [PubMed] [Google Scholar]
- 54.Laskowski R. A., MacArthur M. W., Moss D. A., Thornton J. M. (1996) J. Appl. Crystallogr. 26,283–291 [Google Scholar]