Abstract
The serpinopathies are a family of diseases characterized by the accumulation of ordered polymers of mutant protein within the endoplasmic reticulum. They are a diverse group including α1-antitrypsin deficiency and the inherited dementia familial encephalopathy with neuroserpin inclusion bodies or FENIB. We have used transient transfection of COS7 cells and mouse embryonic fibroblasts, PC12 cell lines that conditionally express wild type and mutant neuroserpin and fly models of FENIB to assess the cellular handling of wild type and mutant serpins. By using a polymer-specific monoclonal antibody, we show that mutant neuroserpin forms polymers after a delay of at least 30 min and that polymers can be cleared in PC12 cell lines and from the brain in a fly model of FENIB. At steady state, the fractions of intracellular polymerogenic G392E mutant neuroserpin in the monomeric and polymeric states are comparable. Inhibition of the proteasome with MG132 reveals that both mutant neuroserpin and α1-antitrypsin are degraded predominantly by endoplasmic reticulum-associated degradation (ERAD). Pharmacological and genetic inhibitions demonstrate that autophagy is responsible for bulk turnover of wild type and mutant serpins, but can be stimulated by rapamycin to compensate for proteasome inhibition. The significance of these findings to the treatment of serpinopathies is discussed.
Many human diseases result from aberrant protein-protein interactions. Often these occur due to improper protein folding and so have been termed the conformational diseases (1). Defective protein folding can lead to the exposure of normally buried hydrophobic residues, thus encouraging non-native interactions. In such cases, unstructured aggregates of protein can accumulate that are toxic to the cell. Such misfolding can either occur constitutively, as a result of coding sequence mutations or be induced by insults such as heat shock or hypoxia. In contrast, some proteins aggregate to form more structured polymers. Many proteins with widely differing primary sequences have been shown to form fibrils through aberrant β-strand linkages. This material forms into highly ordered amyloid fibrils in conditions such as Alzheimer and Huntington diseases and the prion encephalopathies. Another important group of diseases resulting in the formation of high molecular weight-ordered structures is the serpinopathies that result from mutations in members of the serine protease inhibitor (serpin) family of proteins.
Serpins are potent suicide inhibitors of a wide variety of both extra- and intracellular proteases (2). To achieve irreversible inhibition of their target protease, each serpin offers a reactive center loop that acts as a pseudosubstrate for the catalytic site of the protease. This is cleaved, triggering a dramatic conformational change in the serpin and the formation of an inactive complex (3). Mutations within the serpins subvert this mechanism and allow insertion of the reactive center loop of one molecule into the β-sheet of another, an event that can be recursive, leading to the formation of long ordered polymers (4, 5). This can cause disease by either loss-of-function due to the deregulation of proteolysis or a toxic-gain-of-function through the local accumulation of polymers within the endoplasmic reticulum (ER)6 of the cell of synthesis (2).
Aggregation of misfolded protein within the ER is countered by a complex homeostatic pathway, the unfolded protein response (UPR) that aims to adapt the cell to its increased load of ER client protein (6, 7). This response involves a transient attenuation of protein translation and a parallel transcriptional up-regulation of components of the ER folding machinery. In addition, misfolded proteins are targeted for degradation by the ubiquitin-proteasome pathway, requiring their retrotranslocation from the ER lumen back into the cytoplasm, so-called ER-associated degradation (ERAD) (8). If, however, the combined efforts of the UPR and ERAD fail to adapt the cell to its ER synthetic load, an as yet poorly defined process leads to death of the cell by apoptosis (9, 10).
We have described a familial dementia, FENIB, caused by polymerization of neuroserpin within the brains of affected humans (11). This disease demonstrates a remarkable genotype-phenotype correlation, with a number of separate mutations affecting the in vitro rate of polymerization with a concordant increase in disease severity (12). Although rare, affecting only a few families worldwide, FENIB provides a useful model with which to understand the pathogenesis of other more common serpinopathies. Recently, we observed that the accumulation of polymerized mutant neuroserpin within the ER fails to trigger the UPR (13). Instead, an UPR-independent but calcium-dependent signaling pathway leads to the activation of NFκB. A similar response has been described to the accumulation of other folded ER proteins and termed the ER Overload Response (EOR) (14).
Recent reports have suggested that the accumulation of polymers of mutant α1-antitrypsin within the ER of hepatocytes is handled by autophagy (15, 16). Similar findings have been described for cytosolic protein aggregates of Huntingtin, suggesting that diseases caused by protein accumulation may be treated by augmentation of autophagy (17). We were intrigued by the potential to treat FENIB by promoting polymer degradation. We therefore generated mammalian cell lines and Drosophila models (12) of neuroserpin accumulation in order to elucidate the pathways by which polymers are cleared from cells. Here, we demonstrate that autophagy does not show specificity for polymerogenic serpin mutants in neuronal-like PC12 cells, but instead is involved in the degradation of all forms of neuroserpin. In contrast, ERAD selectively degrades mutant neuroserpin and α1-antitrypsin. Of clinical significance, augmentation of constitutive autophagy by treatment with rapamycin can selectively overcome the accumulation of mutant serpins when ERAD is impaired.
EXPERIMENTAL PROCEDURES
Fly Culture
All stocks had the w1118 genetic background and were cultured at 25 °C on standard fly food with dried yeast. Stocks were maintained at 25 °C. Transgene expression was driven with the elavc1555-GAL4 pan-neuronal driver (18) and the GeneSwitch system was employed (19) for polymer washout experiments. Briefly, RU486, Mifepristone (Sigma-Aldrich Co.) was dissolved in 100% (v/v) ethanol at 20 mg/ml (19, 20). This stock solution was diluted in water to 2% (v/v) and used to make yeast paste with 1 ml of RU486 solution to 0.55 g of dried yeast. Yeast paste was streaked onto standard fly food prepared without dried yeast. Expression was induced by placing flies carrying the elavGS driver and the relevant neuroserpin transgene onto the food for 48 h. Induction was stopped by moving flies to normal food with dried yeast.
ELISA
The ELISA protocol for cell and fly extracts was essentially identical, except fly extracts were prepared as follows. Three aliquots of 5 whole flies were homogenized in 300 μl of 150 mm NaCl, 50 mm Tris pH 7.5, 1% v/v Nonidet p40, 5 mm EDTA with EDTA-free protease inhibitor mixture tablets (Roche Applied Science). The samples were homogenized using or tungsten balls and a mixer mill (Retsch, Haan, Germany) in 96-well 1.2-ml storage plates (Abgene, Epsom, UK). The samples were centrifuged, and 70 μl of supernatant was retrieved. High binding surface COSTAR 96-well plates (Corning, NY) were coated overnight with affinity-purified rabbit polyclonal antibodies against neuroserpin (Abcam, Cambridge, UK) at 2 μg/ml in carbonate/bicarbonate buffer (Na2CO3/NAHCO3, pH 9.5). After washing (0.9% (w/v) NaCl, 0.05% (v/v) Tween 20), the plates were blocked for 1 h in blocking buffer (PBS, 0.25% (w/v) bovine serum albumin, 0.05% (v/v) Tween 20). Samples and standards were diluted in blocking buffer and incubated for 2 h. The detection antibodies were a pool of mouse monoclonal antibodies (12) and were used at 333 ng/ml each, to a total concentration of 1 μg/ml or the antipolymer monoclonal antibody (7C6) used at 1 μg/ml, and incubated for 2 h. Rabbit anti-mouse IgG horseradish peroxidase-labeled antibody (Sigma Aldrich) diluted 1:20,000 was used as a secondary antibody and incubated for 1 h. The reaction was developed with TMB liquid substrate (Sigma Aldrich) for 10 min in the dark, and the reaction was stopped with 1 m H2SO4. Absorbance was read at 450 nm on a Thermo-max microplate reader (Molecular Devices, Sunnyvale, CA).
Expression Vectors
Transient transfections were performed with pcDNA3.1 encoding the sequences for neuroserpin (WT, S52R, G392E) and α1-antitrypsin (M, Z). Tet-ON inducible PC12 cells conditionally expressing WT and S52R mutant neuroserpin were generated using pTRE2hyg plasmid (BD Bioscience). In co-transfection experiments, luciferase was expressed from the pTRE2hyg vector (BD Bioscience).
Cell Culture and Transfection
COS7 cells were maintained in DMEM supplemented with 10% v/v fetal bovine serum at 37 °C and 5% CO2 v/v in a humidified incubator. Transfections were performed in 6-well plates that had been precoated with 0.1 mg/ml poly-l-lysine. Typically, 11 × 104 COS7 cells were plated into each well of a 6-well dish and transfected with 2.5 μg of plasmid DNA were introduced into each well mixed with 6.25 μl of LipofectamineLTX (Invitrogen, 15338-100) and 2.5 μl of PLUS reagent (Invitrogen, 11514-015) in serum-free Opti-MEM-Glut culture medium (Invitrogen, 31985-062) following the protocol recommended by Invitrogen. MEFs were cultured similarly, except for transfections when typically, 23 × 104 MEF cells were plated into each well of a 6-well dish and transfected with 4.0 μg of plasmid DNA into each well mixed with 6 μl of Lipofectamin2000 (Invitrogen, 11668-019) and 200 μl DMEM medium (Sigma, D6546). PC12 cells were differentiated into neurons by plating onto glass coverslips pretreated with 0.1 mg/ml poly-l-lysine and 0.1 mg/ml rat tail collagen I, and culturing in DMEM supplemented with 1% (v/v) heat-inactivated horse serum, nonessential amino acids, HEPES buffer, 0.2 units/ml bovine insulin, 200 mg/ml Geneticin, 100 mg/ml Hygromycin B, and 150 ng/ml nerve growth factor for 7 days.
SDS and Non-denaturing PAGE and Western Blot Analysis
The cell pellet from each well of 6-well plates was lysed in 150 μl of Nonidet lysis buffer (150 mm NaCl, 50 mm Tris-Cl, pH 7.5, 1% v/v Nonidet P-40), and 2 ml of culture medium from each well were concentrated 10 times in Vivaspin 2-ml concentrators with a 3000-Da cut-off membrane (Viva-science AG, Hannover, Germany). Samples of 20 μl were mixed with 5 μl 5× loading buffer containing 10% (v/v) β-mercaptoethanol and 4% w/v SDS and analyzed by 10% or 12% w/v SDS-polyacrylamide gel electrophoresis (PAGE). Nondenaturing PAGE was performed on 7.5% (w/v) acrylamide nondenaturing gels using 5× loading buffer without SDS and β-mercaptoethanol. The proteins were transferred from the gels onto Immobilon P membrane (Millipore Corp., Bedford, MA) at 100 V for 1 h for Western blot analysis. 20% v/v methanol was added to the transfer buffer for gels that had been run in SDS. After transfer, the membrane was washed in PBT (PBS plus 0.1% (v/v) Tween 20) and blocked overnight in PBT plus 5% (w/v) dried skimmed milk powder. The following day, the membrane was incubated with appropriate antibody in 5% (w/v) bovine serum albumin stock (in PBS with 0.01% (v/v) sodium azide) for 120 min, washed six times for 5 min with PBT, and then incubated with 1:75,000 anti-rabbit IgG-horseradish peroxidase antibody in PBT-milk for 75 min. The membrane was washed another six times for 5 min with PBT and 10 min in PBS before developing using the ECL SuperSignal West Femto maximum sensitivity substrate (Pierce) and exposed to film.
Metabolic Labeling and Immunoprecipitation
Thirty-six hours after transfection, cells were starved in 1 ml of methionine and cysteine-free DMEM for 1 h, pulsed for 30 min with 1.3 MBq/well Easy TagTM Expre35S Protein Labeling Mix (PerkinElmer) containing 35S-labeled methionine and cysteine, and then harvested or washed with cold PBS and cultured in 1 ml of chase medium (DMEM containing 200 mm methionine and 200 mm cysteine). For the pulse-chase of 3 h, the chase medium was supplemented with 10% (v/v) fetal bovine serum. After the chase period, the culture medium was collected and centrifuged at 2000 rpm and 4 °C for 11 min, and the cells were harvested by adding 0.5 ml/well Nonidet lysis buffer (150 mm NaCl, 50 mm Tris-Cl, pH 7.5, 1% v/v Nonidet P-40) containing a protease inhibitor mixture (Complete; EDTA-free protease inhibitor mixture tablets (Roche Applied Science)), scraping and vortexing for 3 × 3 s, and then centrifuging at 16,000 rpm and 4 °C for 16 min. The supernatants from culture medium and cell lysate samples were precleared with rabbit IgG bound to 50 μl of 50% (v/v) protein A-Sepharose for at least 1 h at 4 °C, and then neuroserpin proteins were immunoprecipitated in parallel either with a purified antibody for total neuroserpin or with a polymer-specific antibody overnight at 4 °C. The specific antibodies were prebound to protein A-Sepharose (50 μl of 50% (v/v) protein A-Sepharose plus 1.5 μg of purified antibody at 4 °C for 2 h). The following day, immunocomplexes were washed four times with cold washing buffer (150 mm NaCl, 50 mm Tris-Cl, pH 7.5, 1% v/v Nonidet P-40) and once with cold PBS. Radiolabeled proteins were recovered 2× SDS-PAGE loading buffer by heating for 5 min at 99 °C, separated on 10% (w/v) acrylamide gels, and detected by autoradiography with a Cyclone phosphorimager (Packard Instrument Co.).
RESULTS
Neuroserpin Forms Intracellular Polymers in a Time-dependent Manner Following Completion of the Polypeptide Chain Biosynthesis
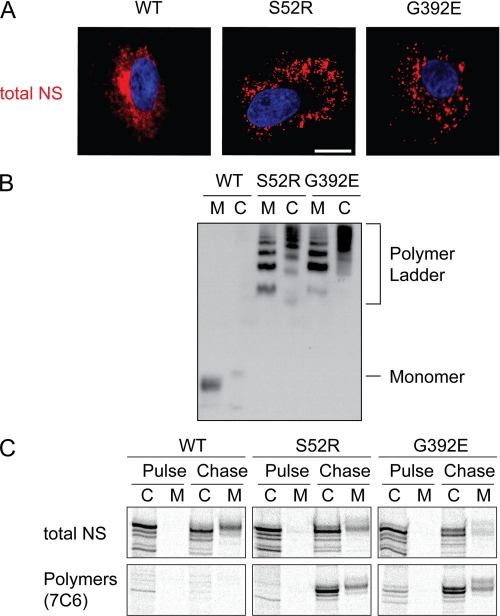
When wild type (WT) neuroserpin is expressed heterologously in COS7 cells, it can readily be detected by immunofluorescence confocal microscopy as reticular staining throughout the cell (Fig. 1A, left panel). We have previously shown that WT neuroserpin co-localizes with both ER and Golgi markers (12). In contrast, polymerogenic mutants, such as S52R and G392E neuroserpin, are retained in punctate structures (Fig. 1A, center and right panels) that we have previously shown to co-localize with ER markers (12). This is related to the formation of polymers that can be visualized by non-denaturing PAGE (Fig. 1B). The polymers isolated from medium sometimes resolve at different sizes compared with the polymers from cells. This is likely to reflect differences in the glycosylation of the extracellular proteins resulting from their transit through the Golgi (21).
FIGURE 1.
Expression of polymerogenic mutants of neuroserpin. A, COS7 cells were transiently transfected with WT, S52R, and G392E neuroserpin expression vectors. Neuroserpin was detected using a specific polyclonal antibody and rhodamine-conjugated secondary antibody. Nuclei were stained with DAPI. Bar, 10 μm. B, COS7 cells were transiently transfected with WT, S52R, and G392E neuroserpin expression vectors. Conditioned medium M and cell lysate C were separated by 7.5% w/v non-denaturing PAGE and immunostained with a polyclonal neuroserpin antibody. Note the formation of polymer ladders in the S52R and G392E mutants within both the cells and the medium. C, COS7 cells expressing WT, S52R, and G392E neuroserpin were pulsed for 30 min with [35S]methionine and chased in excess unlabeled methionine. Samples of cell lysate C and media M were taken at the end of the pulse and following 3 h of chase. Parallel samples were immunoprecipitated with a pan-neuroserpin rabbit polyclonal antibody (total NS; upper panels) and with an anti-neuroserpin polymer mouse monoclonal antibody (7C6, lower panels) and then assessed by 10% w/v SDS-PAGE.
To test if neuroserpin polymers form co-translationally or post-translationally, COS7 cells were transfected with expression vectors for WT, S52R, or G392E neuroserpin. These cells were then pulse-labeled with [35S]methionine/cysteine for 30 min followed by a 3 h chase in the absence of label and in the presence of excess unlabeled methionine and cysteine. Lysates were prepared and subjected to immunoprecipitation using either a total neuroserpin polyclonal antiserum or a polymer selective monoclonal antibody (7C6 (12)). At the end of the pulse, WT neuroserpin was detected solely within the cell lysate using the total neuroserpin antiserum, while after 3 h of chase it could also be detected within the medium (Fig. 1C). No WT neuroserpin could be immunoprecipitated using the polymer selective monoclonal antibody. In contrast, when cells expressing G392E mutant neuroserpin were analyzed, we found that even at the end of the chase period relatively little labeled neuroserpin could be detected within the medium. The S52R mutant, which polymerizes less rapidly and produces a milder clinical phenotype, was secreted into the medium at a level intermediate between WT and G392E neuroserpin. The polymer-specific antibody immunoprecipitated mutant neuroserpin from cell lysates of both S52R- and G392E-expressing cells and this “polymeric” fraction increased dramatically during the chase period. Because all the specimens were lysed simultaneously and treated identically, it is unlikely that these observed differences were caused by post-lysis polymerization and therefore indicate that following its initial synthesis, mutant neuroserpin polymerizes within the ER, and this is associated with impaired secretion. Crucially, this demonstrates for the first time in vivo that polymerization of a mutant serpin occurs post-translationally following a delay of at least 30 min, probably after individual molecules are completely folded.
Neuroserpin Accumulation Requires Continued Protein Synthesis
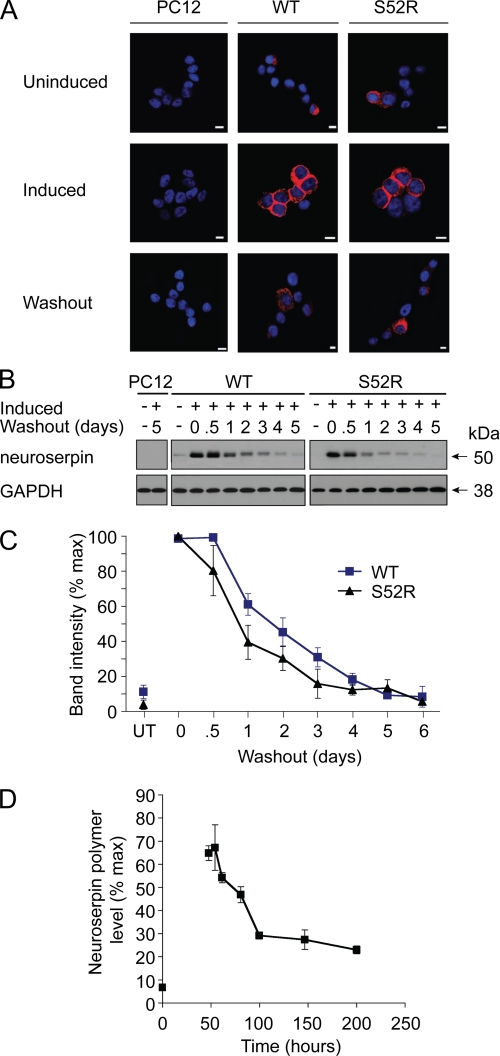
To determine whether intracellular polymers can be cleared from the cell, we induced WT and S52R neuroserpin in PC12 cells by incubating the cell lines with doxycycline for 2 days followed by withdrawal of the drug. We then monitored the persistence of transgenic neuroserpin by both immunofluorescence and immunoblot analysis (Fig. 2, A and B). Neuroserpin immunoreactivity was lost from cells at a similar rate in both WT and S52R genotypes, with decay curves that were statistically indistinguishable (p > 0.5, two way analysis of variance; Fig. 2C).
FIGURE 2.
Neuroserpin levels decay equally for wild type and polymerogenic neuroserpin following cessation of expression. A, Tet-ON WT and S52R PC12 were induced to express neuroserpin with doxycycline for 2 days, fixed, and stained for neuroserpin. Cells were similarly induced followed by doxycycline withdrawal for 5 days. Bar, 10 μm, B, Tet-ON WT and S52R PC12 were induced to express neuroserpin with doxycycline for 2 days followed by a washout for 0.5 to 6 days. Samples were separated by 10% w/v SDS-PAGE and immunoblotted for neuroserpin. GAPDH served as a loading control. C, combined data from three independent repeats of B; untreated samples (UT) show non-induced expression levels; mean ± S.E. D, transgenic GeneSwitch Drosophila melanogaster were induced to express G392E neuroserpin by the addition of RU486 to the feed. After 2 days, flies were transferred onto food lacking RU486. Neuroserpin polymers were measured in fly heads by ELISA; n = 3, mean ± S.E.
To test whether this effect was relevant to neuroserpin polymers in vivo, we generated lines of Drosophila using the GeneSwitch (19) system in which the expression of neuroserpin could be regulated by addition of the drug RU486 to the feed. As we have previously shown for constitutive expression of neuroserpin (12), the most polymerogenic mutant of neuroserpin (G392E) accumulated in fly brains (Fig. 2D). This was shown to be in the form of polymers by using a specific anti-polymer ELISA (Fig. 2D) (12). Furthermore, withdrawal of the drug was followed by a washout of these polymers from the brain. This is the first demonstration that serpin polymers can be cleared from cells in an animal model of a serpinopathy.
Chemical Inhibition of Autophagy Does Not Distinguish between Wild Type and Mutant Serpins in COS7 Cells
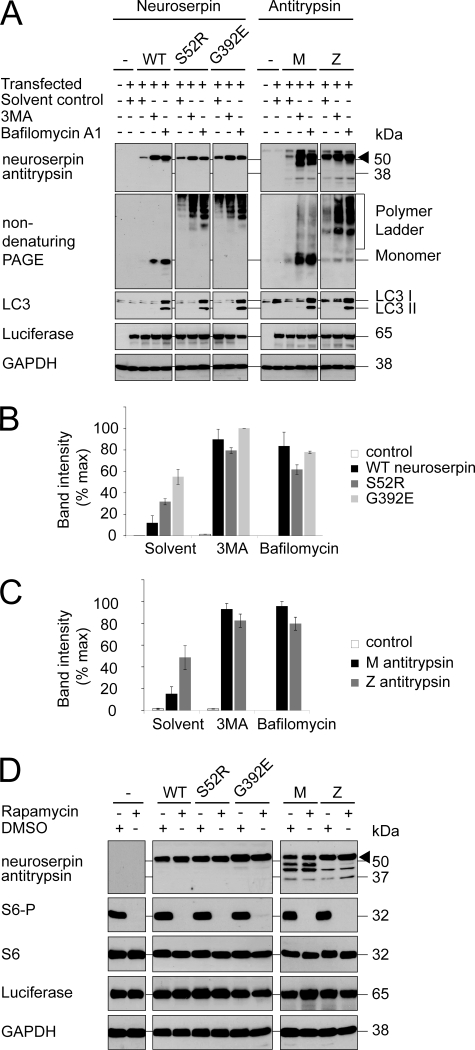
Serpin polymers are relatively stable structures in vitro, yet their levels decayed when synthesis was discontinued. This indicated that polymers must be degraded within the cell. Previous reports have implicated autophagy in the clearance of mutant α1-antitrypsin (15, 16), and so we assessed the effects of agents that modify this process on the clearance of neuroserpin in a COS7 cell model of disease (21). 3MA, a phosphatidylinositol (PI) 3-kinase inhibitor frequently used to block autophagy (22), was added to the growth medium, and then total neuroserpin was analyzed in cell lysates. Whereas 3MA did impair clearance of the neuroserpin signal, it failed to distinguish between WT neuroserpin and either of the polymerogenic mutants (Fig. 3, A and B). We were concerned that these results might have been caused by an effect of 3MA on pathways other than autophagy. For this reason, we also treated cells with bafilomycin A1, an inhibitor of the V-ATPase essential for autophagosome maturation (23). LC3 is a protein involved in autophagasome formation that is lipidated during this process. This lipidation causes an apparent mobility shift of LC3 to a smaller LC3 II form, which is found on incompletely formed autophagasomes. Bafilomycin A1 treatment caused an increase of both LC3 and its processed form LC3 II, confirming the accumulation of immature autophagosomes (Fig. 3A). This treatment increased the levels of both WT and polymerogenic neuroserpin, suggesting that in COS7 cells autophagy was not directed toward the polymerogenic proteins, but instead was involved in constitutive protein turnover (Fig. 3, A and B).
FIGURE 3.
Autophagy degrades both wild type and polymerogenic serpins in COS7 cells. A, COS7 cells were transfected with expression vectors for wild type neuroserpin (WT) or either of two polymerogenic mutants (S52R and G392E) or wild type (M) or polymerogenic mutant (Z) α1-antitrypsin. A luciferase expression vector was used as transfection control. Cells were treated for 1 day with solvent control, 10 mm 3MA, or 400 nm Bafilomycin A1. Samples were subjected to 10% w/v SDS-PAGE and immunoblotted for neuroserpin or α1-antitrypsin (arrowhead). GAPDH served as a loading control and LC3 immunoblot demonstrated LC3 II accumulation during bafilomycin A1 treatment. B, neuroserpin band intensity was quantified and expressed graphically; n = 3, mean ± S.E. C, α1-antitrypsin band intensity was quantified and expressed graphically; n = 3, mean ± S.E. D, cells were transfected as in A and treated either with 200 nm rapamycin or solvent control. The immunoblot demonstrated no changes in the level of neuroserpin or α1-antitrypsin (arrowhead). The lower molecular weight bands detected by the α1-antitrypsin antibody most likely represent degradation products. S6 phosphorylation was abrogated by rapamycin without significant effect on total S6 levels. GAPDH served as loading control and luciferase served as transfection control.
Our findings appeared at odds with previous reports that autophagy is critical to the clearance of polymers for another serpin, α1-antitrypsin (15, 16). We therefore assessed the effects of these pharmacological agents on the clearance of α1-antitrypsin during transient expression. Once again, inhibition of autophagy failed to distinguish between WT (M) antitrypsin and the polymerogenic Z mutant of α1-antitrypsin, but instead increased the level of both proteins to a similar extent (Fig. 3, A and C). Next, we attempted to augment autophagy through inhibition of the mTOR signaling pathway with the selective compound rapamycin (24). Once again, there was no evidence of mutant-selective autophagic degradation, as rapamycin failed to affect the levels of any of the serpins despite efficient mTOR inhibition as reported by loss of S6 phosphorylation (Fig. 3D).
Chemical Inhibition of Autophagy Does Not Distinguish between Wild Type and Mutant Serpins in PC12 Cells
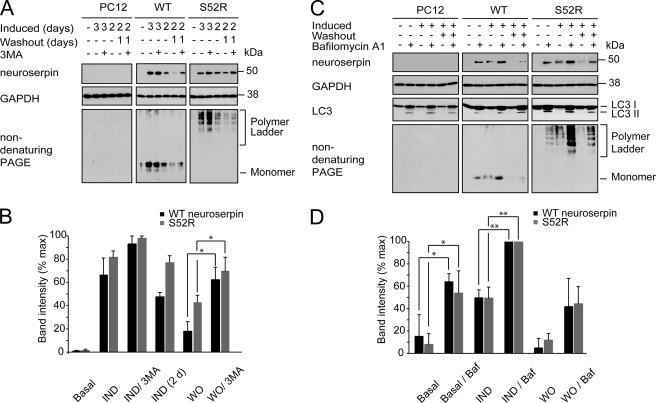
This phenomenon was not limited to COS7 cells, as similar effects on neuroserpin levels were observed with both 3MA and bafilomycin A1 for the PC12 cells lines expressing neuroserpin from an inducible promoter (Fig. 4). Expression of transgenic protein was induced and then allowed to clear by withdrawal of the induction agent. Once again, elevated LC3 and LC3 II levels in the bafilomycin samples confirmed the accumulation of immature autophagosomes (Fig. 4C). Treatment with either agent increased the level of WT and mutant neuroserpin in these cells during the induction phase and delayed the loss of neuroserpin during the washout phase. Indeed, treatment of non-induced WT or mutant-expressing cells with either agent led to neuroserpin accumulation within the cells. This most likely reflected the accumulation of neuroserpin expressed at low levels because of leakiness of the inducible promoter.
FIGURE 4.
3MA and bafilomycin A1 inhibit loss of wild type and polymerogenic neuroserpin in TetON PC12 cell lines. A, Tet-ON WT and S52R PC12 were induced to express neuroserpin with doxycycline for 2 or 3 days followed by 1 day of doxycycline withdrawal. The 2-day induction was included to demonstrate neuroserpin expression levels at the time of 3MA addition. Samples were subjected to 10% w/v SDS-PAGE and immunoblotted for neuroserpin. GAPDH served as a loading control. B, intensity of the neuroserpin band was quantified and expressed graphically; n = 3, mean ± S.E.; *, p < 0.05. C, Tet-ON WT and S52R PC12 were induced to express neuroserpin with doxycycline for 2 days followed by 1 day of doxycycline withdrawal. The cells were treated with bafalomycin A1 as shown. Samples were subjected to 10% w/v SDS-PAGE and immunoblotted for neuroserpin and LC3. GAPDH served as a loading control. D, intensity of the neuroserpin band was quantified and expressed graphically; n = 3, mean ± S.E.; *, p < 0.05; **, p < 0.01. IND, induction; WO, washout; Baf, bafalomycin.
The Proteasome Is Important in the Selectivity of Mutant Serpin Degradation
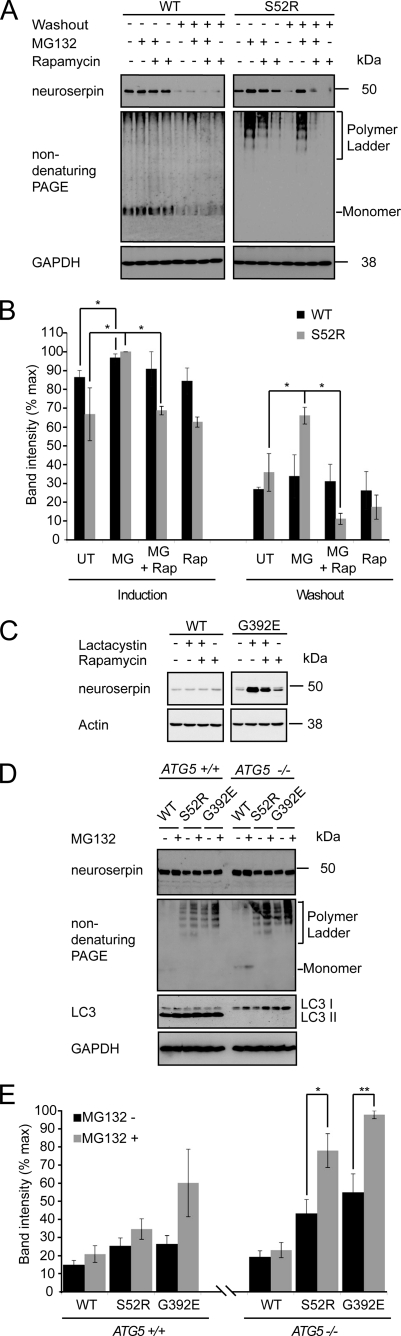
Because we were unable to observe a mutant-specific effect of inhibiting autophagy in either COS7 or PC12 cells, we next assessed the role of ERAD in the degradation of serpin polymers. Tet-On PC12 cell lines were induced to express neuroserpin prior to treatment with the proteasome inhibitor MG132. This led to an increase in the level of each transgenic protein as we have described previously for transient neuroserpin expression in COS7 cells (21). However, the effect of proteasome inhibition was more marked for the cells expressing mutant neuroserpin (Fig. 5, A and B). To confirm these results we then tested the effect of an alternative proteasomal inhibitor, lactacystin, on the G392E polymerogenic mutant of neuroserpin. Once again, inhibition of the proteasome specifically increased mutant protein levels (Fig. 5C).
FIGURE 5.
ER-associated degradation selectively degrades mutant neuroserpin. A, Tet-ON WT and S52R PC12 were induced to express neuroserpin with doxycycline for 2 days followed by withdrawal for 1 day of washout. During either induction or washout periods, cells were treated with 100 nm MG132 (MG) and/or rapamycin 200 nm (Rap). Control cells were left untreated (UT). Cell lysates were subjected to SDS-PAGE and immunoblotted for total neuroserpin using the polyclonal anti-neuroserpin antibody (21). B, combined data from three independent repeats are expressed graphically; mean ± S.E. *, p < 0.05. C, Tet-ON WT and G392E PC12 were induced to express neuroserpin with doxycycline for 2 days. During the final 23 h of induction, cells were treated with 5 μm lactacystin and/or rapamycin 200 nm. Cell lysates were subjected to SDS-PAGE and immunoblotted for total neuroserpin using the polyclonal anti-neuroserpin antibody (21). D, ATG5+/+ or ATG5−/− MEFs were transiently transfected with expression vectors for wild type (WT) or polymerogenic mutants (S52R and G392E) of neuroserpin. Cells were treated with 100 nm MG132 and/or 200 nm rapamycin, and cell lysates were subjected to SDS-PAGE and immunoblotted for total neuroserpin. E, combined data from three independent repeats are expressed graphically; mean ± S.E.*, p < 0.05; **, p < 0.01.
An up-regulation of autophagy has been described in some settings of ER dysfunction, although the signaling pathway linking these two processes has yet to be fully defined (25–27). Despite our finding that ERAD is primarily responsible for the selective clearance of mutant serpins, there remained the potential that autophagy might play a role. We therefore manipulated the autophagic pathway using chemical and genetic means in cells in which ERAD was compromised.
First, cells were induced to express neuroserpin in the presence of MG132. These cells were then treated with rapamycin to augment autophagy. Rapamycin partially rescued the MG132-induced accumulation of neuroserpin, an effect that was more dramatic for the polymerogenic (S52R) mutant (Fig. 5, A and B). The effect became even more apparent during the washout phase. MG132 significantly inhibited the loss of mutant but not WT neuroserpin. Remarkably, this effect was abrogated by the co-administration of rapamycin, while rapamycin had little effect alone (Fig. 5, A and B). Similar effects were seen with lactacystin and the G392E mutant (Fig. 5C).
An important weakness of the pharmacological approach was the potential for off-pathway effects of the chemicals used. To address this concern, we also utilized mouse embryonic fibroblasts (MEFs) that were incapable of autophagy because of targeted deletion of the ATG5 gene (28). ATG5+/+ and ATG5−/− cells were transiently transfected with expression vectors encoding either WT or polymerogenic mutants of neuroserpin and then treated with MG132. Inhibition of the proteasome in ATG5+/+ MEFs caused a modest increase of neuroserpin immunoreactivity, that was most marked for the G392E mutant. However, in the autophagy-deficient ATG5−/− MEFs, both polymerogenic mutants were markedly increased following treatment with MG132 (S52R p < 0.05, G392E p < 0.01; Fig. 5, D and E). While it would not be valid to compare total levels of transiently expressed neuroserpin between the two cell lines, it was apparent the proteasome inhibition had a more dramatic effect in the autophagy-deficient cells. This suggested that, at least in fibroblasts, autophagy cooperates with ERAD to oppose accumulation of the polymerogenic mutants of neuroserpin.
MG132 Increases Intracellular Levels of Both Monomeric and Polymeric Mutant Neuroserpin
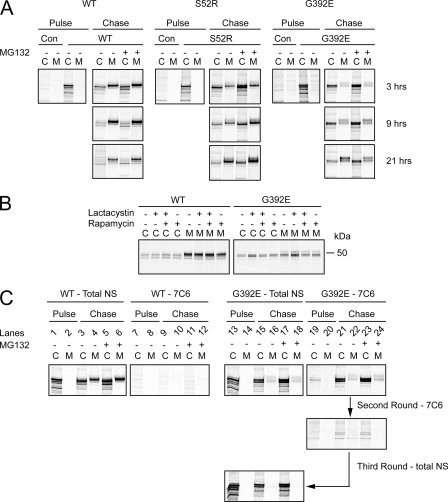
MG132 is well known to induce both the unfolded protein and heat shock responses. We were therefore concerned that these homeostatic responses might affect intracellular levels of transgenic protein through mechanisms other than degradation, e.g. transcription or translation. We therefore pulse-labeled COS7 cells expressing WT or mutant neuroserpin with [35S]methionine and cysteine and then tested the effect of MG132 during the chase period. Inhibition of the proteasome led to persistence of intracellular S52R and G392E neuroserpin and increased secretion of S52R neuroserpin (Fig. 6A). This indicated that MG132 was affecting a post-translational process, most likely ERAD of the transgenic proteins.
FIGURE 6.
MG132 increases intracellular levels of both monomeric and polymeric mutant neuroserpin. A, COS7 cells expressing WT, S52R or G392E neuroserpin, or the parental vector pcDNA3.1 (Con) were metabolically labeled for 30 min with [35S]methionine and cysteine. Cells were treated throughout with 7 μm MG132 and/or 0.5 μm rapamycin or with the drug solvent control. Neuroserpin was immunoprecipitated from cell lysates (C) and conditioned media (M) with the polyclonal (total) neuroserpin antibody then samples were resolved on 10% w/v SDS-PAGE and subjected to autoradiography. B, COS7 cells expressing WT or G392E neuroserpin were metabolically labeled for 30 min with [35S]methionine and cysteine. During the pulse, cells were treated with lactacystin 25 μm and/or rapamycin 0.5 μm. Neuroserpin was immunoprecipitated from cell lysates (C) and conditioned media (M) with the polyclonal (total) neuroserpin antibody then samples were resolved on 10% w/v SDS-PAGE and subjected to autoradiography. C, COS7 cells expressing WT or G392E neuroserpin were metabolically labeled for 30 min with [35S]methionine and cysteine. After 3 h of chase in the presence or absence of MG132, neuroserpin was immunoprecipitated from cell lysates and medium with either the total neuroserpin polyclonal antibody or the polymer-specific monoclonal antibody (7C6). The G392E neuroserpin flow-through was then subjected to a further round of immunoprecipitation with the 7C6 antibody before the remaining neuroserpin was immunoprecipitated with the polyclonal (total) antibody. Samples were resolved on 10% w/v SDS-PAGE and subjected to autoradiography.
Next, we tested the ability of rapamycin to reduce the levels of mutant neuroserpin in metabolically labeled COS7 cells treated with the proteasome inhibitor lactacystin. The effects were similar to those seen by Western blot in Fig. 5, with rapamycin selectively lowering mutant G392E neuroserpin only in cells treated with the proteasome inhibitor (Fig. 6B).
Finally, we wished to determine if the effect of proteasome inhibition was on the levels of mutant monomers, polymers, or both. Once again, we metabolically labeled newly synthesized WT neuroserpin or the highly polymerogenic G392E mutant for 30 min and then chased with unlabeled medium for 3 h to allow formation of polymers. During the chase period, cells were treated with or without MG132. Cell lysates and conditioned media were then subjected to immunoprecipitation either with the polyclonal antibody to detect total neuroserpin or with the polymer-specific 7C6 monoclonal antibody (Fig. 6C). MG132 led to a modest increase in total WT protein both in cell lysates and in the medium. As before, no WT protein was detectable with the polymer-specific antibody. In contrast, in the G392E-expressing cells MG132 caused a marked increase of intracellular neuroserpin, both total and polymeric. To ensure that polymeric neuroserpin had been successfully immunodepleted, a further immunoprecipitation was then performed on the flow-through, which yielded no further neuroserpin. However, when the flow-through was immunoprecipitated with the polyclonal antibody able to recognize all neuroserpin conformers, neuroserpin could be isolated indicating that a substantial proportion of the labeled protein remained in the monomeric form. These data demonstrated that within the cell the levels of polymers and monomers of the polymerogenic mutant neuroserpin were comparable. Treatment with MG132 increased both monomeric and polymeric neuroserpin to a similar extent.
DISCUSSION
In this study, we set out to determine how polymerogenic mutant serpins are cleared from the cell. We observed that neuroserpin polymers form slowly, with a lag period of at least 30 min. We also observed in both inducible Tet-ON PC12 cells and transiently transfected COS7 cells that ERAD is responsible for much of the degradation of polymerogenic mutant serpins, while autophagy appears to degrade both mutant and WT transgenic protein without evidence of selectivity. In contrast, in fibroblasts, inhibition of the proteasome had a proportionally greater effect in autophagy-deficient ATG5−/− cells, indicating that at least in this cell type some of the mutant protein could be cleared selectively by autophagy.
Aberrant protein folding is the cause of many human diseases (7). The serpinopathies comprise an unusual subset of these diseases, because their causative mutations promote the formation of large ordered polymers (1). These polymers are not trafficked, but instead accumulate as punctate structures that co-localize with ER markers (12, 21, 29). It is striking that unlike many abnormally folded ER proteins, retained serpin polymers fail to activate the unfolded protein response (30–32). Large inclusions of neuroserpin polymers form in the brain of patients with FENIB, however, their appearance takes many years, rather than the days required in cultured cells (11, 12, 34). It is likely that the faster rate of polymer accumulation seen in the cultured cells reflects higher expression levels in this model or defects in the degradation of mutant neuroserpin.
The established model of serpin polymerization, whereby the reactive center loop of one molecule inserts into the β-sheet A of another (2), has recently been challenged by a model based on the crystal structure of a dimer of antithrombin formed at low pH (4). This structure showed a larger domain swap that involved a β-hairpin containing the reactive center loop and strand 5A. This mechanism was extrapolated by molecular modeling into longer chains of polymers. The authors argue that this mechanism is likely as serpin polymers formed while the protein was folding rather than from folded protein. We therefore determined if aberrant folding of newly synthesized neuroserpin might allow co-translational polymerization. Our inability to detect polymers by specific immunoprecipitation following metabolic labeling for 30 min, followed by the appearance of polymers after 3 h of chase, demonstrated that full-length polypeptide chains are first synthesized and only then are polymers assembled from pre-existing neuroserpin molecules. This still does not preclude the formation of polymers from incompletely folded subunits, but it does indicate that monomers require at least 30 min of ER residence before being incorporated into polymers.
Previous studies in both yeast (35, 36) and mammalian cells (16, 37, 38) have suggested that ERAD is involved in the degradation of Z α1-antitrypsin. However, the remarkable stability of serpin polymers in vitro has led some to question whether ERAD could adequately handle serpin polymers in vivo. This would require the delivery of serpin monomers back into the cytosol and such disassembly of serpin polymers has not yet been observed in cell or animal models of disease. Autophagy offered an attractive alternative mechanism, since this process is able to degrade large organelles in addition to long-lived proteins (24). Morphologic studies of Z α1-antitrypsin-expressing fibroblast cell lines supported such a role for autophagy, with electron microscopy revealing dilated ER cisternae filled with granular material that co-localized with autophagosome markers (16). Subsequently, the degradation of Z α1-antitrypsin was shown to be delayed in autophagy-deficient ATG5−/− embryonic stem cells and that Z α1-antitrypsin co-localized with LC3-GFP, a marker of autophagosomes (15). It is worth highlighting, however, that in the latter study, degradation of WT α1-antitrypsin was not measured. It is therefore unclear if autophagy degrades heterologously expressed mutant α1-antitrypsin in fibroblasts to a greater or lesser extent compared with WT α1-antitrypsin. Further studies in yeast identified Z α1-antirypsin degradation-deficient (add) mutants, of which one (add3) could be complemented by VPS30/Atg6, a component of a PI 3-kinase complex involved in autophagy (39). Thus it was suggested that separate pathways might be responsible for degrading soluble Z α1-antitrypsin and large aggregates. While heterologous expression in yeast has provided some insights into serpin degradation, we felt it necessary to study neuroserpin degradation in cells with similar physiology to those likely to express the protein in vivo. We observed that, surprisingly, WT and mutant neuroserpins are cleared from cells and fly models of disease with equal efficiency. PC12 cells and neurons package WT neuroserpin into secretory vesicles that undergo regulated exocytosis, while retaining mutant neuroserpins within their ER. We reasoned, therefore, that much of the decay of mutant neuroserpin immunoreactivity likely reflected the degradation of retained proteins rather than secretion. We found that both autophagy and ERAD participate in the degradation of neuroserpin in neuronal-like PC12 cells. However it was ERAD, and not autophagy, that provided specificity for mutant neuroserpin. The finding that polymer formation occurred in vivo following a lag phase of at least 30 min suggested that mutant monomers may be susceptible to classical ERAD for a considerable period of time. It is interesting that we went on to observe in MEFs that inhibition of the proteasome had a far less dramatic effect, except in ATG5−/− MEFs in which the autophagic pathway was disabled genetically. This most likely reveals an important cell-type specific difference in serpin degradation and emphasizes the importance of studying each serpin in a cell type appropriate to its synthesis in vivo.
Autophagy has been shown to be up-regulated during ER stress (25, 26) but there is little evidence for ER stress signaling in the serpinopathies (30–32). Autophagy plays a major role in organelle turnover and this may explain why its deficiency in ATG5−/− cells is associated with the accumulation of serpin polymers. One possible interpretation of our findings is that proteasomal degradation selectively targets mutant monomers, while autophagy is responsible for bulk turnover. This model would help reconcile the competing evidence that both autophagy and ERAD play important roles in serpin clearance. It also fits well with the observation that alternate pathways appear to be responsible for degrading soluble Z α1-antitrypsin and large aggregates in a yeast model (39). This is notable, since these mutants fail to activate an UPR and may indicate that their targeting for ERAD involves a signal distinct from that required by the UPR or that their degradation is so efficient that activation of the UPR is avoided. During these experiments, we were mindful that although MG132 is frequently used as a proteasome inhibitor, it also has some inhibitory activity against lysosomal cathepsins. Our observation that MG132 treatment led to a more dramatic increase of mutant neuroserpin in ATG5−/− mutant cells was reassuring in this regard, because it indicated that the MG132 effect was not mediated through impaired autophagolysosome function.
We have shown that while inhibition of autophagy, either through pharmacological or genetic means, led to an increase in wild type and mutant serpin levels, augmenting autophagy with rapamycin failed to improve serpin clearance under basal conditions. This may indicate that autophagic turnover of proteins within the ER was functioning maximally in the system used and that inhibition of mTOR signaling was unable to increase the rate further. However, when proteasome impairment led to the accumulation of serpin polymers, rapamycin treatment did rescue the clearance of the accumulated polymerogenic mutants. The reason why autophagic clearance could be induced in that setting is not clear. Perhaps additional signals are generated during such treatments that are able to activate autophagy over-and-above mTOR inhibition, or else the accumulated mutant protein might become more accessible to autophagic clearance either by being relocated to a subdomain of the ER (29) or perhaps some of the protein was returned to the cytosol by retrotranslocation. A link between proteasome function, longevity and aging has been suggested (40) and aging has been show to perturb 26S proteasome assembly (33). It is therefore tempting to speculate that if impaired proteasome function with age contributes to the late accumulation of intracellular polymers, then augmentation of autophagy with agents such as rapamycin might provide clinical benefit.
Taken together, our findings demonstrate that the degradation of polymerogenic mutant serpins involves both specific ERAD and nonspecific autophagy. Although autophagic protein turnover may play a minor part in mutant protein degradation, its importance can be revealed when ERAD fails. At that point the augmentation of autophagy can help rescue the clearance of these toxic species.
Acknowledgments
We thank Prof. Karin Römisch (Saarland University, Germany) for helpful comments on the manuscript. The ATG5−/− MEFs were a kind gift from Prof. Noboru Mizushima, Tokyo Medical and Dental University.
This work was supported in part by grants from the Medical Research Council, Engineering and Physical Sciences Research Council, the Alzheimer Research Trust, and the Papworth National Health Service Trust.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- ERAD
- endoplasmic reticulum-associated degradation
- PBS
- phosphate-buffered saline
- DMEM
- Dulbecco's modified Eagle's medium
- WT
- wild type
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Carrell R. W., Lomas D. A. (1997) Lancet 350,134–138 [DOI] [PubMed] [Google Scholar]
- 2.Lomas D. A., Mahadeva R. (2002) J. Clin. Invest. 110,1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntington J. A., Read R. J., Carrell R. W. (2000) Nature 407,923–926 [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki M., Li W., Johnson D. J., Huntington J. A. (2008) Nature 455,1255–1258 [DOI] [PubMed] [Google Scholar]
- 5.Gooptu B., Lomas D. A. (2008) J. Exp. Med. 205,1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8,519–529 [DOI] [PubMed] [Google Scholar]
- 7.Marciniak S. J., Ron D. (2006) Physiol. Rev. 86,1133–1149 [DOI] [PubMed] [Google Scholar]
- 8.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7,766–772 [DOI] [PubMed] [Google Scholar]
- 9.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) Genes Dev. 18,3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12,982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R. L., Shrimpton A. E., Holohan P. D., Bradshaw C., Feiglin D., Collins G. H., Sonderegger P., Kinter J., Becker L. M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D. A., Yerby M. S., Shaw C. M., Gooptu B., Elliott P. R., Finch J. T., Carrell R. W., Lomas D. A. (1999) Nature 401,376–379 [DOI] [PubMed] [Google Scholar]
- 12.Miranda E., MacLeod I., Davies M. J., Pérez J., Römisch K., Crowther D. C., Lomas D. A. (2008) Hum. Mol. Genet. 17,1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies M. J., Miranda E., Roussel B. D., Kaufman R. J., Marciniak S. J., Lomas D. A. (2009) J. Biol. Chem. 284,18202–18209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pahl H. L. (1999) Physiol. Rev. 79,683–701 [DOI] [PubMed] [Google Scholar]
- 15.Kamimoto T., Shoji S., Hidvegi T., Mizushima N., Umebayashi K., Perlmutter D. H., Yoshimori T. (2006) J. Biol. Chem. 281,4467–4476 [DOI] [PubMed] [Google Scholar]
- 16.Teckman J. H., Perlmutter D. H. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279,G961–G974 [DOI] [PubMed] [Google Scholar]
- 17.Ravikumar B., Duden R., Rubinsztein D. C. (2002) Hum. Mol. Genet. 11,1107–1117 [DOI] [PubMed] [Google Scholar]
- 18.Lin D. M., Goodman C. S. (1994) Neuron 13,507–523 [DOI] [PubMed] [Google Scholar]
- 19.Osterwalder T., Yoon K. S., White B. H., Keshishian H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98,12596–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latouche M., Lasbleiz C., Martin E., Monnier V., Debeir T., Mouatt-Prigent A., Muriel M. P., Morel L., Ruberg M., Brice A., Stevanin G., Tricoire H. (2007) J. Neurosci. 27,2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miranda E., Römisch K., Lomas D. A. (2004) J. Biol. Chem. 279,28283–28291 [DOI] [PubMed] [Google Scholar]
- 22.Seglen P. O., Gordon P. B. (1982) Proc. Natl. Acad. Sci. U.S.A. 79,1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. (1998) Cell Struct. Funct. 23,33–42 [DOI] [PubMed] [Google Scholar]
- 24.Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Nat. Rev. Drug Discov. 6,304–312 [DOI] [PubMed] [Google Scholar]
- 25.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Mol. Cell. Biol. 26,9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) J. Biol. Chem. 281,30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yorimitsu T., Klionsky D. J. (2007) Autophagy 3,160–162 [DOI] [PubMed] [Google Scholar]
- 28.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432,1032–1036 [DOI] [PubMed] [Google Scholar]
- 29.Granell S., Baldini G., Mohammad S., Nicolin V., Narducci P., Storrie B., Baldini G. (2008) Mol. Biol. Cell 19,572–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham K. S., Le A., Sifers R. N. (1990) J. Biol. Chem. 265,20463–20468 [PubMed] [Google Scholar]
- 31.Hidvegi T., Schmidt B. Z., Hale P., Perlmutter D. H. (2005) J. Biol. Chem. 280,39002–39015 [DOI] [PubMed] [Google Scholar]
- 32.Lawless M. W., Greene C. M., Mulgrew A., Taggart C. C., O'Neill S. J., McElvaney N. G. (2004) J. Immunol. 172,5722–5726 [DOI] [PubMed] [Google Scholar]
- 33.Vernace V. A., Arnaud L., Schmidt-Glenewinkel T., Figueiredo-Pereira M. E. (2007) FASEB J. 21,2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takao M., Benson M. D., Murrell J. R., Yazaki M., Piccardo P., Unverzagt F. W., Davis R. L., Holohan P. D., Lawrence D. A., Richardson R., Farlow M. R., Ghetti B. (2000) J. Neuropathol. Exp. Neurol. 59,1070–1086 [DOI] [PubMed] [Google Scholar]
- 35.Scott C. M., Kruse K. B., Schmidt B. Z., Perlmutter D. H., McCracken A. A., Brodsky J. L. (2007) Mol. Biol. Cell 18,3776–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCracken A. A., Kruse K. B. (1993) Mol. Biol. Cell 4,729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu D., Teckman J. H., Omura S., Perlmutter D. H. (1996) J. Biol. Chem. 271,22791–22795 [DOI] [PubMed] [Google Scholar]
- 38.Teckman J. H., Burrows J., Hidvegi T., Schmidt B., Hale P. D., Perlmutter D. H. (2001) J. Biol. Chem. 276,44865–44872 [DOI] [PubMed] [Google Scholar]
- 39.Kruse K. B., Brodsky J. L., McCracken A. A. (2006) Mol. Biol. Cell 17,203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C. M., Xu C. F., Neubert T. A., Mor A., Philips M. R., Ron D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]