Abstract
Benzo[c]phenanthrene dihydrodiol epoxide (B[c] PhDE) is well known as an important environmental chemical carcinogen that preferentially modifies DNA in adenine residues. However, the molecular mechanism by which B[c]PhDE induces tumorigenesis is not fully understood. In this report, we demonstrate that DNA mismatch repair (MMR), a genome maintenance system, plays an important role in B[c]PhDE-induced carcinogensis by promoting apoptosis in cells treated with B[c]PhDE. We show that purified human MMR recognition proteins, MutSα and MutSβ, specifically recognized B[c]PhDE-DNA adducts. Cell lines proficient in MMR exhibited several-fold more sensitivity to killing than cell lines defective in either MutSα or MutLα by B[c]PhDE; the nature of this sensitivity was shown to be due to increased apoptosis. Additionally, wild-type mice exposed to B[c]PhDE had intestinal crypt cells that underwent apoptosis significantly more often than intestinal crypt cells found in B[c]PhDE-treated Msh2–/– or Mlh1–/– mice. These findings, combined with previous studies, suggest that the MMR system may serve as a general sensor for chemical-caused DNA damage to prevent damaged cells from mutagenesis and carcinogenesis by promoting apoptosis.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAH) are widely distributed in our environment as by-products of tobacco smoke and fuel combustion. Many of these environmental pollutants are metabolically activated to highly reactive PAH diol epoxide derivatives, which can covalently bind to native DNA to implement their mutagenic and carcinogenic activity (1). These PAH diol epoxides exist as a pair of diastereoisomers, designated as syn- or anti-derivatives. Each diastereoisomer in turn can be further resolved into two enantiomers, specified as (+) and (–). Depending on the manner by which it reacts with DNA, each enantiomer can form either a trans- or cis-DNA adduct. Benzo[c]phenanthrene 3,4-dihydrodiol-1,2 epoxide (B[c]PhDE) is one of the most potent mutagenic and carcinogenic PAH compounds (2–5). Unlike most of the PAH epoxides, e.g. benzo[a]pyrene dihydrodiol epoxides (B[a]PDE), which react preferentially with the guanine residue (6–8), B[c]PhDE reacts preferentially with adenine residues in DNA (2,9,10).
Although PAH adducts are highly mutagenic and carcinogenic, they can be removed by DNA repair systems such as the nucleotide excision repair and base excision repair pathways (for reviews see 11,12). Recently, it has been demonstrated that the DNA mismatch repair (MMR) pathway plays a critical role in mediating DNA damage-induced apoptosis (13). MMR is a critical mutation avoidance system that is well known for its tumor suppression function by correcting mismatches generated during normal DNA metabolism (14,15). In Escherichia coli, the MMR pathway is dependent on MutS, MutL and MutH proteins (reviewed in 16). In eukaryotic cells, MutS homologs and MutL homologs function as heterodimeric complexes (17,18). It has been shown recently that cells deficient in either MutS or MutL complexes are highly resistant to cytotoxic effects of many chemicals or chemical carcinogens, including B[a]PDE, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), aminofluorene derivatives and cisplatin (reviewed in 13,18,19). MutS complexes have been shown to specifically bind to DNA adducts caused by these DNA-damaging agents (13). These findings indicate that besides mismatch correction, the MMR pathway stabilizes genomic integrity through a second route: signaling cells with severe DNA damage to undergo apoptosis (13).
To understand if the MMR pathway can mediate DNA damage-induced apoptosis caused by B[c]PhDE, cells with different MMR backgrounds were analyzed for their ability to respond to B[c]PhDE. We demonstrate here that B[c]PhDE induces apoptotic cell death in both human cells and mice, and that the apoptotic response requires both functional MutS complexes and MutL complexes. We also show that human MutS heterodimeric complexes, MutSα and MutSβ, efficiently recognize DNA containing a single B[c]PhDE-adducted adenine residue. These observations indicate that like many DNA adducts that are modified at guanine residues, B[c]PhDE-dA DNA adducts can also be recognized and processed by the MMR system.
MATERIALS AND METHODS
Cell lines and nuclear extracts
Lymphoblastoid lines TK6 and MT1 were grown in RPM1 1640 medium (Gibco) supplemented with 10% fetal bovine serum (HyClone) as described (20). Colorectal tumor cell lines HCT116 and HCT116-Chr.3 were grown in McCoy’s 5A medium with 10% fetal bovine serum. HeLa cells were purchased from the National Cell Culture Center (Minnesota). Nuclear extracts from HeLa and MT1 cells were prepared as described (21).
MutSα and MutSβ purification
MutSα (MSH2–MSH6) or MutSβ (MSH2–MSH3) were purified from nuclear extracts of HeLa and MT1 cells, respectively, as described (22–24). Unlike HeLa cells, which possess both MutSα and MutSβ, MT1 cells are defective in MSH6 (25) and only express MutSβ. Briefly, the 36–65% ammonium sulfate fractions of nuclear extracts were first applied onto a single-strand DNA cellulose column, and fractions containing MutSα or MutSβ were detected by band-shift assay (see below). These fractions were pooled and further purified through Mono Q and Superose 200 FPLC column chromatography. The resulting proteins were nearly homogenous as judged by Coomassie Brilliant Blue staining after SDS–polyacrylamide gel electrophoresis (PAGE) (data not shown). The individual subunits of these two heterodimers were confirmed by western blots using antibodies against MSH2, MSH3 or MSH6 (not shown).
Substrate construction and band shift analysis
Oligodeoxynucleotide 5′-ACCGCCGGCCAGGAGGAGTAC-3′ containing a single (+)-trans-anti- or (–)-trans-anti-B[c]PhDE at the bold A residue was synthesized, purified and characterized as described (26). The B[c]PhDE-containing oligomers were annealed to the centrally located complementary sequence of a 50mer (5′-GCAGATCTGGCCTGGTA CTCCTCCTGGGCGGCG GTTAACAGTACGTAGTC-3′, which was purchased from Invitrogen and purified by PAGE) and ligated with a 15mer oligonucleotide (5′-GACTACGTACTGTTA-3′) to the modified oligonucleotide (Fig. 1). The ligated product was elongated using the 50mer as a template in the presence of [α-32P]dCTP and the other three dNTPs and the Klenow fragment of DNA polymerase I as described (27). The resulting 50mer DNA duplexes containing B[c]PhDE adducts were purified by PAGE as described (28). Several 50mer control duplexes containing no B[c]PhDE adducts were also similarly constructed, which included an A-T homoduplex, an A-C heteroduplex, a CA di-nucleotide (nt) and a CACA tetra-nt insertion/deletion heteroduplex. These duplexes contained the same sequence as the B[c]PhDE-containing duplex except for the heterologies, which were placed at the same location as the B[c]PhDE adduct. Band shift assays were performed as described (27) in 25 µl reactions containing 0.5 pmol of 32P-labeled oligonucleotide duplex, 25–100 ng of MutSα or MutSβ, 0.4 µg of circular double-stranded f1MR3 bacterial phage DNA (competitor DNA), 10 mM HEPES–KOH (pH 7.5), 110 mM KCl, 1 mM EDTA, 1 mM DTT and 4% glycerol. Reaction mixtures were incubated on ice for 20 min followed by addition of 5 µl of 50% sucrose (w/v). The samples were then fractionated at room temperature in a 6% non-denaturing polyacrylamide gel in 6.7 mM Tris–acetate (pH 7.5) and 1 mM EDTA with buffer recirculation. Bands were detected by autoradiography.
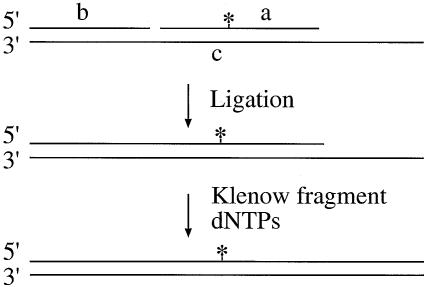
Figure 1.
Construction of 50mer deoxyoligonucleotide containing a single B[c]PhDE-dA adduct. B[c]PhDE-containing 21mer (oligonucleotide a) was ligated with oligonucleotide b after they were annealed to a complementary 50mer (oligonucleotide c). The ligated oligomer was then elongated to 50 nucleotides using oligonucleotide c as the template. The 50mer homoduplex (A-T) and heteroduplex containing an A-C mismatch, a 2-nt insertion/deletion (ID) mispair, or a 4-nt ID mispair were also constructed using similar procedures. The star represents B[c]PhDE adduct.
Treatment of cell lines with B[c]PhDE
The desired amount of B[c]PhDE (Chemsyn Science Laboratories, Lenexa, KS) was dissolved in a small amount of dimethyl sulfoxide and was added to cells suspended in culture medium. Cells were then incubated at 37°C in the presence of 5% CO2 for 1 h. The exposure was terminated by centrifuging and resuspending the cells in fresh medium. The final concentration of dimethyl sulfoxide was always <0.1% (v/v) and did not contribute to toxicity. The survival of carcinogen-treated or untreated cells was determined by clonogenic analysis as previously described (23). Treated TK6 and MT1 cells were plated in 96-well plates at a density of 1–3 cells/well in 0.2 ml of medium in the presence of γ-irradiated TK6 cells as feeder cells (1 × 105 cells/well), which were irradiated (137Cs) for a total of 4.2 kRad. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2 until clone formation was visible. Treated HCT116 and HCT116-Chr.3 cells were plated in 6-well plates (∼50 cells/plate) and cultured at 37°C in a 5% CO2 atmosphere. After incubation for 2 weeks, clones were counted under a microscope.
B[c]PhDE-induced apoptosis in cell lines was determined by the analysis of DNA fragmentation using agarose gel electrophoresis and the terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) method as previously described (23). Cells were incubated in medium with or without 0.25 µM B[c]PhDE at 37°C for 1 h and released in fresh medium. After 12 and 24 h of culture, an aliquot of the cells was harvested and DNA was isolated from these cells, electrophoresized through 1.6% agarose gels and visualized under UV light in the presence of ethidium bromide. The remaining cells were fixed with 1% (w/v) paraformaldehyde in phosphate-buffered saline, and labeled by propidium iodide (PI) and FITC-dUTP in the presence of TdT, which were provided in the APO-DIRECT™ kit (PharMingen, San Diego, CA). The cells were then analyzed by flow cytometry.
Apoptosis analysis in mice
Mice with a complete inactivation of Msh2 (a generous gift from Tak Mak) or Mlh1 (a generous gift from Michael Liskay) were previously described (29,30). Mice (wild type, Msh2–/– and Mlh1–/–) were given an intraperitoneal (i.p.) injection of B[c]PhDE (0.2 µmol/kg) and were sacrificed at hour 0, 3, 6, 12 and 24 after injection. Small intestines were fixed and embedded in paraffin. Sections (5 µm thick) were then prepared and stained with hematoxylin and eosin (H&E) for histological analysis or stained with ApopTag Peroxidase Kits (Intergen Co., Purchase, NY) for apoptotic analysis. A minimum of 50 half-crypts were scored per animal as described (31) under a microscope.
RESULTS
Recognition of B[c]PhDE adducts by MutSα and MutSβ
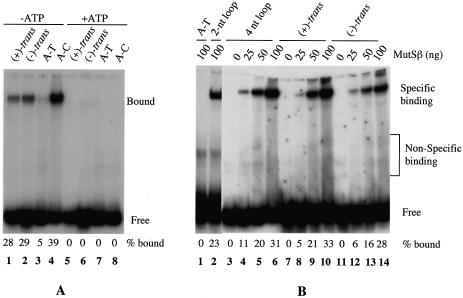
Recognition of DNA containing chemically modified guanine adducts by MutS homologs has been documented (13). To determine if these MMR proteins can recognize DNA adducts in adenine residues, MutSα and MutSβ were tested for their ability to bind a 50mer oligodeoxynucleotide duplex containing a single (+)-trans-anti-B[c]PhDE-dA adduct or (–)-trans-anti-B[c]PhDE-dA adduct. As shown in Figure 2A, MutSα efficiently binds to oligonucleotide duplexes containing each of the B[c]PhDE adducts (lanes 1 and 2). Although the affinity of MutSα for the B[c]PhDE substrates was less than for an A-C mismatched substrate (Fig. 2A, lane 4) under the experimental conditions, it was significantly higher than the affinity to a homoduplex, whose interaction with MutSα was almost undetectable (Fig. 2A, lane 3). Like MutSα, MutSβ demonstrated a high affinity for each of the B[c]PhDE substrates (Fig. 2B, lanes 7–14), while it poorly interacted with the homoduplex control (Fig. 2B, lane 1). However, unlike MutSα, MutSβ appears to bind equally well to the B[c]PhDE-containing substrates and heteroduplexes containing a 2-nt (Fig. 2B, lane 2) or 4-nt (Fig. 2B, lanes 3–6) loop, as judged by the fact that ∼30% of duplexes in each case were shifted by 100 ng MutSβ (lanes 6, 10 and 14). The binding of both MutSα and MutSβ to the B[c]PhDE adducts was totally inhibited in the presence of 1 mM ATP (Fig. 2A, lanes 5–8, and data for MutSβ not shown). Inhibition of the interaction between MutS homologs and heteroduplexes by ATP has been documented (22,32), and could be due to the fact that binding of ATP to MutS homologs and/or the subsequent hydrolysis of ATP induce conformational changes of MutS homologs and promote translocation of the proteins along the DNA molecules (32–34). Our gel-shift results indicate that the MMR initiation factors are capable of recognizing and interacting with DNA damage induced by B[c]PhDE in a manner by which they interact with other DNA adducts (reviewed in 13) and heteroduplexes (22,32).
Figure 2.
Binding of MutSα and MutSβ to DNA containing B[c]PhDE-dA adducts. Unless otherwise indicated, gel-shift assays were performed in 25 µl reactions containing 0.5 pmol 32P-labeled oligonucleotide duplexes, MutSα (100 ng) or MutSβ (25–100 ng). After 20 min incubation on ice, 5 µl 50% sucrose was added. The samples were fractionated at room temperature through a 6% non-denaturing polyacrylamide gel in 6.7 mM Tris–acetate (pH 7.5) and 1 mM EDTA with buffer recirculation. When present, ATP is at a final concentration of 1 mM. (A) Interaction of MutSα with B[c]PhDE-dA substrates. (B) Interaction of MutSβ with B[c]PhDE-dA substrates. (+)-trans, (+)-trans-anti-B[c]PhDE; (–)-trans, (–)-trans-anti-B[c]PhDE; A-T, oligonucleotide substrate containing no mismatches; A-C, oligonucleotide substrate containing an A-C mismatch; 2-nt loop, oligonucleotide substrate containing two unpaired (or ID) nucleotides; 4-nt loop, oligonucleotide substrate containing four unpaired nucleotides.
Mismatch repair proficient cells are sensitive to B[c]PhDE cytotoxicity
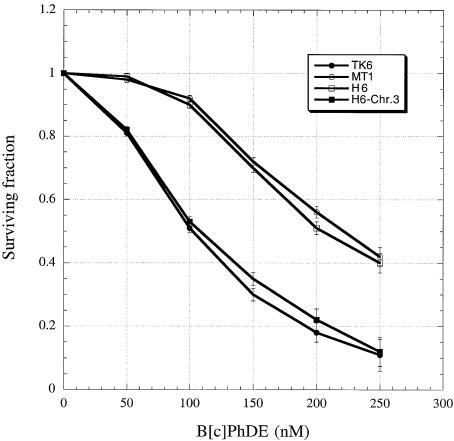
Given the specific interaction of human MutS homologs with DNA containing B[c]PhDE adducts, it is conceivable that the MMR pathway could influence the biological impact of B[c]PhDE cytotoxicity. Thus, the cytotoxic effects of exposure to B[c]PhDE were determined in MMR-proficient and -deficient cells. The MMR-deficient cell lines used were MSH6-defective MT1 (defective in MutSα) and MLH1-defective HCT116 (defective in all MutL heterodimers); the corresponding wild-type cell lines used were TK6 and HCT116-Chr.3. MT1 was derived from lymphoblastoid cell line TK6 by frameshift mutagenesis and selected by its tolerance to N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (35); HCT116-Chr.3 was derived from colorectal tumor cell line HCT116 by transfer of chromosome 3 that carries the wild-type MLH1 gene (36). Cells were treated with various concentrations of B[c]PhDE and subjected to clonogenic analysis. As shown in Figure 3, both MMR-proficient cell lines (TK6 and HCT116-Chr.3) exhibited lower levels (2–3-fold lower) of clonal survival than their repair-deficient counterparts (MT1 and HCT116), respectively, following exposure to B[c]PhDE treatment. These results indicate that cytotoxicity of B[c]PhDE requires functional MutS and MutL homologs. The sensitivity of MMR-proficient cells to other DNA damage agents, such as MNNG, cisplatin and B[a]PDE, has been previously reported (reviewed in 13), and our observations with B[c]PhDE in this study are consistent with previous studies using other DNA damage agents.
Figure 3.
Sensitivity of MMR-proficient cells to cytotoxicity of B[c]PhDE. Clonogenic survival of MMR-proficient (TK6 and HCT116–3-6) and MMR-deficient (MT1 and HCT116) cells following exposure to B[c]PhDE. Cells were incubated at 37°C in medium with or without (control) B[c]PhDE, as indicated, for 1 h, and then cultured in fresh medium without the chemical. The survival of both B[c]PhDE-treated or untreated cells was determined by clonogenic analysis as described in the Materials and Methods.
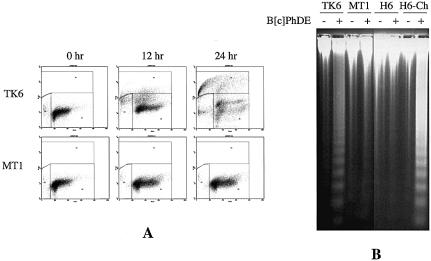
To further determine the nature of the B[c]PhDE sensitivity, cells were treated with 0.25 µM of B[c]PhDE and analyzed for apoptotic cell death using both TUNEL and classical DNA fragmentation analyses. Results of TUNEL analysis are shown in Figure 4A. In this assay, cells undergoing DNA fragmentation/breaking were labeled with FITC-dUTP by terminal deoxynucleotidyl transferase (TdT) and were believed to be apoptotic cells (positive in TUNEL assay). At 24 h after the treatment, ∼50% of TK6 cells were TUNEL-positive while no significant increase of TUNEL-staining cells in MT1 was observed. Similar results were also obtained in cell line pairs HCT116 and HCT116-Chr.3 (data not shown). The B[c]PhDE-induced apoptosis of MMR-proficient cells was confirmed by direct visualization of DNA fragmentation, a hallmark of apoptosis (37), as judged by the fact that DNA fragmentation was observed in MMR-proficient TK6 and HCT116-Chr.3 cells, but not in MMR-deficient MT1 and HCT116 cells in response to B[c]PhDE treatment (Fig. 4B). These results confirm that cellular sensitivity to B[c]PhDE cytotoxicity is via MMR-dependent apoptosis.
Figure 4.
B[c]PhDE-induces MMR-dependent apoptosis. (A) TUNEL analysis. Cells were treated with 0.25 µM B[c]PhDE at 37°C for 1 h, cultured in fresh medium for various times as indicated, and subjected to TUNEL analysis as described in Materials and Methods. (B) DNA fragmentation analysis. Cells were treated with B[c]PhDE (0.25 µM) for 1 h and cultured in fresh medium for 24 h. Genomic DNA was isolated by protease K digestion and phenol/chloroform extractions, fractionated through 1.5% agarose gels and visualized under UV light in the presence of ethidium bromide.
B[c]PhDE-induced apoptosis in mouse small intestine requires MSH2 or MLH1
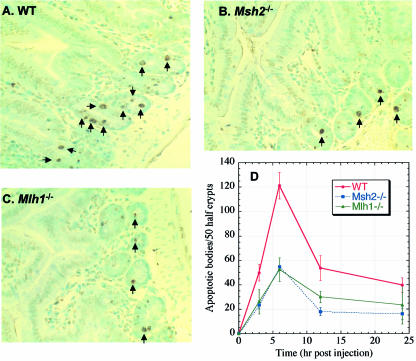
To determine if MMR-mediated apoptosis in response to chemical carcinogen treatment in cell lines actually reflects what occurs in vivo, apoptotic cell death in the small intestine of wild-type and Msh2–/– or Mlh1–/– mice was determined after exposure to B[c]PhDE. Apoptosis was scored within the crypts of the small intestine as described (38). Mice (wild type, Msh2–/– and Mlh1–/–) were given an i.p. injection of B[c]PhDE and were sacrificed at hour 0, 3, 6, 12 and 24 after injection. At each time point, a minimum of three animals was used and the small intestine was removed and fixed. Histological sections were stained using the ApopTag Peroxidase Kits (Intergen Co.) to determine the extent of apoptosis, and a minimum of 50 half-crypts were scored per animal. In this assay, nuclei in apoptotic cells are stained brown. Examples of this analysis are shown in Figure 5. Measurable increases in intestine apoptosis were observed in wild-type mice, with the peak induction of apoptosis at 6 h (Fig. 5D). However, the apoptotic response in Msh2–/– or Mlh1–/– mice was greatly diminished (Fig. 5D, also compare B and C with A). These observations strongly suggest that, like in cell lines, MMR-dependent apoptosis in response to B[c]PhDE-induced DNA damage also occurs in vivo.
Figure 5.
B[c]PhDE-induced apoptosis in mouse small intestine. Mice (wild type, Msh2–/– or Mlh1–/–) were injected with B[c]PhDE (0.2 µmol/kg) and sacrificed at hour 0, 3, 6, 12 or 24 after injection. Paraffin sections of small intestines were analyzed for apoptosis by ApopTag Peroxidase Kits (Intergen Co., Purchase, NY). For each point, a minimum of three mice were used and 50 half-crypts were scored under a microscopy.
DISCUSSION
It has been shown that cells deficient in either MutSα or MutL complexes are highly resistant to the cytotoxic effects of many chemical compounds as compared to cells proficient in the repair pathway (reviewed in 13,18). These compounds include B[a]PDE, alkylating agents, aminofluorene derivatives, hydrogen peroxide, cisplatin, etc. (20,23,39). A common feature of these compounds is to preferentially modify DNA at guanine residues. The DNA adducts caused by these guanine-damaging agents have been shown to be specifically recognized by MutS and its eukaryotic homologs (reviewed in 13). B[c]PhDE is one of the most potent environmental chemical carcinogens, and its mutagenic and carcinogenic activities have been well documented (4,5), and it preferentially modifies adenine residues in DNA (2,9,10). Our results in this study show that like guanine-specific chemical compounds, B[c]PhDE triggers MSH2- and MLH1-dependent apoptosis; this dependency was demonstrated both in human cell lines and in live mice. Clonogenic analysis of human cell lines indicate that MMR-proficient cells (TK6 and HCT116-Chr.3) are ∼3-fold more sensitive to B[c]PhDE treatment than MMR-deficient cells (MT1 and HCT116) (Fig. 4); and mice with the wild-type Msh2 or Mlh1 gene display a measurable increase in apoptosis in the small intestine in response to B[c]PhDE treatment as compared with Msh2- or Mlh1-knockout mice (Fig. 5). These results are in agreement with previous studies in cell lines in response to guanine-specific damaging agents (reviewed in 13,18), further confirming that MMR proteins play an important role in the DNA damage response.
We also demonstrate that human MutS homologs specifically recognize DNA containing B[c]PhDE-dA adducts, suggesting that the MMR system may recognize damaged adenine residues in a similar way by which it recognizes damaged guanine residues (reviewed in 13). These findings support the idea that besides mismatch correction, MMR proteins function as a general DNA damage sensor to promote apoptosis (13,18).
It is known that DNA damage caused by chemical carcinogens can be repaired by the nucleotide excision repair pathway (11,12). A recent in vitro study demonstrated that O6-methyl guanine could be removed by the MMR system in a manner dependent on a strand break in the same strand as the adduct (40), indicating that MMR can also process chemically adducted DNA damage in a strand-specific manner. However, given the pre-requisition for a strand break, removal of the chemically induced DNA adducts by the eukaryotic MMR system in vivo may not be practical. Extensive studies in eukaryotic MMR do not support the existence of a eukaryotic homolog of E.coli MutH, an activity required to create a strand break for MMR initiation in E.coli. Therefore, it is believed that MMR in eukaryotic cells is coupled with DNA replication, where strand breaks required to direct MMR are available. If this is the case, the MMR system will not be able to remove adducts located in the template strand, and may induce a futile repair cycle (41,42). The cycle may be the event that triggers DNA damage-induced apoptosis (13,18).
Despite the fact that cells defective in MMR are highly resistant to many DNA damaging agents, these cells also exhibit an increased rate of mutation after drug treatment (20,43–46), a phenomenon that is believed to initiate tumorigenesis (47). Although we did not determine the rate of B[c]PhDE-induced tumorigenesis of MMR wild-type and knockout mice, several studies have shown that homozygous MMR knockout mice are hypersensitive to alkylating agent-induced mutations and tumor development (43,48–50). Therefore, the MMR-dependent apoptosis in response to DNA damage is likely to prevent tumorigenesis by eliminating cells that contain severe DNA damage from growing (13,18).
How the processing of DNA adducts by the MMR system induces apoptosis is not fully understood. Recent studies have indicated that both p53 and its related protein p73 are implicated in MMR-dependent apoptosis, as phosphorylation of p53 and/or p73 is closely associated with the process (40,51). ATM and c-Abl appear to be the kinases that phosphorylate these proteins during the damage response (51). Physical interactions between MMR proteins (e.g. MutSα and MutLα) and proteins involved in the DNA damage-signaling network (e.g. ATM and p73) have been recently identified (52,53). These observations indicate that MMR-dependent apoptosis in response to DNA damage involves a signaling cascade. Understanding the signaling cascade requires further investigation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Michael Liskay and Tak Mak for Mlh1 and Msh2 mice, respectively, and Steve Presnell for comments on the manuscript. This work was supported by grants from the National Cancer Institute and the Madeline F. James endowment from the University of Kentucky Markey Cancer Center to G.M.L.
REFERENCES
- 1.Hall M. and Grover,P.L. (1990) Policyclic aromatic hydrocarbons: metabolism, activation and tumor initiation. Academic Press, San Diego, CA. [Google Scholar]
- 2.Agarwal R., Canella,K.A., Yagi,H., Jerina,D.M. and Dipple,A. (1996) Benzo[c]phenanthrene-DNA adducts in mouse epidermis in relation to the tumorigenicities of four configurationally isomeric 3,4-dihydrodiol 1,2-epoxides. Chem. Res. Toxicol., 9, 586–592. [DOI] [PubMed] [Google Scholar]
- 3.Bigger C.A., Strandberg,J., Yagi,H., Jerina,D.M. and Dipple,A. (1989) Mutagenic specificity of a potent carcinogen, benzo[c]phenanthrene (4R,3S)-dihydrodiol (2S,1R)-epoxide, which reacts with adenine and guanine in DNA. Proc. Natl Acad. Sci. USA, 86, 2291–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidel A., Oesch,F. and Steinberg,P. (1995) Malignant transformation of the liver tumour precursor cell line OC/CDE 22 by the four stereoisomeric fjord region 3,4-dihydrodiol 1,2-epoxides of benzo[c]phenanthrene. Carcinogenesis, 16, 2111–2115. [DOI] [PubMed] [Google Scholar]
- 5.Amin S., Krzeminski,J., Rivenson,A., Kurtzke,C., Hecht,S.S. and el-Bayoumy,K. (1995) Mammary carcinogenicity in female CD rats of fjord region diol epoxides of benzo[c]phenanthrene, benzo[g]chrysene and dibenzo[a,l]pyrene. Carcinogenesis, 16, 1971–1974. [DOI] [PubMed] [Google Scholar]
- 6.Meehan T., Straub,K. and Calvin,M. (1977) Benzo[alpha]pyrene diol epoxide covalently binds to deoxyguanosine and deoxyadenosine in DNA. Nature, 269, 725–727. [DOI] [PubMed] [Google Scholar]
- 7.Straub K.M., Meehan,T., Burlingame,A.L. and Calvin,M. (1977) Identification of the major adducts formed by reaction of benzo(a)pyrene diol epoxide with DNA in vitro. Proc. Natl Acad. Sci. USA, 74, 5285–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne M.R., Beland,F.A., Harvey,R.G. and Brookes,P. (1976) The reaction of (+/–)-7α, 8β-dihydroxy-9β, 10β-epoxy- 7,8,9,10-tetrahydrobenzo(a)pyrene with DNA. Int. J. Cancer, 18, 364–368. [DOI] [PubMed] [Google Scholar]
- 9.Dipple A., Pigott,M.A., Agarwal,S.K., Yagi,H., Sayer,J.M. and Jerina,D.M. (1987) Optically active benzo[c]phenanthrene diol epoxides bind extensively to adenine in DNA. Nature, 327, 535–536. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal R., Yagi,H., Jerina,D.M. and Dipple,A. (1996) Benzo[c]phenanthrene 3,4-dihydrodiol 1,2-epoxide adducts in native and denatured DNA. Carcinogenesis, 17, 1773–1776. [DOI] [PubMed] [Google Scholar]
- 11.Sancar A. (1994) Mechanisms of DNA excision repair. Science, 266, 1954–1956. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 13.Li G.-M. (1999) The role of mismatch repair in DNA damage-induced apoptosis. Oncol. Res., 11, 393–400. [PubMed] [Google Scholar]
- 14.Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 15.Kolodner R.D. and Alani,E. (1994) Mismatch repair and cancer susceptibility. Curr. Opin. Biotechnol., 5, 585–594. [DOI] [PubMed] [Google Scholar]
- 16.Modrich P. (1991) Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet., 25, 229–253. [DOI] [PubMed] [Google Scholar]
- 17.Kolodner R.D. and Marsischky,G.T. (1999) Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev., 9, 89–96. [DOI] [PubMed] [Google Scholar]
- 18.Li G.M. (2003) DNA mismatch repair and cancer. Front Biosci., 8, D997–1017. [DOI] [PubMed] [Google Scholar]
- 19.Bellacosa A. (2001) Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ., 8, 1076–1092. [DOI] [PubMed] [Google Scholar]
- 20.Li G.-M., Wang,H. and Romano,L.J. (1996) Human MutSα specifically binds to DNA containing aminofluorene and acetylaminofluorene adducts. J. Biol. Chem., 271, 24084–24088. [PubMed] [Google Scholar]
- 21.Kat A., Thilly,W.G., Fang,W.H., Longley,M.J., Li,G.M. and Modrich,P. (1993) An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl Acad. Sci. USA, 90, 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes J., Clark,S. and Modrich,P. (1990) Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl Acad. Sci. USA, 87, 5837–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond J.T., Li,G.M., Longley,M.J. and Modrich,P. (1995) Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science, 268, 1909–1912. [DOI] [PubMed] [Google Scholar]
- 24.Wu J., Gu,L., Wang,H., Geacintov,N.E. and Li,G.-M. (1999) Mismatch repair processing of carcinogen-DNA adducts triggers apoptosis. Mol. Cell. Biol., 19, 8292–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genschel J., Littman,S.J., Drummond,J.T. and Modrich,P. (1998) Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem., 273, 19895–19901. [DOI] [PubMed] [Google Scholar]
- 26.Papadopoulos N., Nicolaides,N.C., Liu,B., Parsons,R.E., Palombo,F., D’Arrigo,A., Markowitz,S., Willson,J.K.V., Kinzler,K., Jiricny,J. et al. (1995) Mutations in GTBP in genetically unstable cells. Science, 268, 1914–1917. [DOI] [PubMed] [Google Scholar]
- 27.Steinbrecher T., Becker,A., Stezowski,J.J., Oesch,F. and Seidel,A. (1993) Synthesis of oligodeoxynucleotides containing diastereomeric diol epoxide-N6-deoxy-adenosine adducts of polycyclic aromatic hydrocarbons. Tetrahedron Lett., 34, 1773–1774. [Google Scholar]
- 28.Zou Y., Liu,T.M., Geacintov,N.E. and Van Houten,B. (1995) Interaction of UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG(–)-anti-BPDE. Biochemistry, 34, 13582–13593. [DOI] [PubMed] [Google Scholar]
- 29.Reitmair A.H., Schmits,R., Ewel,A., Bapat,B., Redston,M., Mitri,A., Waterhouse,P., Mittrucker,H.W., Wakeham,A., Liu,B. et al. (1995) MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nature Genet., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- 30.Baker S.M., Plug,A.W., Prolla,T.A., Bronner,C.E., Harris,A.C., Yao,X., Christie,D.M., Monell,C., Arnheim,N., Bradley,A. et al. (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet., 13, 336–342. [DOI] [PubMed] [Google Scholar]
- 31.Clarke A.R., Gledhill,S., Hooper,M.L., Bird,C.C. and Wyllie,A.H. (1994) p53 dependence of early apoptotic and proliferative responses within the mouse intestinal epithelium following γ-irradiation. Oncogene, 9, 1767–1773. [PubMed] [Google Scholar]
- 32.Gradia S., Acharya,S. and Fishel,R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell, 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- 33.Allen D.J., Makhov,A., Grilley,M., Taylor,J., Thresher,R., Modrich,P. and Griffith,J.D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J., 16, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishel R. (1998) Mismatch repair, molecular switches and signal transduction. Genes Dev., 12, 2096–2101. [DOI] [PubMed] [Google Scholar]
- 35.Goldmacher V.S., Cuzick,R.A. and Thilly,W.G. (1986) Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosourea and N-methyl-N′-nitro-nitrosoguanidine. J. Biol. Chem., 261, 12462–12471. [PubMed] [Google Scholar]
- 36.Koi M., Umar,A., Chauhan,D.P., Cherian,S.P., Carethers,J.M., Kunkel,T.A. and Boland,C.R. (1994) Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N- nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation [published erratum appears in Cancer Res. (1995) 55, 201]. Cancer Res., 54, 4308–4312. [PubMed] [Google Scholar]
- 37.Wyllie A.H. (1981) Cell death: a new classification separating apoptosis from necrosis. Chapman and Hall, New York, NY.
- 38.Toft N.J., Winton,D.J., Kelly,J., Howard,L.A., Dekker,M., te Riele,H., Arends,M.J., Wyllie,A.H., Margison,G.P. and Clarke,A.R. (1999) Msh2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc. Natl Acad. Sci. USA, 96, 3911–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickman M.J. and Samson,L.D. (1999) Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl Acad. Sci. USA, 96, 10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duckett D.R., Bronstein,S.M., Taya,Y. and Modrich,P. (1999) hMutSα- and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl Acad. Sci. USA, 96, 12384–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karran P. and Bignami,M. (1992) Self-destruction and tolerence in resistance of mammalian cells to alkylation damage. Nucleic Acids Res., 20, 2933–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Modrich P. (1997) Strand-specific mismatch repair in mammalian cells. J. Biol. Chem., 272, 24727–24730. [DOI] [PubMed] [Google Scholar]
- 43.Claij N., van der Wal,A., Dekker,M., Jansen,L. and te Riele,H. (2003) DNA mismatch repair deficiency stimulates N-ethyl-N-nitrosourea-induced mutagenesis and lymphomagenesis. Cancer Res., 63, 2062–2066. [PubMed] [Google Scholar]
- 44.Xu X.S., Narayanan,L., Dunklee,B., Liskay,R.M. and Glazer,P.M. (2001) Hypermutability to ionizing radiation in mismatch repair-deficient, Pms2 knockout mice. Cancer Res., 61, 3775–3780. [PubMed] [Google Scholar]
- 45.Sansom O.J., Toft,N.J., Winton,D.J. and Clarke,A.R. (2001) Msh-2 suppresses in vivo mutation in a gene dose and lesion dependent manner. Oncogene, 20, 3580–3584. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S., Lloyd,R., Bowden,G., Glickman,B.W. and de Boer,J.G. (2001) Msh2 DNA mismatch repair gene deficiency and the food-borne mutagen 2-amino-1-methy1-6-phenolimidazo [4,5-b] pyridine (PhIP) synergistically affect mutagenesis in mouse colon. Oncogene, 20, 6066–6072. [DOI] [PubMed] [Google Scholar]
- 47.Loeb L.A., Loeb,K.R. and Anderson,J.P. (2003) Multiple mutations and cancer. Proc. Natl Acad. Sci. USA, 100, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin X., Zhou,H., Liu,L. and Gerson,S.L. (1999) Transgenic expression of human MGMT blocks the hypersensitivity of PMS2-deficient mice to low dose MNU thymic lymphomagenesis. Carcinogenesis, 20, 1667–1673. [DOI] [PubMed] [Google Scholar]
- 49.Qin X., Liu,L. and Gerson,S.L. (1999) Mice defective in the DNA mismatch gene PMS2 are hypersensitive to MNU induced thymic lymphoma and are partially protected by transgenic expression of human MGMT. Oncogene, 18, 4394–4400. [DOI] [PubMed] [Google Scholar]
- 50.Yoshino M., Nakatsu,Y., te Riele,H., Hirota,S., Kitamura,Y. and Tanaka,K. (2002) Additive roles of XPA and MSH2 genes in UVB-induced skin tumorigenesis in mice. DNA Repair (Amst.), 1, 935–940. [DOI] [PubMed] [Google Scholar]
- 51.Gong J.G., Costanzo,A., Yang,H.Q., Melino,G., Kaelin,W.G.,Jr, Levrero,M. and Wang,J.Y. (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature, 399, 806–809. [DOI] [PubMed] [Google Scholar]
- 52.Brown K.D., Rathi,A., Kamath,R., Beardsley,D.I., Zhan,Q., Mannino,J.L. and Baskaran,R. (2003) The mismatch repair system is required for S-phase checkpoint activation. Nature Genet., 33, 80–84. [DOI] [PubMed] [Google Scholar]
- 53.Shimodaira H., Yoshioka-Yamashita,A., Kolodner,R.D. and Wang,J.Y. (2003) Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl Acad. Sci. USA, 100, 2420–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]