Abstract
YVH1 was one of the first eukaryotic dual specificity phosphatases cloned, and orthologues posses a unique C-terminal zinc-coordinating domain in addition to a cysteine-based phosphatase domain. Our recent results revealed that human YVH1 (hYVH1) protects cells from oxidative stress. This function requires phosphatase activity and the zinc binding domain. This current study provides evidence that the thiol-rich zinc-coordinating domain may act as a redox sensor to impede the active site cysteine from inactivating oxidation. Furthermore, using differential thiol labeling and mass spectrometry, it was determined that hYVH1 forms intramolecular disulfide bonds at the catalytic cleft as well as within the zinc binding domain to avoid irreversible inactivation during severe oxidative stress. Importantly, zinc ejection is readily reversible and required for hYVH1 activity upon returning to favorable conditions. This inimitable mechanism provides a means for hYVH1 to remain functionally responsive for protecting cells during oxidative stimuli.
Human YVH1 (hYVH12; also known as DUSP12) is a member of the dual specificity phosphatase (DUSP) subfamily of protein-tyrosine phosphatases (PTPs) (1, 2). It is constructed of an N-terminal DUSP catalytic domain and a unique C-terminal zinc coordinating domain (3). Poor characterization and lack of mitogen-activated protein kinase targeting motifs further classify this enzyme as an atypical DUSP (1). YVH1 orthologues exhibit high evolutionary conservation and similar domain organization (3). Deletion of the yvh1 gene in yeast disrupts normal growth processes (4), whereas insertion and expression of the hyvh1 gene is capable of restoring a normal yeast growth phenotype (3). Amplification of the dusp12/hyvh1 gene has been reported in multiple sarcomas, implicating a role for hYVH1 in human disease (5–7).
Recently, deletion studies from our laboratory have shown that the C-terminal zinc binding domain of hYVH1 is not essential for intrinsic phosphatase activity in vitro; however, it is required for interaction with the ATPase domain of heat shock protein 70 (8). Similarly, overexpression of wild type hYVH1 but not catalytically dead or zinc coordinating domain deletion mutants prevents cell death induced by Fas receptor activation, heat shock, and hydrogen peroxide (H2O2) (8). Despite these findings, current information on hYVH1 enzymatic and physiological functions remains limited.
PTPs and DUSPs share similar active site architecture and catalytic mechanism, characterized by the conserved HCX5R(S/T) motif (9, 10). The unique microenvironment within the HCX5R(S/T) motif reduces the pKa value of the active site cysteine, enhancing both nucleophilicity and oxidation susceptibility (11, 12). Stimulated or constituent generation of ROS can result in oxidative second messenger signaling responses capable of transient and reversible post-translational inactivation of both PTPs and DUSPs through oxidation of the catalytic cysteine (13–15).
This oxidative susceptibility and modification varies among PTPs and DUSPs, a likely consequence of slight variations in active site conformations or mediated through unique regulatory domains (16–18). Accumulating evidence suggests that redox-mediated oxidation of PTPs is a dynamic modification that can differentially regulate PTPs (13, 19). Sulfenic acid, cyclic sulfenamide, and disulfide bond formation have all been shown to facilitate stable, reversible active site modifications among various PTPs and DUSPs (12, 14, 20). Furthermore, evidence suggests that oxidation predominantly and rapidly targets the active site cysteine, whereas other cysteinyl residues remain in the reduced state (15, 20).
This study investigated the relationship between the zinc-coordinating C-terminal domain and the catalytic domain of hYVH1 during oxidative conditions. We provide data suggesting that the zinc binding domain can serve as a reducing agent during oxidative stress to impede the oxidation of the active site cysteine. Increased exposure to oxidative conditions readily induces disulfide bond formation within the zinc-coordinating and catalytic domains, resulting in concomitant zinc ejection and enzymatic inactivation. Zinc ejection is readily reversible and required for hYVH1 activity upon returning to reducing conditions. Thus, we propose a mechanism for phosphatase active site protection through the intrinsic redox buffering capacity of this unique zinc binding domain.
EXPERIMENTAL PROCEDURES
Cell Culture and Annexin V Assay
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum at 37 °C and 5% CO2. Transient transfection was carried out at 70% confluence using FuGENE 6 HD (Roche Applied Science) according to the manufacturer's protocol. For H2O2 treatments, HeLa cells were exposed to the indicated concentrations for 1 h at 37 °C. Cell viability was determined using a fluorescein isothiocyanate-conjugated annexin V kit (Vybrant Apoptosis Assay Kit 3; Molecular Probes) according to the manufacturer's instructions and as described previously (8). Cells were also treated with Hoechst 33342 dye and viewed using fluorescence microscopy. Approximately 500 cells were counted per experiment using the Northern Eclipse software program, and apoptotic cells were detected by a positive annexin V stain. The percentage of apoptotic cells in the total sample population as determined by Hoechst staining was calculated. The data shown are from three independent experiments averaged together.
Recombinant Protein Expression and Purification
Bacterial expression of pGEX/hYVH1 constructs and protein purification were as previously described (3, 8). Production of the C-terminal zinc-coordinating domain deletion mutant, hYVH1ΔCT1, was previously described (8). Recombinant proteins were eluted with 50 mm Tris-HCl, 150 mm NaCl, 2.5 mm CaCl2, 0.1% 2-mercaptoethanol, 80 units of thrombin (Sigma), pH 8, at 4 °C on glutathione-agarose (Sigma) to remove the glutathione S-transferase tag. Purified protein samples were concentrated and exchanged into metal-free 50 mm Tris-HCl, 50 mm NaCl, 1 mm dithiothreitol, 0.25 mm phenylmethylsulfonyl fluoride, pH 7.4, using Amicon Ultra-4 centrifugal filters (Millipore) at 4 °C. Metal-free buffers were passed through a Chelex-100 column (Sigma) to remove contaminating metals. Protein concentration was determined against standard bovine serum albumin as per the Bradford assay, and samples were aliquoted and stored at −80 °C until use. The purity of all proteins in this study was ∼80–90%, as judged by SDS-PAGE analysis.
Oxidation-induced Enzyme Inactivation, Thiol Loss, and Zinc Ejection
Phosphatase activity was monitored at 30 °C using the artificial substrate analog 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP; Invitrogen). Protein samples were exchanged into preheated (30 °C), metal-free 50 mm Tris-HCl, 50 mm NaCl, pH 7.4, using 0.5 ml of Zeba Desalt Spin Columns (Pierce), followed by immediate treatment with H2O2. At defined time points, aliquots were mixed with 0.9 mm DiFMUP in 50 mm Tris-hydroxyaminomethane, 50 mm bis-Tris, 150 mm NaCl, pH 6, similar to previously described methods (12). Samples were loaded into a 96-well FluroNunc plate (Sigma), and initial velocity data were recorded (excitation, 358 nm; emission, 450 nm). Data were expressed as percentage of activity of reduced hYVH1 samples in 5 mm dithiothreitol (DTT; Sigma). Specific activity values were calculated and compared for both fully reduced hYVH1 and hYVH1ΔCT1. As a comparative control, phosphatase activity of the archetypical DUSP, vaccinia H1-related (VHR; Biomol), was monitored under similar conditions. Similarly, at defined time points of oxidation, samples were incubated with 20 mm DTT for 30 min to quench oxidation, followed by the addition of DiFMUP as above to assess the reversibility of activity. Thiol quantitation was accomplished using a modified dithionitrobenzoic acid (DTNB; Sigma) assay similar to one described (21). Aliquots were treated for 10 min with defined concentrations of H2O2 and then incubated for 5 min with ∼50 units of catalase. Reactions were mixed with 500 μm DTNB in 50 mm Tris-HCl, 50 mm NaCl, pH 7.4, for 30 min, and absorbance at 412 nm was monitored (DTNB, ϵ = 14,150 liters/mol·cm). Zinc release was monitored using a 4-(2-pyridylazo)resorcinol/parachloromercuribenzoic acid (PAR/PCMB; Sigma)-based assay as described (22). Aliquots were taken at defined time points and mixed with metal-free 100 μm PAR in 40 mm potassium phosphate buffer, pH 7.5, and absorbance at 500 nm was monitored (PAR, ϵ = 66,000 liters/mol·cm). The addition of 50 μm PCMB in 100 μm PAR solution elicited complete zinc release, and the total absorbance was recorded. Data were expressed as percentage of total zinc (free zinc + protein-bound zinc).
Probing Thiol Status Using MALDI-TOF Mass Spectrometry
A differential thiol labeling strategy similar to one previously described (22) was utilized to assess thiol status in response to oxidation. Full-length hYVH1 and hYVH1ΔCT1 were oxidized with a 100-fold molar excess of H2O2 for 1.5 h, precipitated with 10% trichloroacetic acid (w/v), and washed three times with 100% cold acetone. Pellets were resuspended in denaturing buffer (6 m urea, 200 mm Tris-HCl, 10 mm EDTA, pH 8.5) supplemented with 100 mm iodoacetamide (Sigma), shaking for 1 h at 25 °C. Each sample was divided and precipitated as above. Samples were resuspended in denaturing buffer in the presence or absence of 10 mm DTT, shaking for 45 min at 25 °C, followed by the addition of 100 mm N-ethylmaleimide (NEM; Sigma), shaking for 1 h at 25 °C. Samples were again precipitated as above, resuspended in 50 mm ammonium bicarbonate, and digested with either trypsin (Promega) or endoproteinase Glu-C (Roche Applied Science). Aliquots were collected at various time points, quenched with 1% formic acid, and stored at −20 °C for future MALDI-TOF analysis. To further assist identification of interpeptide disulfide bond formation, a tryptic digest of oxidized, iodoacetamide-labeled, full-length hYVH1 was modified using the Ettan CAF MALDI sequencing kit (Amersham Biosciences) as per the manufacturer's protocol, without lysine modification. The resulting peptides have a sulfonic acid group (136 Da) added to all free amines. All digests were mixed 1:1 with 10 mg/ml matrix solution (α-cyano-4-hydroxycinnamic acid in 60% acetonitrile, 1% formic acid; Sigma) on the target plate. Peptide mass fingerprints and tandem mass spectrometry (MS/MS) using postsource decay (PSD) was performed on selected parent ions as described previously (8) and compared with in silico fragmentation using the Protein Prospector software (available on the World Wide Web).
Metal-free Enzymatic Activity, Reversibility of Zinc Binding, and Thiol Oxidation State Analysis
Protein samples were exchanged into metal-free 50 mm Tris-HCl, 50 mm NaCl, pH 7.4, as above and then treated for 10 min with or without a 2.5 molar excess of PCMB/total thiol content. Samples were exchanged as above to remove excess PCMB and free zinc, incubated with or without 5 mm DTT for ∼30 min, and then mixed with 0.9 mm DiFMUP and analyzed as above. Similarly, full-length hYVH1 was treated as above or with a 100-fold molar excess of H2O2 for 1.5 h to elicit zinc ejection. Samples treated with PCMB were incubated in the presence or absence of a slight excess of DTT for ∼10 min without previous buffer exchange to facilitate reversible zinc binding. Samples oxidized with H2O2 were incubated in 50 units of catalase and then in the presence or absence of a slight excess of DTT without previous buffer exchange. All samples were mixed with 100 μm PAR, and the absorbance at 500 nm was monitored. Thiol oxidation state was monitored using the electrophilic reagent 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-Cl; Sigma), as described (12, 23). Full-length hYVH1 was exchanged, oxidized, and quenched as above and then incubated with 1 mm NBD-Cl for 1 h. Samples were again exchanged as above and compared with a non-oxidized, NBD-Cl-treated control by monitoring the absorbance at 250–530 nm.
RESULTS
hYVH1-mediated Protection against Hydrogen Peroxide-induced Cell Death
Our previous work demonstrated that hYVH1 can potently defend cells from oxidative insults, such as heat shock, Fas receptor activation, and H2O2 (8). Moreover, a catalytically inactive mutant or a zinc domain deletion variant failed to protect, indicating that the phosphatase activity and the zinc binding domain are required for the cell survival function of hYVH1.
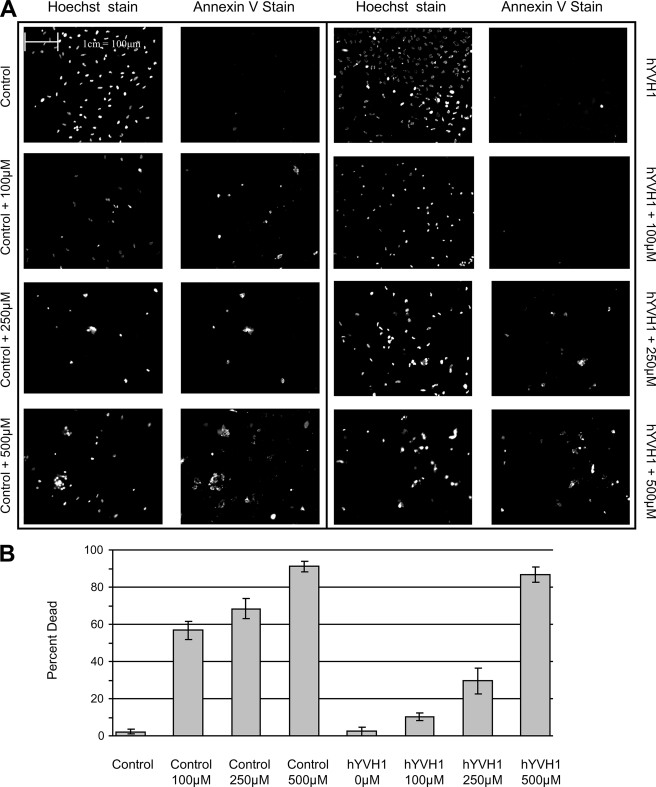
To test the extent of the hYVH1 cytoprotective ability, we subjected HeLa cells to increasing amounts of H2O2. Cells expressing hYVH1 were substantially more resistant to H2O2 cytotoxicity at 100 and 250 μm final concentrations compared with control cells, displaying ∼27- and 7-fold less cell death, respectively (Fig. 1). However, at 500 μm H2O2, hYVH1 failed to protect cells to any significant extent. These results suggest that hYVH1 can protect cells up to a threshold level of oxidative stress, above which there may be a mechanism in place to down-regulate the cell survival ability of hYVH1.
FIGURE 1.
hYVH1 protects cells from oxidative stress. HeLa cells transiently transfected with either empty vector (Control) or hYVH1 and were treated with H2O2 (100, 250, or 500 μm) for 1 h at 37 °C. A, cell viability was detected using fluorescein isothiocyanate-conjugated annexin V as per the manufacturer's protocol, and total cell number was determined using Hoechst 33342 dye. B, graphic representation from three independent experiments expressing cell death as a percentage of total cell number determined using fluorescein isothiocyanate-conjugated annexin V and Hoechst 33342 nuclear stain. Error bars, S.D.
hYVH1 Resists Oxidative Inactivation in Vitro
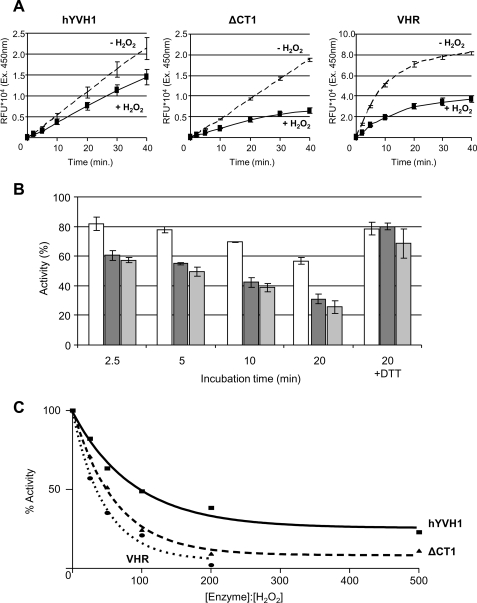
It is believed that the susceptibility of PTPs and DUSPs to oxidative conditions occurs predominantly through oxidation of the nucleophilic cysteine residue within the active site due to its low pKa value. Since hYVH1 has a zinc binding domain containing seven zinc-coordinating Cys residues, we hypothesized that this domain may reduce the susceptibility of redox-induced inactivation at the active site Cys. As a control, the prototypical DUSP, VHR, was used. VHR shares 25% sequence identity with hYVH1 in the DUSP catalytic domain and has been shown to be sensitive to redox-induced inactivation through formation of a stable sulfenic acid (12). To assess oxidative resistance, wild type hYVH1, zinc domain deletion mutant (hYVH1ΔCT1), and VHR were subjected to H2O2 and then assayed for phosphatase activity toward the exogenous substrate DiFMUP. Under reducing conditions, the activity of VHR was significantly greater than that of both hYVH1 and hYVH1ΔCT1. In comparison, removal of the zinc coordinating domain did not show significant deviation in activity compared with wild type under reducing conditions (Fig. 2A). In stark contrast, exposure to H2O2 at a molar ratio of 1:100 rapidly inactivates VHR (by ∼45% after 2.5 min and ∼60% after 10 min). Meanwhile, hYVH1 displayed greater resistance to the oxidative conditions, displaying activity loss of ∼25% after 2.5 min and ∼35% after 10 min (Fig. 2, A and B). The hYVH1ΔCT1 variant experienced activity loss similar to that of VHR, suggesting that the inherent resistance to inactivation by hYVH1 is mediated by the zinc binding domain. The addition of excess DTT, postoxidation, resulted in nearly complete regeneration (∼80%) of phosphatase activity among all enzymes (Fig. 2B). Furthermore, wild type hYVH1 demonstrated similar resistant behavior when titrated across an increasing molar ratio of H2O2 when compared with VHR and hYVH1ΔCT1 (Fig. 2C). Collectively, these results demonstrate that the zinc binding domain provides hYVH1 the capacity to impede redox-induced inactivation.
FIGURE 2.
Analysis of hYVH1 inactivation by H2O2. A and B, inactivation effects of H2O2 on DUSP catalytic activity. A, initial velocity raw data comparing hYVH1, hYVH1ΔCT1, and VHR after incubation at 30 °C in a 1:100 molar ratio of H2O2 or 5 mm DTT for 10 min. B, designated enzymes (□, hYVH1; ■, hYVH1ΔCT1; ■, VHR) were incubated as above for the defined times and then monitored for relative changes in activity. Inactivation and reactivation is expressed as percentage of fully reduced enzyme without H2O2. C, similarly, designated enzymes were oxidized at the defined molar ratios for 20 min and then assayed for remaining activity. Data were fit to a one-phase exponential decay equation using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA). All data analyses represent the mean of at least three independent determinations. Error bars, S.D.
Hydrogen Peroxide-mediated Thiol Oxidation Leads to Zinc Release
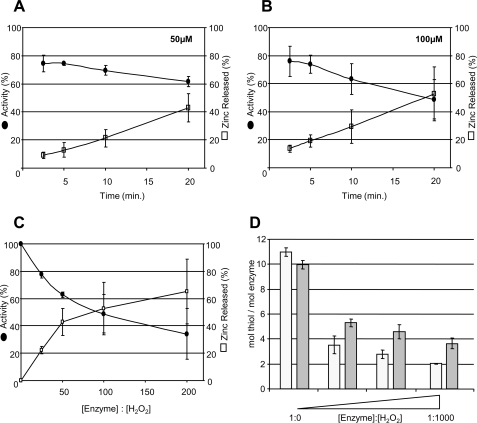
The above findings suggest that the thiol-rich, C-terminal zinc coordinating domain may be involved in resisting oxidative inactivation of hYVH1. To examine the effects of oxidative conditions on zinc coordination, we performed a PAR/PCMB assay to monitor zinc release in response to oxidative conditions. At ratios of H2O2 similar to those examined above, zinc was readily released from hYVH1 (Fig. 3, A–C). Moreover, the loss of phosphatase activity seemed to parallel the observed rate of zinc ejection at lower concentrations of H2O2 (Fig. 3, A and B). In all cases, spontaneous thiol oxidation and zinc release was determined to be negligible over the defined experimental times. These results suggest that zinc ejection and activity loss during increasing levels of oxidative stress is probably due to thiol oxidation within both the zinc binding and active site domains. Since multiple Cys residues must become oxidized before zinc ejection is detected, the above trend also suggests that the zinc coordinating cysteines may be preferentially oxidized over the active site Cys.
FIGURE 3.
hYVH1 zinc release, inactivation, and thiol depletion by H2O2 exposure. A and B, plots of hYVH1 time-dependent enzymatic inactivation (●) and zinc release (□). Enzyme was incubated in increasing molar ratios of H2O2 (A, 1:50; B, 1:100) for the defined times and then analyzed for changes in activity or zinc release, as described under “Experimental Procedures.” C, similarly, hYVH1 was oxidized at the defined molar ratios for 20 min and then assayed for remaining activity and bound zinc. D, thiol oxidation of hYVH1. hYVH1 (□) and hYVH1C/S (■) were incubated with increasing molar ratios of H2O2 (1:0, 1:100, 1:500, and 1:1000) for 10 min, and then thiol content was analyzed using the DTNB assay, as described under “Experimental Procedures.” Data were normalized to the concentration of enzyme. All data analyses represent the mean of at least three independent determinations. Error bars, S.D.
To examine thiol oxidation status more directly, a DTNB-based assay was performed. The catalytically dead mutant (hYVH1 C115S) was also analyzed to obtain some insight into the oxidation state of the active site cysteine residue. From primary sequence analysis, wild type hYVH1 and the C115S mutant contain 11 and 10 cysteine residues, respectively, consistent with observed findings of total thiol content/mol of protein (Fig. 3D). Upon treatment with increasing H2O2 concentrations, rapid loss of multiple free thiols was observed within 10 min as compared with untreated control. If oxidation of the active site cysteine to a cyclic sulfenamide, sulfenic, sulfinic, or sulfonic acid occurred, we would expect an equal number of free thiols between wild type hYVH1 and the C115S mutant. However, experimental evidence demonstrated that upon oxidation, wild type hYVH1 harbored approximately one less free thiol than the C115S mutant, suggesting that the active site cysteine may be participating in intramolecular disulfide bond formation during oxidative stress (Fig. 3D).
Oxidation of hYVH1 Results in Intramolecular Disulfide Bond Formation
The above data suggest that the active site Cys of hYVH1 may be capable of forming an intramolecular disulfide bond. This ability would represent a mechanism to avoid irreversible oxidation during even extreme oxidative stress.
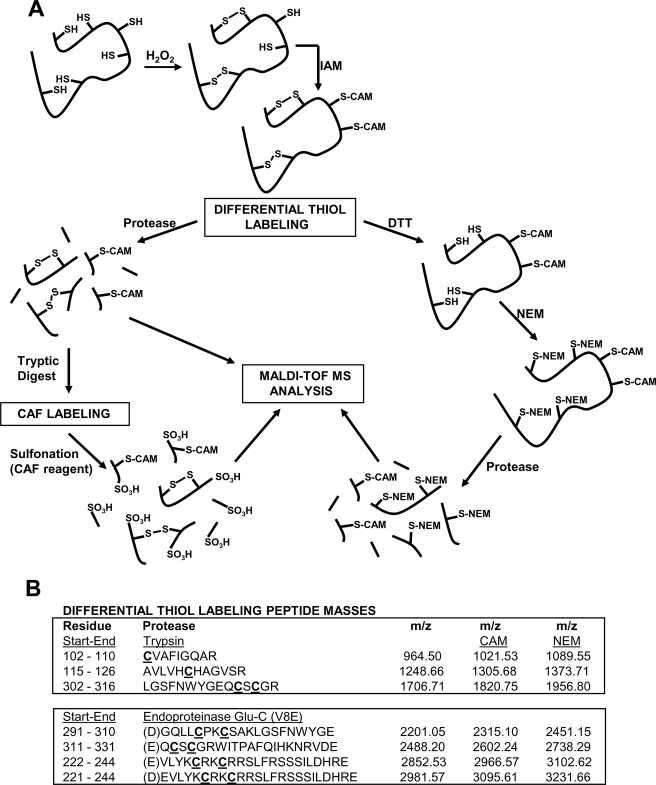
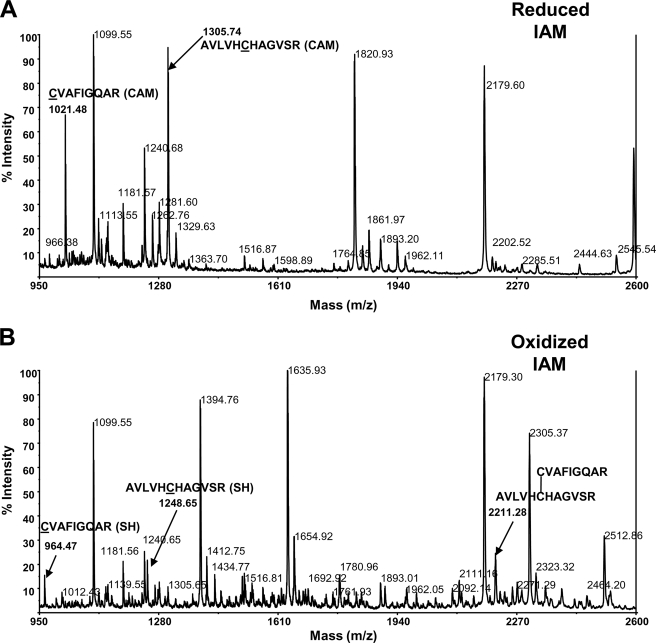
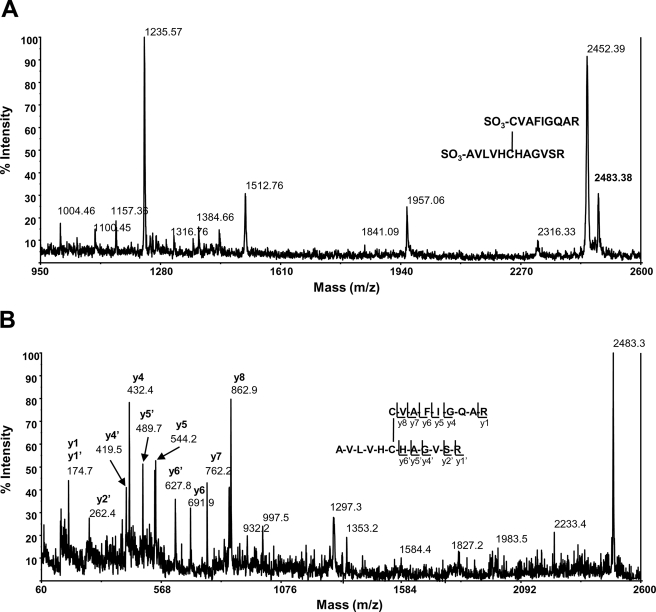
Therefore, to further confirm the DTNB data, thiol bonding status of both reduced and oxidized hYVH1 was analyzed using MALDI-TOF MS through the application of differential thiol labeling methodology (Fig. 4A). Mass fingerprints of reduced and oxidized, carbamidomethylated (CAM) hYVH1 tryptic digests were obtained and compared for target peptide identification (Fig. 4B). Reduced CAM hYVH1 displayed the characteristic active site peak at m/z 1305.74 along with that of an N-terminal proximal thiol at m/z 1021.48. No significant amounts of unmodified peptides were observed (Fig. 5A). Oxidized CAM hYVH1 displayed almost undetectable levels of both of these target peptides; however, both the active site and proximal thiol peptides appeared in their reduced form at m/z 1248.65 and 964.47, respectively. Interestingly, a peak at m/z of 2211.28 appeared, which is ∼2 Da less than the resultant sum of these target peptides, suggesting formation of an intramolecular disulfide bond (Fig. 5B). The remaining tryptic digest was sulfonated using CAF reagent labeling to facilitate unambiguous identification of this unknown peptide. An intramolecular disulfide-bonded peptide upon sulfonation would be doubly modified at both N termini, causing a mass shift of 272 Da, which was detected for this peptide (m/z 2483.38) (Fig. 6A). Unambiguous identification was accomplished using MS/MS PSD analysis of this parent ion, and the formation of an intramolecular disulfide bond between the active site cysteine and the N-terminal proximal cysteine was confirmed (Fig. 6B).
FIGURE 4.
MALDI-TOF MS approach to probe hYVH1 thiol redox status. A, schematic diagram of the differential thiol labeling strategy utilized to identify Cys-specific modifications of hYVH1 in response to H2O2-mediated oxidation. B, table of target hYVH1 Cys containing tryptic and glutamic peptides.
FIGURE 5.
Mass fingerprint analysis of hYVH1 tryptic peptides. A, reduced, carbamidomethylated hYVH1 mass fingerprint. Highlighted are the active site and N-terminal proximal thiol peptides at m/z 1305.74 and 1021.48, respectively. B, oxidized, carbamidomethylated hYVH1 mass fingerprint. Highlighted are the reduced active site and N-terminal proximal thiol peptides at m/z 1248.65 and 964.27, respectively. Carbamidomethylated peaks are largely reduced or absent. Also highlighted is a putative intramolecular disulfide peak at m/z 2211.28 of these target peptides.
FIGURE 6.
Mass fingerprint and MS/MS PSD analysis of CAF-labeled hYVH1 tryptic peptides. A, oxidized, carbamidomethylated, CAF-labeled hYVH1 mass fingerprint. Highlighted is the doubly CAF-labeled putative disulfide peak at m/z 2483.38. B, MS/MS analysis of the m/z peak at 2483.38. The y ion fragment series correspond to the two target peptides AVLVHCHAGVSR and CVAFIGQAR, denoting unambiguous identification of active site disulfide bond formation.
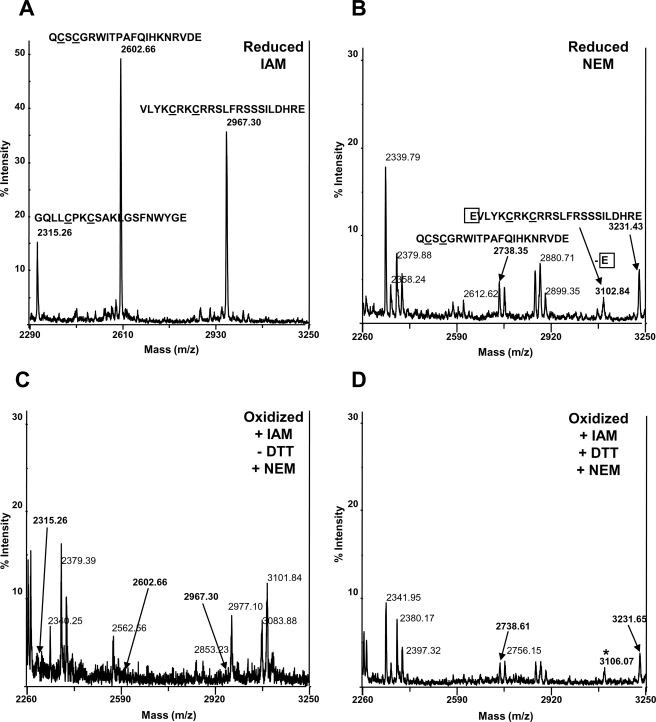
Similarly, mass fingerprints of reduced CAM or NEM hYVH1 Glu-C digests were obtained and compared for target peptide identification (Fig. 4B). Oxidized CAM hYVH1 was subjected to differential thiol labeling, as described under “Experimental Procedures,” and mass fingerprints of all Glu-C digests were recorded. Reduced CAM or NEM digests displayed characteristic peaks, which were assigned to vicinal thiol-containing peptides of the C-terminal zinc binding domain (Fig. 7, A and B). Upon oxidation, a loss of CAM m/z peaks 2315.26, 2602.66, and 2967.30 is observed, whereas formation of one NEM peptide at m/z 3101.84 is evident, suggesting that some reduced thiols exist during oxidation (Fig. 7C). Oxidized, differentially labeled, CAM hYVH1 resulted in the appearance of target NEM peaks at m/z 2738.61, 3231.65, and 3106.07, which correspond to the reduced NEM digest (Fig. 7, D and B). These observations suggest that in addition to the ability of the active site Cys to form an intramolecular disulfide bond, the zinc-coordinating Cys residues in the C-terminal domain are putatively involved in a distinct disulfide bonding network. In both cases, higher oxidation states of the thiolate moieties were not observed, further confirming predominant formation of disulfide bonds within both domains.
FIGURE 7.
Differential thiol labeling mass fingerprints of hYVH1 C-terminal glutamic peptides. A, reduced, carbamidomethylated hYVH1 mass fingerprint. Highlighted are three vicinal thiol pair peptides within the zinc-coordinating domain, m/z 2315.26, 2602.66, and 2967.30. B, reduced, N-ethylmaleimidyl hYVH1 mass fingerprint. Highlighted are two of the vicinal thiol pair peptides, m/z 2738.35, 3102.84, and 3231.43. C, oxidized, carbamidomethylated hYVH1 mass fingerprint. Highlighted is the absence of characteristic peaks at m/z 2315.26, 2602.66, 2967.30 and the presence of a peak at m/z 3101.84. D, differential thiol labeled hYVH1 mass fingerprint. Highlighted is the reappearance of peaks at m/z 2738.61, 3231.65, and 3106.07.
Zinc Coordination is Reversible and Required for Optimal Enzymatic Activity
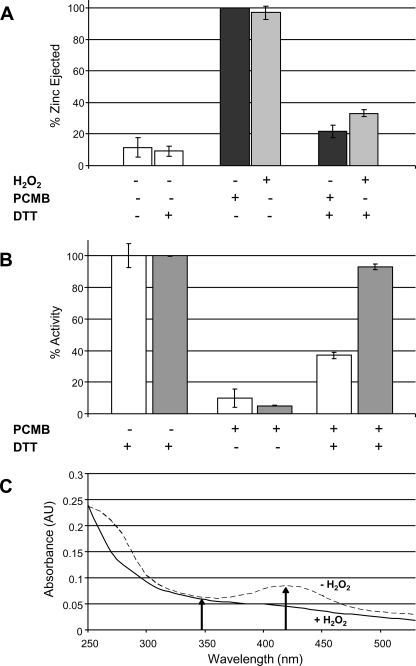
Disulfide bond formation within the zinc-coordinating domain would allow this domain to bind zinc under reducing conditions and form disulfide bonds under oxidative conditions in a reversible regulatory fashion. To assess if the C-terminal domain is capable of potentially governing regulatory events, reversibility of zinc coordination was tested. Treatment with either the thiol-reactive reagent PCMB or H2O2 results in essentially complete zinc ejection in the absence of reducing reagents. This phenomenon is readily and rapidly reversible to near completion (∼85%) upon the addition of reducing agents (Fig. 8A). Interestingly, upon returning to reducing conditions, zinc rebinding seems to be a necessity for complete catalytic recovery. Complete zinc ejection and removal from the system leaves hYVH1 with only ∼40% of activity in comparison with zinc-bound hYVH1 when reducing conditions are restored (Fig. 8B). Notably, the C-terminal zinc domain deletion variant hYVH1ΔCT1 retains full activity under similar conditions. This suggests that once the C-terminal cysteines are rereduced, zinc coordination is required to prevent disturbing the phosphatase domain.
FIGURE 8.
Reversible zinc coordination and effects on enzymatic activity. A, reversible zinc coordination of hYVH1 under reducing conditions. Complete zinc ejection via H2O2 (■) or PCMB (■) is rapidly reversible upon subsequent treatment with a slight excess of reducing agent. B, effects of zinc coordination on enzymatic activity. Treatment of hYVH1 (□) or hYVH1ΔCT1 (■) with PCMB reversibly modifies all thiol residues, resulting in zinc ejection. Reduction with DTT after desalting restored hYVH1ΔCT1 with full activity, whereas hYVH1 remained partially inactive. C, probing oxidized hYVH1 for sulfenic acid modifications. Oxidation of hYVH1 results in loss of free thiols and preferential formation of reversible cystine bonds, not cysteinyl sulfenic acids. Data analyses (A and B) represent the mean of at least three independent determinations. Error bars, S.D. Data analysis (C) represents an individual trial of three independent experiments.
The above results suggest that oxidation of hYVH1 is a reversible event. Non-enzymatic, reversible oxidation of thiols can only readily occur at lower thiol oxidation states, which include disulfide bonds and sulfenic acids. To further confirm disulfide bond formation upon oxidation of hYVH1, NBD-Cl was used to probe for the presence of sulfenic acid modifications. Upon interaction with a free thiol, the formed adduct displays a λmax of 420 nm; however, interaction with a sulfenic acid results in formation of an adduct having a λmax of 347 nm, ultimately allowing for spectral identification between these species (23). When NBD-Cl was used to probe hYVH1, no visible evidence of sulfenic acid formation was observed upon oxidation, confirming that disulfide bond formation is the predominant oxidative modification of hYVH1 cysteine thiols (Fig. 8C).
DISCUSSION
This current study has added insight to our initial findings that hYVH1 acts as a cell survival phosphatase in response to oxidative stress conditions. Moreover, we have revealed that the zinc binding domain is critical for hYVH1 to resist oxidative inactivation of its phosphatase activity. Although other phosphatases have shown the ability to resist oxidation of their active sites by various mechanisms (16–18, 20), this is the first report to demonstrate the involvement of a dynamic zinc coordinating domain in facilitating oxidative resistance for a phosphatase.
The prototypical DUSP, VHR, was found to have higher activity than hYVH1 toward the artificial substrate DiFMUP under reducing conditions. However, in the presence of H2O2, VHR lost activity readily, whereas hYVH1 still maintained 60% of its original activity after 20 min of exposure. Inactivation was readily reversible upon treatment with excess reductant, which is consistent with multiple other studies of PTPs and DUSPs (11, 12). This result highlights the sensitivity of typical PTPs, such as VHR, to redox conditions and suggests that hYVH1 may be able to maintain activity during times of oxidative stress when other PTPs are rendered inactive. The cytoprotective nature of hYVH1-expressing cells also supports this claim (Fig. 1). Why hYVH1 has lower intrinsic phosphatase activity in vitro under reducing conditions compared with VHR is puzzling, given that it possesses all of the classical invariant residues known to be critical for catalytic activity. It may be that hYVH1 is specific for its physiological substrate or that it is functionally slow in terms of turnover rate to better suit being active under cellular stress conditions. We are currently pursuing the three-dimensional structure of hYVH1 by x-ray crystallography to gain more insight into this issue and examine the structural relationship between the catalytic domain and the zinc-binding region.
Another intriguing result was the observation that ejection of zinc in response to high levels of H2O2 correlated with a progressive decrease in phosphatase activity. The zinc binding domain of hYVH1 contains two zinc-coordinating regions with seven of the eight coordinating amino acids being Cys residues (3). These Cys residues that coordinate the zinc ions may act as a redox buffer, becoming oxidized preferentially over the active site Cys. This hypothesis is supported by comparing the released zinc and remaining activity plots shown in Fig. 3, where there seems to be a correlation between rate of zinc ejection and rate of inactivation. Ejection of zinc is not released and detected until sequential oxidation of multiple Cys residues occurs, whereas enzymatic inactivation is limited to the active site cysteine alone. Thus, this finding opens up the possibility that the zinc binding domain of hYVH1 may serve as a redox sink capable of protecting the active site Cys up to a certain threshold through preferential oxidation of zinc-coordinating cysteines.
To examine the extent of active site Cys oxidation, we measured free thiol content in response to increasing H2O2. The active site mutant (C/S) was used for comparison. The control conditions produced the expected result, detecting the 11 Cys residues that are present in wild type hYVH1. The hYVH1 C/S mutant was measured at the expected 10 Cys residues (Fig. 3D). Under increasing oxidative conditions, hYVH1 C/S lost seven more free thiols, which correlate nicely to the seven coordinating Cys residues in the zinc binding domain. We were expecting under these conditions that wild type and the C/S mutant would contain the same number of reactive free thiols due to oxidation of the active site Cys. Instead, we reproducibly detected approximately one less free thiol than in the C/S mutant. This result raised the possibility that the active site cysteine participated in the formation of an intramolecular disulfide bond upon oxidation, resulting in one less reactive thiol for analysis via DTNB. Since the C115S mutant lacks this capability, an additional reactive thiol would remain available for detection with DTNB.
Further efforts to definitively demonstrate the thiol redox status of the multiple cysteine residues in hYVH1 were conducted using MALDI-TOF MS (Fig. 4). The target active site and an N-terminal cysteine-containing peptide were identified by mass fingerprint analysis of reduced, carbamidomethylated hYVH1 (Fig. 5A). Upon oxidation, both characteristic peaks were lost; however, the presence of reduced forms of each peptide appeared along with an unknown peak at m/z 2211.28 (Fig. 5B). No higher oxidation states of these Cys residues were observed, and the presence of reduced forms supports the idea of functional catalysis under oxidative conditions. Notably, the putative disulfide mass of these peptides is 2211.17, very close to the unknown peptide observed after oxidation. To decipher the identity of the unknown peptide, the tryptic digest was CAF-labeled to enhance and simplify PSD analysis. Initial evidence of an interpeptidyl disulfide bond is a doubly labeled moiety, shown as m/z 2483.38 in Fig. 6A, with unambiguous identification by MS/MS PSD (Fig. 6B). Mass spectrometry analysis of the C-terminal deletion variant hYVH1ΔCT1 also identified this intramolecular disulfide bond formation upon oxidation (supplemental Fig. 1). These findings strongly support our initial studies that suggest sustainable activity under oxidative conditions as well as the formation of an intramolecular disulfide bond within the active site.
To probe the thiol status of the C-terminal zinc-coordinating cysteines, Glu-C peptides were identified by mass fingerprint analysis. Both CAM and NEM reduced peptides were identified as reference controls (Fig. 7, A and B). Only two characteristic peptides could be identified in the NEM mass fingerprint (Fig. 7B). After oxidation, all characteristic peaks denoting zinc finger vicinal thiol pairs were lost; however, a peak at m/z 3101.84 appeared, possibly denoting a reduced vicinal thiol pair after oxidation (Fig. 7C). Furthermore, higher oxidation states of these peptides were not readily observed. Differential labeling resulted in the appearance of all NEM peptides previously identified in the reference mass fingerprint. A peak broadening at m/z 3102.84 and 3106.07 appeared, which may be due to peptide overlap of a singly and doubly modified vicinal thiol pair at residues 221–244 and 222–244, respectively (Figs. 7D and 4B). Supporting evidence was found when tryptic peptides were analyzed following the differential thiol labeling. We identified a disulfide bond involving the extreme C-terminal vicinal thiol pair (residues 302–316) (Fig. 4B and supplemental Fig. 2). Overall, these data further support the working hypothesis that the C-terminal zinc-coordinating domain is capable of acting as a redox buffer via oxidation of vicinal thiol pairs, resulting in disulfide bond formation. Also, the presence of partially and fully reduced thiol pairs under oxidative conditions supports the idea that this region can potentially act as a reducing agent in promotion of a catalytically active phosphatase.
The capacity of hYVH1 to form disulfide bonds strongly supports the idea that enzymatic inactivation and zinc coordination are reversible phenomena. Upon treatment with a micromolar concentration of reducing agent, zinc-free hYVH1 was capable of rapid, recoordination of ∼85–90% total zinc (Fig. 8A). Interestingly, examining the activity of zinc-free hYVH1 in the presence of reducing agents resulted in a marked decrease in activity of hYVH1 (∼60%), whereas the C-terminal deletion variant hYVH1ΔCT1 (∼10%) was unaffected (Fig. 8B). These findings suggest that both a reduced active site and proper zinc coordination are necessary for full activity in vitro during reducing conditions, highlighting the importance of rapid rebinding of zinc following recovery from oxidative conditions. This reversible capacity of zinc binding and disulfide bond formation suggests a dynamic regulatory mechanism under conditions of cellular oxidative stress (Fig. 9).
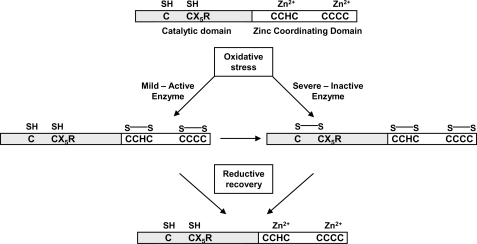
FIGURE 9.
Proposed model of sustainable activity under conditions of oxidative stress. Under mild oxidative conditions, zinc is ejected due to thiols in the zinc binding region serving as reducing agents, allowing the active site cysteine to remain reduced and catalytically competent. Severe oxidative conditions supersede the reducing power of the C-terminal thiols. However, the active site cysteine is protected from irreversible thiol oxidation (e.g. sulfonic acid) by disulfide bond formation with a proximal cysteine residue. Once reducing conditions are restored, the active site cysteine thiol is recovered while the C-terminal thiols rapidly recoordinate zinc.
The precise biological role of hYVH1 remains unclear, and currently, no substrate has been identified for this phosphatase. Since the phosphatase activity of hYVH1 is required for its cytoprotective function (8), identification of its phospho-target during oxidative stress conditions is imperative for acquiring mechanistic understanding of how hYVH1 can protect cells from various insults. Also, it has been recently suggested that YVH1 is a novel 60 S ribosome biogenesis factor in yeast (24). If hYVH1 plays a similar role in human cells, it will be interesting to investigate if the redox-sensing function of hYVH1 proposed in this study regulates the rate of ribosome biogenesis during cellular stress. Various substrate trap-mass spectrometry efforts and hYVH1-mediated ribosome dynamics studies are ongoing to further understand the role of hYVH1 in cellular regulation.
Altogether, the discovery of this inherent redox defense mechanism has provided valuable insights into the protective value of the zinc binding domain of hYVH1. This unique regulatory mechanism also sheds further light on the ability of certain cysteine-based phosphatases to cope with the destructive consequences of oxidative environments.
Supplementary Material
Acknowledgments
We thank Colleen Mailloux for construction of the pGEX/hYVH1ΔCT1 plasmid and Marianne Ilbert for PAR/PCMB protocols.
This study was supported by Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant 298468 (to P. O. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- hYVH1
- human YVH1
- DUSP
- dual specificity phosphatase
- PTP
- protein-tyrosine phosphatase
- MALDI
- matrix-assisted laser desorption ionization
- TOF
- time of flight
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- PSD
- postsource decay
- CAM
- carbamidomethylated
- NEM
- N-ethylmaleimidyl
- CAF
- chemically assisted fragmentation
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DiFMUP
- 6,8-difluoro-4-methylumbelliferyl phosphate
- VHR
- vaccinia H1-related
- DTT
- dithiothreitol
- DTNB
- dithionitrobenzoic acid
- PAR
- 4-(2-pyridylazo)resorcinol
- PCMB
- parachloromercuribenzoic acid
- NBD-Cl
- 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole.
REFERENCES
- 1.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117,699–711 [DOI] [PubMed] [Google Scholar]
- 2.Denu J. M., Stuckey J. A., Saper M. A., Dixon J. E. (1996) Cell 87,361–364 [DOI] [PubMed] [Google Scholar]
- 3.Muda M., Manning E. R., Orth K., Dixon J. E. (1999) J. Biol. Chem. 274,23991–23995 [DOI] [PubMed] [Google Scholar]
- 4.Beeser A. E., Cooper T. G. (2000) J. Bacteriol. 182,3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendrzyk F., Korshunov A., Benner A., Toedt G., Pfister S., Radlwimmer B., Lichter P. (2006) Clin. Cancer Res. 12,2070–2079 [DOI] [PubMed] [Google Scholar]
- 6.Kresse S. H., Berner J. M., Meza-Zepeda L. A., Gregory S. G., Kuo W. L., Gray J. W., Forus A., Myklebost O. (2005) Mol. Cancer 4,39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S. K., Chu W. S., Hale T. C., Wang X., Craig R. L., Wang H., Shuldiner A. R., Froguel P., Deloukas P., McCarthy M. I., Zeggini E., Hasstedt S. J., Elbein S. C. (2006) Diabetes 55,2631–2639 [DOI] [PubMed] [Google Scholar]
- 8.Sharda P. R., Bonham C. A., Mucaki E. J., Butt Z., Vacratsis P. O. (2009) Biochem. J. 2,391–401 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z. Y., Wang Y., Dixon J. E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91,1624–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denu J. M., Dixon J. E. (1995) Proc. Natl. Acad. Sci. U.S.A. 92,5910–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grzyska P. K., Kim Y., Jackson M. D., Hengge A. C., Denu J. M. (2004) Biochemistry 43,8807–8814 [DOI] [PubMed] [Google Scholar]
- 12.Denu J. M., Tanner K. G. (1998) Biochemistry 37,5633–5642 [DOI] [PubMed] [Google Scholar]
- 13.Meng T. C., Fukada T., Tonks N. K. (2002) Mol. Cell 9,387–399 [DOI] [PubMed] [Google Scholar]
- 14.Tonks N. K. (2005) Cell 121,667–670 [DOI] [PubMed] [Google Scholar]
- 15.Lou Y. W., Chen Y. Y., Hsu S. F., Chen R. K., Lee C. L., Khoo K. H., Tonks N. K., Meng T. C. (2008) FEBS J. 275,69–88 [DOI] [PubMed] [Google Scholar]
- 16.Groen A., Lemeer S., van der Wijk T., Overvoorde J., Heck A. J., Ostman A., Barford D., Slijper M., den Hertog J. (2005) J. Biol. Chem. 280,10298–10304 [DOI] [PubMed] [Google Scholar]
- 17.Ross S. H., Lindsay Y., Safrany S. T., Lorenzo O., Villa F., Toth R., Clague M. J., Downes C. P., Leslie N. R. (2007) Cell. Signal. 19,1521–1530 [DOI] [PubMed] [Google Scholar]
- 18.Weibrecht I., Böhmer S. A., Dagnell M., Kappert K., Ostman A., Böhmer F. D. (2007) Free Radic. Biol. Med. 43,100–110 [DOI] [PubMed] [Google Scholar]
- 19.Fox G. C., Shafiq M., Briggs D. C., Knowles P. P., Collister M., Didmon M. J., Makrantoni V., Dickinson R. J., Hanrahan S., Totty N., Stark M. J., Keyse S. M., McDonald N. Q. (2007) Nature 447,487–492 [DOI] [PubMed] [Google Scholar]
- 20.Chiarugi P., Fiaschi T., Taddei M. L., Talini D., Giannoni E., Raugei G., Ramponi G. (2001) J. Biol. Chem. 276,33478–33487 [DOI] [PubMed] [Google Scholar]
- 21.Baker L. M., Baker P. R., Golin-Bisello F., Schopfer F. J., Fink M., Woodcock S. R., Branchaud B. P., Radi R., Freeman B. A. (2007) J. Biol. Chem. 282,31085–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilbert M., Horst J., Ahrens S., Winter J., Graf P. C., Lilie H., Jakob U. (2007) Nat. Struct. Mol. Biol. 14,556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis H. R., Poole L. B. (1997) Biochemistry 36,15013–15018 [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Chang A. (2009) Genetics 181,907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.