Abstract
The role of Hedgehog (Hh) signaling as a developmental pathway is well established. Several recent studies have implicated a role for this pathway in multiple cancers. In this study we report that expression of GLI1 and osteopontin (OPN) increase progressively with the progression of melanoma from primary cutaneous cancer to metastatic melanoma in clinically derived specimens. We have further determined that OPN is a direct transcriptional target of GLI1. We have observed that OPN expression is stimulated in the presence of Hh ligands and inhibited in the presence of the Smoothened (SMO) inhibitor, cyclopamine. Transcriptional silencing of GLI1 negatively impacts OPN expression and compromises the ability of cancer cells to proliferate, migrate, and invade in vitro and interferes with their ability to grow as xenografts and spontaneously metastasize in nude mice. These altered attributes could be rescued by re-expressing OPN in the GLI1-silenced cells, suggesting that OPN is a critical downstream effector of active GLI1 signaling. Our observations lead us to conclude that the GLI1-mediated up-regulation of OPN promotes malignant behavior of cancer cells.
The Hedgehog (Hh)2 pathway that has a central role in developmental patterning (ontogeny) and in maintenance of stem or progenitor cells in many adult tissues (1) has been demonstrated to be active in multiple cancer types (2, 3). Active Hh signaling is also reported to influence the tumor stromal microenvironment (4) and support stem cells in the tumor in an undifferentiated, proliferative state (2, 5).
Hh signaling in mammalian cells is mediated by the GLI family of zinc finger transcription factors comprising GLI1, GLI2, and GLI3. GLI1 is a strong transcriptional activator; GLI2 has both activator and repressor functions; and GLI3 is mostly a repressor (reviewed in Ref. 6). In the Hh ligand-dependent pathway, in the absence of the ligand, Desert hedgehog, Indian hedgehog (IHH), or Sonic hedgehog (SHH), the Hh signaling pathway is inactive, GLI1 is sequestered in the cytoplasm and repressed for its transcription activity (7–9). Binding the Hh ligands to the receptor patched-1 or patched-2 (PTCH1 or -2) changes the GLI code (6): GLI1 is activated by release from a large protein complex and translocates to the nucleus to function as a transcriptional activator (10, 11).
Signaling via the Hh pathway plays a determinative role in the development of the dorsal brain, near the sites of origin of melanogenic precursors (12). Stecca et al. (13) have reported that the Hh pathway is required for normal proliferation of human melanocytes in vitro and for proliferation and survival of human melanoma in vivo. Activation of Hh signaling results in transcriptional activation of the expression of several genes including insulin-like growth factor-binding protein, cyclin D2, and osteopontin (OPN) (11). In the present study, we report that the expression levels of GLI1 and OPN are significantly elevated in surgically excised metastatic melanoma specimens compared with surgically obtained basal and squamous cell carcinomas and primary melanoma samples. We further characterize the functional relevance of this association. Our observations from this study suggest that the Hh pathway acts via OPN to regulate malignant behavior of cancer cells. Thus, our study has identified a clinically relevant relationship between OPN and Hh signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
Tissues of primary and metastatic melanoma resected from patients were cultured and maintained as previously described (14). The melanoma cell lines, MCC012A and MCC012F, used in this study were established from two subcutaneous metastatic nodules from the same patient. Cells were grown in a Dulbecco's modified minimum essential medium, F-12 mixture (1:1) (Invitrogen) supplemented with 5% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 200 μm sodium pyruvate (Invitrogen), and 20 μm non-essential amino acids (Invitrogen). All cells were maintained in a humidified 5% CO2 environment. MDA-MB-435 cells were also cultured under similar conditions.
Generation of Stable Transfectants
Endogenous GLI1 from MDA-MB-435 cells was silenced using shRNAs (short hairpin RNA) cloned into pSuperior.neo+gfp plasmid (OligoEngine, Seattle, WA) (see Table 1). We also generated stable vector-only and non-targeting (scrambled control) transfectants. Stable transfectants were selected on medium supplemented with 500 μg/ml Geneticin (Invitrogen). The four GLI1-knocked down clones chosen for this study were based on the extent of GLI1 knockdown and were termed KO1, KO2, KO3, and KO4. The expression of OPN was restored in the KO1 cells by transfecting with pcDNA3.1-OPN (kindly provided by Dr. Ann Chambers, London Heath Sciences Center). A corresponding vector-only transfectant was also generated. Transfectants were selected on medium containing Geneticin (500 μg/ml) and hygromycin (750 μg/ml).
TABLE 1.
Details of the region in GLI1 transcript that was targeted by the shRNAs used to assess efficacy of silencing GLI1
Serum-free conditioned medium harvested from ∼3.0 × 106 cells after 24 h was assayed for OPN by immunoblotting. To test the inhibitory effect of cyclopamine on Hh pathway activation, cells were cultured in Dulbecco's modified minimum essential medium supplemented with 0.5% fetal bovine serum and treated for the indicated time intervals with dimethyl sulfoxide (vehicle control), 10 and 20 μm cyclopamine (Sigma).
Western Blotting Analysis
Whole cell lysates were collected in Nonidet P-40 buffer (150 mm NaCl, 50 mm Tris, 1% Nonidet P-40). Isolation of cytosolic and nuclear fractions was done as previously reported (15). Total protein (30 μg) was resolved by SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. Membranes were immunoblotted overnight at 4 °C with antibodies to OPN (catalog number 905-629; Assay Designs, Ann Arbor, MI), GLI1 (sc-20687; Santa Cruz Biotechnology, Santa Cruz, CA), CD44 (HCAM) (sc-7946; Santa Cruz Biotechnology), vimentin (sc-32322; Santa Cruz Biotechnology), N-cadherin (catalog 18-0224; Invitrogen), SNAI2 (catalog H00006591-M02; Novus Biologicals, Littleton, CO), SHH (sc-1194; Santa Cruz Biotechnology), or PTCH1 (sc-6149; Santa Cruz Biotechnology). Equal loading was confirmed with anti-β-actin (Sigma) antibody. The purity of cytosolic and nuclear fractions was confirmed with anti-β-tubulin (catalog 2146; Cell Signaling, Danvers, MA) or anti-HDAC1 (catalog 2062; Cell Signaling) antibodies, respectively. Secreted OPN was assessed by loading an equal quantity of protein from the serum-free conditioned medium. Corresponding horseradish peroxidase-conjugated secondary antibodies were used for detection; blots were developed with SuperSignal enhanced chemiluminescence substrate (Pierce) and imaged using a Fuji LAS3000 imager.
Expression Constructs
OPN promoter activity was assessed by a luciferase reporter assay using the human OPN promoter construct (OPN-352) cloned into pGL3-basic vector (Promega) (16). The putative GLI1 binding site (17) (5′-TGCTGAATGCCCATCCC-3′) in the OPN promoter was disrupted using an inside-out PCR and replaced with a NotI site using primers: forward, 5′-CTCAGCGGCCGCTAATAAATGAAAAAGC-3′ and reverse, 5′-GTTAGCGGCCGCTGAGAGTTCCAGGAAG-3′. The resultant construct, referred to as OPN-352Mut has a mutated GLI1-binding site.
Luciferase Assay
Cells (40,000) were transfected with pGL3-OPN-352 or pGL3-OPN-352Mut in combination with pLNCX or pLNCX-GLI1 (kindly obtained from Dr. Jingwu Xie, University of Texas Medical Branch, Galveston, TX) as previously described (16). Empty pGL3 vector was used as control. Hh ligands were added to the well 6 h prior to harvesting the cells (∼33 h of initiation of transfection) for assay. Readings were normalized to total protein content. Each parameter was studied in triplicate and the experiment repeated at least 3 times. The data are represented as percent luciferase activity, which was derived as a percent of the relative light units in treated groups compared with the untreated groups.
Quantitative RT-PCR (qRT-PCR)
cDNA was generated using High Capacity Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA). Real time PCR was performed using a Bio-Rad iQ5 Real-Time Detection system (Bio-Rad). All reactions were done as three independent replicates. All assays were done using the TaqMan Gene Expression Assays from Applied Biosystems. OPN (SPP1: Hs 00959010_m1) transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (Hs 99999905_m1) levels (δCT), which was used to calculate changes in OPN expression (2−δδCT). To analyze the effect of cyclopamine treatment on OPN expression, untreated samples were set as calibrator (control) and compared with their respective treated samples.
GLI1 and SHH (Hs 00179843_m1) expressions were also similarly assessed with glyceraldehyde-3-phosphate dehydrogenase as an endogenous control. To analyze the knockdown effectiveness, “scrambled transfectants” of MDA-MB-435 was set as calibrator.
Chromatin Immunoprecipitation Assay
MDA-MB-435 cells were utilized for chromatin immunoprecipitation using the ChIP-IT Express enzymatic kit (Active Motif, Carlsbad, CA) following the manufacturer's protocol using GLI1 (N-16) X TransCruz antibody (Santa Cruz Biotechnology; sc-6153 X). The recovered DNA was PCR amplified using primers: forward, 5′-GTTTTTCCCTACTTTCTCCC-3′ and reverse, 5′-CCAAAAACGCACACACAC-3′ to amplify a 145-bp segment of the OPN promoter containing the putative GLI1 binding site. The specificity of the pull-down was confirmed by amplifying a region ∼1 kb upstream from the PCR product containing the GLI1 site tested. The primers used were: 5′-TTCCCCCTACCAAATGTTCA-3′ and 5′-TGCTGCAAAAGTAATTGTGGTT-3′. The PCR generates a 151-bp product. This segment lacks a predicted GLI1-binding site.
In Vitro Proliferation Assay
Cells (5000) of each cell type were seeded per well in separate 96-well plates. Cells were allowed to grow in complete medium for 6 days. Every day after initial seeding cells were harvested by trypsinization and counted in a hemocytometer. Counting for each cell type for each day was done in triplicate.
Motility Assay
These experiments were performed as previously outlined (18). Images were acquired at T0, the reading at the initial time, and at T10 (10 h later). The experiment was conducted in duplicate and cell motility was calculated as (T0 − T10)/T0, which represents the rate of movement over a 10-h period. For OPN add-back experiments the cells were pretreated for 12 h with 100 ng/ml human OPN (recombinant R & D Systems) and the assay conducted in the presence of OPN. Each experimental group was assessed in duplicate and data were recorded in three fields per well. Thus, we recorded and analyzed six data points per experimental group.
Invasion and Migration Assays
These experiments were performed as previously outlined (18) using a modified Boyden chamber assay. OPN add-back experiments for migration and invasion were conducted as described above with an additional step of pre-treating the cells for 24 h with rOPN. OPN was added to the upper and lower chambers (100 ng/ml) to ensure the OPN was present during the entire duration of the experiment. Each experimental group was assessed as three independent replicates.
In Vivo Assay
One million (100 μl) cells were injected into the third mammary fat pad of 6-week-old female athymic nude mice (Harlan Sprague-Dawley, Indianapolis, IN). Orthogonal tumor measurements were taken twice a week. The mean tumor diameter was calculated by taking the square root of the product of orthogonal measurements. Spontaneous metastasis was monitored as previously described (18). Eight mice were used for each group and the entire experiment was repeated once. All animals were maintained under the guidelines of the National Institutes of Health and University of South Alabama. All protocols were approved and evaluated by the Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical differences between groups were assessed using the Mann-Whitney test, t test, or analysis of variance, using GraphPad Prism 4 software. Statistical significance was determined if the analysis reached 95% confidence. The precise p values are listed in the corresponding figure legends. In all figures the error bars represent mean ± S.E.
RESULTS
Expression Levels of GLI1 and OPN Increase with the Development and Progression of Melanoma
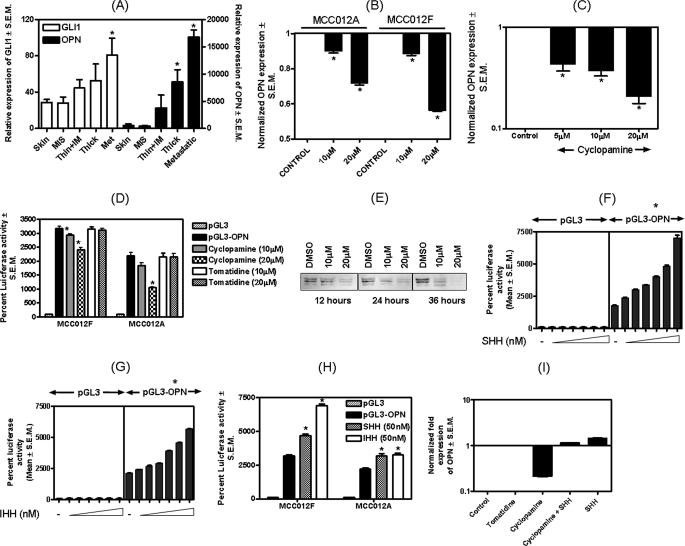
Activation of the Hh pathway results in nuclear translocation of GLI1 transcription factors and up-regulation of target genes. Microarray analysis of genes that were differentially regulated by the Hh pathway revealed that OPN expression was up-regulated (11). We profiled clinically derived primary cutaneous cancers and melanoma specimens by gene expression analysis and reported that the expression of OPN was increased 67.3-fold in metastatic melanoma samples when compared with primary cancer samples (14). We queried this data set for the changes in the expression of GLI1 and OPN with disease progression. As seen in Fig. 1A the expression of GLI1 and OPN increase with the progression of the disease to metastatic melanoma. Specifically, the expression of GLI1 notably increases in thin (up to 1.5 mm in Brelsow thickness) and intermediate (up to 4.0 mm in Brelsow thickness) melanoma specimens and continues to increase as the condition progresses to thick melanoma (>4 mm in Breslow thickness) and beyond into metastatic melanoma. The increase in GLI1 expression in the metastatic melanoma specimens is significantly higher (p < 0.05) as compared with the melanoma in situ (MIS) specimens. In parallel, the expression of OPN also increases as the MIS progresses to thin/intermediate melanoma and beyond into thick and metastatic melanoma. The levels of OPN expression in the thick and metastatic melanoma specimens are significantly greater compared with the corresponding OPN levels in the MIS specimens (p = 0.018 and 0.0018, respectively). It is important to note that whereas the relative expression of GLI1 in metastatic melanoma averages (±S.E.) 80.6 ± 18.5 units, that of OPN peaks at 16,760 ± 1324 units. This is a relevant finding because it underscores the fact that small changes in expression of the transcription factor, GLI1, correlates with changes of a large magnitude in the levels of OPN.
FIGURE 1.
The Hh pathway transcriptionally regulates OPN. A, levels of GLI1 and OPN are increased in primary cutaneous cancer and metastatic melanoma. Gene microarray analysis (utilizing a Human Genome U133 Plus 2.0 array from Affymetrix, Inc.) was used to compare 40 metastatic melanoma samples, composed of 22 bulky, macroscopic (replaced) lymph node metastases, 16 subcutaneous and 2 distant metastases (adrenal and brain), to 16 primary cutaneous melanoma specimens (14). The expression levels of GLI1 and OPN increase progressively beyond the stage of MIS through the stage of metastatic melanoma. Thin, thin melanomas (<1.5 mm in Breslow thickness); IM, intermediate thickness (between 1.5 and 4.0 mm in Breslow thickness); thick, melanomas (that are >4.0 mm in Breslow thickness). The left y axis denotes the scale for GLI1 expression and the right y axis corresponds to OPN levels. As compared with MIS, the increase in GLI1 in the metastatic melanoma samples is statistically significant (p = 0.020). The increase in OPN expression in the thick and metastatic melanoma specimens is statistically significant compared with the corresponding OPN levels in the MIS specimens (p = 0.018 and 0.0018, respectively). B and C, inhibition of the Hh pathway negatively impacts OPN expression in MCC012A, MCC012F, and MDA-MB-435 cells. B, cyclopamine treatment significantly (* indicates p < 0.0001) decreases the levels of OPN mRNA (assessed by qRT-PCR) in MCC012A and MCC012F cells. C, cyclopamine significantly (* indicates p < 0.0001) decreases the levels of OPN mRNA in a dose-dependent manner in MDA-MB-435 cells. Specifically, cells were treated with the indicated concentrations of cyclopamine in Dulbecco's modified minimum essential medium, F-12 supplemented with 0.5% fetal bovine serum. This medium was replaced with fresh cyclopamine-containing medium after 12 h. Cells were harvested for assay after 24 h of cyclopamine treatment. RNA was assessed by real-time RT-PCR for OPN transcript levels. D, cyclopamine causes a dose-dependent decrease in the OPN promoter activity (200 ng of pGL3-OPN transfected) in MCC012A and MCC012F cell lines. In the MCC012F cells, at the doses tested (10 and 20 μm), cyclopamine caused a significant (p = 0.042 and 0.002, respectively) decrease in OPN promoter activity. In the MCC012A cell line, cyclopamine (10 μm) caused a noticeable, but not significant (p = 0.06) decrease in OPN promoter activity. Treatment with 20 μm cyclopamine caused a significant decrease (p = 0.0006) in OPN promoter activity. Tomatidine had no effect on the promoter activity of OPN. E, levels of OPN in the secretome were decreased upon treatment with cyclopamine. MDA-MB-435 cells were grown for the indicated time in 0 (dimethyl sulfoxide (DMSO) control), 10, and 20 μm cyclopamine. The conditioned media were assayed for OPN. F and G, the Hh ligands stimulate OPN promoter activity. Cells (MDA-MB-435) were transfected with the OPN promoter construct (200 ng) and treated with increasing concentrations of either SHH (F) or IHH (G). The asterisk above the graph indicates that the activity of the OPN promoter was significantly (p < 0.0001) higher than that of the corresponding control (untreated) group for all concentrations of IHH and SHH tested. H, triggering the Hh pathway by treatment with the ligands, SHH and IHH, results in a significant increase in OPN promoter activity in metastatic melanoma cell lines, MCC012A (p = 0.0004 for SHH and p < 0.0001 for IHH treatments) and MCC012F (p = 0.0078 for SHH and p = 0.0032 for IHH). I, SHH was able to rescue the inhibitory effects of cyclopamine on OPN transcript levels. MDA-MB-435 cells (1 million) were treated with cyclopamine or tomatidine (20 μm). After 12 h, the medium of one cyclopamine-treated set was replaced with medium containing recombinant SHH (100 nm). The experiment was terminated after 24 h of the start of the initial cyclopamine treatment. RNA was assessed by real-time RT-PCR for OPN transcript levels. Error bars represent mean ± S.E.
OPN Is Transcriptionally Up-regulated by the Hh Pathway
To determine whether OPN expression is regulated by Hh pathway signaling, we decided to study three metastatic melanoma-derived cell lines, MCC012A, MCC012F, and MDA-MB-435. To establish autocrine Hh signaling it is imperative that the cell lines express the Hh pathway products, including the receptor PTCH, the ligand SHH, and the transcription factor GLI1. As seen in supplemental Fig. S1, all three melanoma cell lines express Hh pathway members indicating that the cell lines are capable of autocrine Hh signaling. The expression of these Hh members is significantly greater (p < 0.0001) in metastatic melanoma cell lines compared with that in the primary melanoma-derived cell line, MCC013. All the three metastatic melanoma cell lines also express significantly higher levels of OPN (p < 0.0001) compared with MCC013 (supplemental Fig. S1D).
We tested the effect of the Hh inhibitor, cyclopamine, (19) on OPN levels. As seen in Fig. 1, B and C, cyclopamine significantly (p < 0.05) decreases the levels of OPN mRNA in a dose-dependent manner in two metastatic melanoma-derived cell lines, MCC012A and MCC012F, and in MDA-MB-435 cells (p < 0.0001), suggesting that blocking the Hh pathway interferes with the transcription of OPN. Cyclopamine treatment also decreases the activity of the OPN promoter in a dose-dependent manner (Fig. 1D). In contrast, tomatidine, the structural analog of cyclopamine, had no effect on the promoter activity of cyclopamine.
The decreases in the levels of OPN mRNA are also reflected in the decreased levels of OPN protein in the secretome of cyclopamine-treated cells (Fig. 1E). This effect was more pronounced at time intervals of 24 and 36 h post-treatment, when a lower concentration of cyclopamine was also able to inhibit OPN. In contrast to the inhibitory effect of cyclopamine, treatment of MDA-MB-435 cells with SHH and IHH ligands (Fig. 1, F and G) significantly (p < 0.0001) up-regulated the promoter activity of OPN in a dose-dependent manner. Similarly, SHH and IHH caused a significant up-regulation in promoter activity of OPN in MCC012A (p < 0.01) and MCC012F (p < 0.005) (Fig. 1H). SHH was also able to reverse and rescue the inhibitory effects of cyclopamine on the levels of the OPN transcript thereby re-instating Hh signaling (Fig. 1I).
The Transcription Factor GLI1 Up-regulates OPN
Signaling via the Hh pathway culminates in the transcription of target genes by the GLI transcription factors. We tested the role of transcription factor GLI1 in mediating the effects of the Hh pathway on OPN. We scanned the promoter region of human OPN (up to 1 kb upstream of transcription start site) for GLI1-binding sites using TFSEARCH and identified a putative GLI1 binding site at position −243 to −259 (5′-TGCTGAATGCCCATCCC-3). As shown in Fig. 2A, co-transfection of a GLI1 expressing construct with an OPN promoter construct (OPN-352; encompassing the −352 to +112 region) brought about a significant (p < 0.0001) increase in the activity of the OPN promoter. The putative GLI1-binding site in the OPN promoter differs from the consensus GLI1-binding site by 3 nucleotides as shown in Fig. 2A. We abolished this site from the OPN-352 promoter and replaced it with a NotI site, keeping the distance from the transcription site unchanged (we confirmed that in this process no other transcription factor-binding site was generated). This mutant OPN construct, OPN-352Mut, was unable to respond to GLI1, indicating that this site on the OPN promoter was critical to its ability to be activated by transcription factor GLI1 (Fig. 2A). Additionally, OPN-2Mut is refractory to the effects of SHH and IHH. As seen in Fig. 2B, whereas OPN-352 (bearing the GLI1-recognition site) shows a notable increase (p < 0.0001) in promoter activity in the presence of stimulation by SHH and IHH, OPN-352Mut is immune to the potentially activating effects of SHH and IHH.
FIGURE 2.
The OPN promoter bears a GLI1-binding site that responds to GLI1. A, mutating the putative GLI1-binding site in the OPN promoter makes the OPN promoter insensitive to the effects of GLI1. The OPN promoter (+112 to −352 (pGL3-OPN-352)) was significantly activated (p < 0.0001) in response to GLI1. Cells (MDA-MB-435) were transfected with either 100 ng of pGL3, pGL3-OPN-352, or pGL3-OPN-352Mut and 300 ng of pLNCX or pLNCX-GLI1. Empty pGL3 vectors (devoid of promoter) co-transfected with empty pLNCX vectors served as control. The inset box outlines the consensus GLI1-binding site and defines the GLI1-binding site in the OPN promoter. The underlined nucleotides are distinct from the ones in the consensus site. The GLI1-binding site in the OPN promoter is abolished in OPN-352Mut; the bases in bold have been altered to change from a GLI1-binding site to a NotI restriction enzyme site. Asterisk indicates that the activation of the promoter activity (pGL3-OPN-352) is statistically significant (p < 0.0001) compared with pGL3 alone. B, pGL3-OPN-352 shows a significant (p < 0.0001) activation in the activity in the presence of Hh ligands. In contrast to pGL3-OPN-352, pGL3-OPN-352Mut is resistant to the effects of Hh ligands. C, ChIP assay in MDA-MB-435 cells, showing that GLI1 interacts with the OPN promoter. The antibodies used for immunoprecipitation are indicated. Lane 1, PCR using primers encompassing the GLI1-binding site; lanes 2 and 3, PCR using a kit provided the ChIP-positive control and ChIP-negative primers, respectively; and lane 4, PCR using primers amplifying a region of the OPN promoter that is ∼1 kb upstream of the GLI1-binding site. Error bars represent mean ± S.E.
To determine whether GLI1 physically associates with the OPN promoter, we immunoprecipitated cross-linked chromatin from MDA-MB-435 cells with an anti-GLI1 antibody and amplified the region of the OPN promoter that bears the GLI1 recognition sequence (Fig. 2C), implying that GLI1 associates with the OPN promoter. We also controlled specificity of the ChIP assay by performing PCR of the chromatin immunoprecipitate using primers located ∼1 kb upstream of the GLI1 recognition sequence (16) in the OPN promoter. The absence of a product using these primers confirms specificity of the pull-down. Thus, our data shows that OPN is transcriptionally activated by GLI1.
Knockdown of Endogenous GLI1 Blunts the Malignant Behavior of Tumor Cells
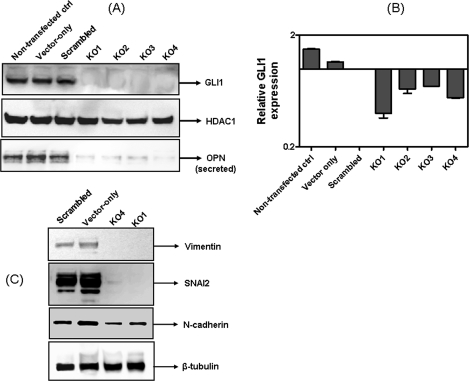
To evaluate the functional effects of active Hh signaling we generated stable cell lines that were knocked down for GLI1 expression by RNA interference. We assessed the efficacy of three shRNA constructs for silencing GLI1 expression (Table 1). Notably, the two shRNA constructs that demonstrated effective GLI1 silencing, shRNA-1 and shRNA-2, overlap in the region they target. Using vector construct GLI1 shRNA-1 (supplemental Fig. S2A), we generated four clones stably silenced for GLI1 expression. All four clones also showed significantly reduced OPN expression. Of the four stable clones, clones KO1 and KO4 expressed the least amount of GLI1 followed by clones KO2 and KO3 (Fig. 3, A and B). KO1 and KO4 were used for further detailed studies.
FIGURE 3.
shRNA to GLI1 abrogates expression of OPN and brings about a partial reversal of EMT. A, MDA-MB-435 cells were stably transfected with shRNA to GLI1. Clones KO1 to KO4 were stably silenced for GLI1 and show notably reduced OPN expression. B, real time RT-PCR shows that the expression of GLI1 mRNA in KO1 to KO4 was notably lower than the controls (vector-only and scrambled transfected). C, vector-only, scrambled transfectants and KO1 and KO4 cells were immunoblotted for the expression of markers of EMT (vimentin, SNAI2, and N-cadherin). β-Tubulin serves as a loading control. Error bars represent mean ± S.E.
The Hh pathway has been reported to influence the expression of signature proteins that mediate epithelial-mesenchymal transition (EMT). Hence we examined GLI1-knocked down clones KO1 and KO4 for the status of these signature markers. As seen in Fig. 3C, expression of vimentin, SNAI2, and N-cadherin were notably decreased in KO1 and KO4 suggesting loss of the mesenchymal phenotype in GLI1-knocked down cells. We were unable to document concomitant expression of E-cadherin in KO1 and KO4 (data not shown).
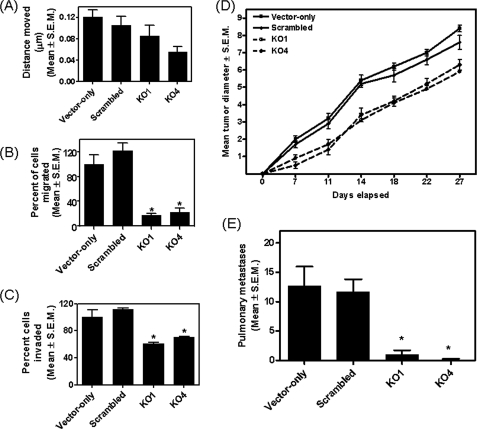
We then tested GLI1-silenced cells for their in vitro attributes of aggressiveness, viz. cell migration, invasion, and motility. Although GLI1 silencing had no significant effect on cell motility measured with a scratch assay (Fig. 4A), cells in which GLI1 has been silenced showed statistically significant (p < 0.0001) decreases in cell migration and invasion measured in modified Boyden chamber assays (Fig. 4, B and C). There was no statistically significant (p > 0.05) effect of GLI1 silencing on cell proliferation in vitro (supplemental Fig. S2B). To examine the effect of GLI1 knockdown on the ability of cells to grow tumors as xenografts, we injected GLI1-silenced cells and the corresponding vector-only and scrambled control cells in athymic nude mice. Although there was no change in the tumor take rate, there was slower growth rate of GLI1-silenced cells up to day 11 (Fig. 4D); the rate of growth subsequent to day 11 was similar between control and silenced cells. The implications of these observations are discussed below. In general, cells silenced for GLI1 showed a significantly (p < 0.005) slower growth of tumors over the monitored time course. This was also reflected in the significantly (p < 0.005) decreased numbers of pulmonary metastases (Fig. 4E) resulting from spontaneous metastasis of the injected cells. In summary, the results suggest that GLI1 silencing has little or no effect on proliferation or primary xenograft growth, but has a marked effect on metastasis. Thus, expression of GLI1 plays a functionally important role in the malignant behavior of tumor cells.
FIGURE 4.
Silencing endogenous GLI1 expression diminishes attributes of motility, invasion, migration, and proliferation and negatively impacts tumorigenicity and metastasis. A, abrogating GLI1 expression (KO1 and KO4) reduced the ability of cells to move and fill in a wound in the cell monolayer (p > 0. 05). Silencing endogenous expression of GLI1 significantly decreases the ability of cells to migrate (B) (p < 0.0001) across gelatin-coated filters and invade (C) (p < 0.0001) through Matrigel. The readings of KO1 and KO4 were compared with the corresponding scrambled control-transfected cells to determine statistical significance. In all cases, the vector-only cells were comparable with the scrambled control cells (p > 0.1). D, GLI1-silenced cells were compromised for their tumorigenicity. Tumor measurements are represented as mean tumor diameter ± S.E. As compared with the scrambled control cells both KO1 (p = 0.0028) and KO4 (p = 0.0018) formed significantly slower growing tumors. E, the GLI1 KO1 (p = 0.0012) and KO4 (p = 0.0005) cells were significantly impaired in their ability to form spontaneous metastases. Error bars represent mean ± S.E.
OPN Mediates the Effect of GLI1 on Malignant Cell Behavior
To determine the role of OPN in mediating GLI1 effects we restored the effects of OPN in GLI1-KO cells. We assessed this in two ways: (a) we treated the GLI1-knocked down cells with recombinant OPN and (b) we stably transfected the GLI1-knocked down cells with a plasmid construct expressing human OPN (Fig. 5A). We then monitored these cells in vitro for properties of migration, invasion, and motility. When the GLI1-knocked down cells were cultured in the presence of OPN, KO1 and KO4 cells were restored for the ability to migrate (p < 0.0001) (Fig. 5B) and invade through Matrigel® (Fig. 5C) in much larger numbers (p < 0.005) compared with untreated cells. Motility of GLI1-silenced cells was also restored in KO1 cells (p < 0.05) and KO4 cells in the presence of recombinant OPN (p > 0.05) (Fig. 5D). The levels of the OPN receptor, CD44, were comparable in the vector-only and KO cells (supplemental Fig. S2C), implying that both cell types should be receptive and responsive to OPN.
FIGURE 5.
Restoring the availability of OPN-initiated signaling in GLI1-silenced cells reinstates their motility and ability to migrate and invade. A, immunoblot representing the restored OPN in cells that have been stably knocked down for GLI1 (GLI1 KO; and consequently express decreased levels of OPN). GLI1 KO cells were stably transfected with empty vector, pcDNA3.1, or pcDNA3.1 expressing OPN. B, migration assay: compared with the respective untreated cells, the OPN-treated cells migrate in significantly larger numbers (p < 0.0001). As compared with KO1, the KO/OPN stable transfectants migrate in significantly greater numbers (p < 0.0001). C, invasion through Matrigel. As compared with the respective untreated cells, the OPN-treated cells invade in significantly larger numbers (KO1, p < 0.0001; and KO4, p = 0.0038). In contrast to KO1, the KO/OPN stable transfectants invade in significantly larger numbers (p < 0.0001). D represents the results of the wound healing assay. Although the KO/OPN and KO1 + OPN cells are able to move significantly faster than the respective control KO1 cells (p = 0.0269 and p = 0.0066, respectively), the motility of KO4 cells treated with OPN follows a similar fast trend (p = 0.15). (KO1 + OPN and KO4 + OPN represent experimental conditions wherein the cells were cultured in OPN-containing medium for 24 h and assayed in the presence of OPN. KO/OPN represents the GLI1-knocked down cells that have been stably transfected with OPN.) E, restoration of OPN in GLI1-silenced cells results in enhanced ability of the cells to grow as xenografts in athymic nude mice. Tumor measurements are represented as mean tumor diameter ± S.E. As compared with the vector-only (pSUPERIOR) and Gli1-silenced (KO1) cells, the two clones, KO1/OPN.5 and KO1/OPN.8, both formed significantly faster growing tumors (p < 0.05). Error bars represent mean ± S.E.
Similarly, stably restoring the expression of OPN (Fig. 5A) in the GLI1-knocked down cells reinstated the ability of the cells to chemotactically migrate through a filter (8 μm pores) (Fig. 5B), invade through Matrigel (Fig. 5C), and restore the ability of the cells to move laterally (in a scratch motility/wound healing assay) (Fig. 5D). Restoration of OPN expression in GLI1-silenced (KO1) cells caused the cells to form rapidly growing tumors in mice (Fig. 5E). As compared with KO1 cells transfected with empty vector (KO1-pcDNA3), the two clones restored for OPN, viz. KO1/OPN.5 and KO1/OPN.8, formed tumors that grew faster than control cells. This implied that regulation of OPN by the Hh pathway is functionally critical to the malignant properties of cancer cells.
DISCUSSION
The role of the Hh pathway has been well documented in several cancer histotypes (13, 20–25). The activities of this pathway have been attributed to several mediators, such as platelet-derived growth factor (26), fibroblast growth factor (27), bone morphogenic protein (28), Notch (29), and Wnt (30), which have been identified as Hh target genes in various models. However, to date, few universal target genes have been identified across different systems and much work still needs to be done to determine how Hh overexpression contributes to tumorigenesis.
Our studies indicate that signaling via the Hh pathway can transcriptionally up-regulate OPN, an oncogene that has been widely reported to promote tumorigenesis, tumor progression, and metastasis in several cancer types. The regulation of OPN by the transcription factor GLI1 is integral to the malignant behavior of cancer cells as evidenced by the impaired ability of tumor cells to migrate, invade, and grow in vivo as xenografts when endogenous GLI1 expression is silenced. OPN is a secreted protein that influences multiple downstream signaling events that allows cancer cells to resist apoptosis, invade through extracellular matrix, evade host immunity (31), and influence growth of indolent tumors (32). OPN induces integrin (33–40) and CD44-mediated migration (41) via hepatocyte growth factor, its receptor, Met (42, 43), and epidermal growth factor (42, 43) and enhances the invasive ability of cells by inducing the expression of proteases such as MT1-matrix metalloproteinase, matrix metalloproteinase-2 (44, 45), and urokinase plasminogen activator (46). Clinically, OPN expression is up-regulated in several malignancies including breast cancer, melanoma, prostate cancer, colorectal cancer, and head and neck cancer (47). We reported that OPN constitutes a component of the secretome of several melanoma-derived cell lines (14) and is expressed in metastatic breast cancer cell lines (18). Independent studies have also reported an increase in levels of OPN in melanoma-derived cell lines (48, 49). Recent findings from our laboratory have revealed that expression of OPN is 13-fold higher when comparing thin melanomas to metastatic melanomas.3 Our studies have revealed that the expression of GLI1 increases notably as cutaneous cancer progresses from a stage of melanoma in situ to intermediate and thick melanoma to metastatic melanoma. The increase in GLI1 expression is paralleled by an increase in OPN levels. Overall, our observations underscore the role of enhanced Hh signaling via increased GLI1 transcriptional activity in potentiating the malignant behavior of melanoma cells and contributing to disease progression. Our findings are in contrast to those reported by Stecca et al. (13) who did not find changes in expression levels of GLI1 in a limited set of sporadic human melanomas versus nevi. Nonetheless, they found that melanomas showed characteristically activated Hh signaling. The active Hh signaling may be due to ligand-dependent mechanisms, cell-intrinsic ligand-independent mechanisms, or cross-talk between the oncogenic Ras/Akt pathway and Hh/GLI signaling resulting in enhancements of GLI1 transcriptional activity (13).
The Hh pathway has been reported to be aberrantly active in several cancer types, including breast cancer (50–52) and melanoma (13, 53). Hh pathway components were detected in nevi, melanoma, and lymph node metastases of melanoma (13). Enforced expression of GLI1 induced the expression of Snail (54), whereas blockade of Hh signaling by the inhibitor cyclopamine suppressed pancreatic cancer invasion and metastasis by inhibiting EMT (55). Our study is in agreement with these reports because we have documented a loss of mesenchymal markers by abrogating GLI1 expression. EMT-related genes (N-cadherin, OPN, and osteonectin) have been reported to contribute to the promotion of the metastatic phenotype in primary cutaneous malignant melanomas by supporting specific adhesive, invasive, and migratory properties (56). Moreover, these findings support our observations showing that GLI1 silencing attenuates malignancy-associated attributes, such as invasion, migration, and motility.
Interestingly, our results show that GLI1 silencing retards the tumor (xenograft) rate only in the early phase. After day 11 post-injection, the growth of GLI1-silenced tumors proceeded at the same rate as that of controls. This data can have multiple implications. First, it is likely that over time, a revertant population outgrew the GLI1-silenced cells. These revertants may have either lost the effects of RNA interference or may have by-passed the requirement for GLI1 signaling. In this case, the cells may have utilized other signaling pathways to up-regulate OPN. Second, trace levels of OPN secreted by GLI1-silenced cells (Fig. 3A) can accumulate in the local microenvironment from the growing tumor and may have stimulated cell growth. Overall, GLI1 silencing had a pronounced effect on tumor malignancy in vivo by reducing metastasis.
We have used the MDA-MB-435 cell line as a model to address and investigate our hypothesis because it has been reported to produce abundant levels of OPN (16, 18, 46, 57–59). This cell line has been at the center of controversy, which has called its origin into question, with the common consensus that it is of melanocytic origin (60–63) despite the fact that it produces milk proteins (64), and is the most reliable and widely used cell line model to study spontaneous metastasis following xenograft establishment in the mammary fat pads. Our study in this model system, which endogenously expresses high levels of OPN, supports a role for the Hh pathway in regulating malignant cell behavior. Moreover, our findings can have implications on multiple cancer histotypes that overexpress OPN (65).
Our findings can have two implications. First, it is interesting that the Hh ligands and OPN, the signaling intermediate of the active Hh pathway, are secreted molecules. This allows them to influence the behavior of cells in the tumor microenvironment. OPN has been documented to influence the behavior of cells in a paracrine manner. Although serum OPN influences the migratory behavior of melanoma cells (66) and tumor-derived OPN inhibits nitric-oxide synthase activity of macrophages (67), OPN produced by fibroblasts is able to influence growth of pre-neoplastic cells (68). Thus, it is likely that active Hh signaling in a subset of cancer cells can potentially be amplified by secretion of OPN into the tumor microenvironment. The secreted OPN, in turn, can promote malignant behavior in neighboring cancer cells, regardless of the status of the Hh pathway. Second, whereas OPN is capable of long-range signaling, the secreted Hh ligand proteins participate in short-range signaling and can move many cell diameters from their source of production and often control developmental outcomes in a concentration-dependent manner (10). For example, during ventral spinal cord patterning, SHH forms a ventral-to-dorsal gradient with different concentrations specifying distinct pools of neural progenitors (69). It is likely that such a situation also prevails in a tumor; in which case, the Hh ligands produced by a subpopulation of cells within a tumor can trigger activation of the pathway in the recipient cell.
Metastatic melanoma continues to be a devastating disease with grim prognosis and a very limited availability of FDA approved agents for its treatment. The Hh pathway has been the subject of intense investigation as a therapeutic target with inhibitors in clinical trials. Our study draws attention for these Hh inhibitors to be explored for treating metastatic melanoma with the prospect that OPN may serve as a surrogate marker of monitoring the effectivity of Hh inhibitors.
Supplementary Material
This work was supported by United States Department of Defense IDEA Award 07-1-0400 (to L. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
B. J. Metge, S. Liu, A. I. Riker, O. Fodstad, R. S. Samant, and L. A. Shevde, manuscript in preparation.
- Hh
- Hedgehog
- ChIP
- chromatin immunoprecipitation
- IHH
- Indian Hedgehog
- MIS
- melanoma in situ
- OPN
- osteopontin
- PTCH
- Patched
- SHH
- Sonic Hedgehog
- shRNA
- short hairpin RNA
- EMT
- epithelial-mesenchymal transition
- KO
- knock-out
- qRT
- quantitative real-time RT-PCR.
REFERENCES
- 1.Beachy P. A., Karhadkar S. S., Berman D. M. (2004) Nature 432,324–331 [DOI] [PubMed] [Google Scholar]
- 2.Jiang J., Hui C. C. (2008) Dev. Cell 15,801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evangelista M., Tian H., de Sauvage F. J. (2006) Clin. Cancer Res. 12,5924–5928 [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S., Frolova N., Sadlonova A., Novak Z., Steg A., Page G. P., Welch D. R., Lobo-Ruppert S. M., Ruppert J. M., Johnson M. R., Frost A. R. (2006) Cancer Biol. Ther. 5,674–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J., Leong N. L., Mung J. C., Hidaka C., Lu H. H. (2008) Osteoarthritis Cartilage 16,70–82 [DOI] [PubMed] [Google Scholar]
- 6.Ruiz i Altaba A., Mas C., Stecca B. (2007) Trends Cell Biol. 17,438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz i Altaba A. (1998) Development 125,2203–2212 [DOI] [PubMed] [Google Scholar]
- 8.Bai C. B., Stephen D., Joyner A. L. (2004) Dev. Cell 6,103–115 [DOI] [PubMed] [Google Scholar]
- 9.Tyurina O. V., Guner B., Popova E., Feng J., Schier A. F., Kohtz J. D., Karlstrom R. O. (2005) Dev. Biol. 277,537–556 [DOI] [PubMed] [Google Scholar]
- 10.Ingham P. W., McMahon A. P. (2001) Genes Dev. 15,3059–3087 [DOI] [PubMed] [Google Scholar]
- 11.Yoon J. W., Kita Y., Frank D. J., Majewski R. R., Konicek B. A., Nobrega M. A., Jacob H., Walterhouse D., Iannaccone P. (2002) J. Biol. Chem. 277,5548–5555 [DOI] [PubMed] [Google Scholar]
- 12.Le Douarin N. M., Dupin E. (2003) Curr. Opin. Genet. Dev. 13,529–536 [DOI] [PubMed] [Google Scholar]
- 13.Stecca B., Mas C., Clement V., Zbinden M., Correa R., Piguet V., Beermann F., Ruiz I., Altaba A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104,5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riker A. I., Enkemann S. A., Fodstad O., Liu S., Ren S., Morris C., Xi Y., Howell P., Metge B., Samant R. S., Shevde L. A., Li W., Eschrich S., Daud A., Ju J., Matta J. (2008) BMC Med. Genomics 1,13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadowski H. B., Gilman M. Z. (1993) Nature 362,79–83 [DOI] [PubMed] [Google Scholar]
- 16.Samant R. S., Clark D. W., Fillmore R. A., Cicek M., Metge B. J., Chandramouli K. H., Chambers A. F., Casey G., Welch D. R., Shevde L. A. (2007) Mol. Cancer 6,6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinzler K. W., Vogelstein B. (1990) Mol. Cell. Biol. 10,634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevde L. A., Samant R. S., Paik J. C., Metge B. J., Chambers A. F., Casey G., Frost A. R., Welch D. R. (2006) Clin. Exp. Metastasis 23,123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J. K., Taipale J., Young K. E., Maiti T., Beachy P. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,14071–14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clement V., Sanchez P., de Tribolet N., Radovanovic I., Ruiz i Altaba A. (2007) Curr. Biol. 17,165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thayer S. (2004) Clin. Adv. Hematol. Oncol. 2,17, 20,, 63 [PubMed] [Google Scholar]
- 22.Thayer S. P., di Magliano M. P., Heiser P. W., Nielsen C. M., Roberts D. J., Lauwers G. Y., Qi Y. P., Gysin S., Fernández-del Castillo C., Yajnik V., Antoniu B., McMahon M., Warshaw A. L., Hebrok M. (2003) Nature 425,851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins D. N., Berman D. M., Baylin S. B. (2003) Cell Cycle 2,196–198 [PubMed] [Google Scholar]
- 24.Watkins D. N., Berman D. M., Burkholder S. G., Wang B., Beachy P. A., Baylin S. B. (2003) Nature 422,313–317 [DOI] [PubMed] [Google Scholar]
- 25.Karhadkar S. S., Bova G. S., Abdallah N., Dhara S., Gardner D., Maitra A., Isaacs J. T., Berman D. M., Beachy P. A. (2004) Nature 431,707–712 [DOI] [PubMed] [Google Scholar]
- 26.Xie J., Aszterbaum M., Zhang X., Bonifas J. M., Zachary C., Epstein E., McCormick F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98,9255–9259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X., Lewandoski M., Meyers E. N., Liu Y. H., Maxson R. E., Jr., Martin G. R. (2000) Nat. Genet. 25,83–86 [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Carroll T. J., McMahon A. P. (2002) Development 129,5301–5312 [DOI] [PubMed] [Google Scholar]
- 29.Hallahan A. R., Pritchard J. I., Hansen S., Benson M., Stoeck J., Hatton B. A., Russell T. L., Ellenbogen R. G., Bernstein I. D., Beachy P. A., Olson J. M. (2004) Cancer Res. 64,7794–7800 [DOI] [PubMed] [Google Scholar]
- 30.Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T., Gumucio D. L. (2005) Development 132,279–289 [DOI] [PubMed] [Google Scholar]
- 31.Bellahcène A., Castronovo V., Ogbureke K. U., Fisher L. W., Fedarko N. S. (2008) Nat. Rev. Cancer 8,212–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAllister S. S., Gifford A. M., Greiner A. L., Kelleher S. P., Saelzler M. P., Ince T. A., Reinhardt F., Harris L. N., Hylander B. L., Repasky E. A., Weinberg R. A. (2008) Cell 133,994–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu D. D., Hoyer J. R., Smith J. W. (1995) Ann. N.Y. Acad. Sci. 760,312–314 [DOI] [PubMed] [Google Scholar]
- 34.Hu D. D., Lin E. C., Kovach N. L., Hoyer J. R., Smith J. W. (1995) J. Biol. Chem. 270,26232–26238 [DOI] [PubMed] [Google Scholar]
- 35.Bayless K. J., Meininger G. A., Scholtz J. M., Davis G. E. (1998) J. Cell Sci. 111,1165–1174 [DOI] [PubMed] [Google Scholar]
- 36.Smith L. L., Cheung H. K., Ling L. E., Chen J., Sheppard D., Pytela R., Giachelli C. M. (1996) J. Biol. Chem. 271,28485–28491 [PubMed] [Google Scholar]
- 37.Denda S., Reichardt L. F., Müller U. (1998) Mol. Biol. Cell 9,1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liaw L., Lindner V., Schwartz S. M., Chambers A. F., Giachelli C. M. (1995) Circ. Res. 77,665–672 [DOI] [PubMed] [Google Scholar]
- 39.Seftor R. E., Seftor E. A., Hendrix M. J. (1999) Cancer Metastasis Rev. 18,359–375 [DOI] [PubMed] [Google Scholar]
- 40.Varner J. A., Nakada M. T., Jordan R. E., Coller B. S. (1999) Angiogenesis 3,53–60 [DOI] [PubMed] [Google Scholar]
- 41.Katagiri Y. U., Sleeman J., Fujii H., Herrlich P., Hotta H., Tanaka K., Chikuma S., Yagita H., Okumura K., Murakami M., Saiki I., Chambers A. F., Uede T. (1999) Cancer Res. 59,219–226 [PubMed] [Google Scholar]
- 42.Tuck A. B., Elliott B. E., Hota C., Tremblay E., Chambers A. F. (2000) J. Cell. Biochem. 78,465–475 [DOI] [PubMed] [Google Scholar]
- 43.Tuck A. B., Hota C., Wilson S. M., Chambers A. F. (2003) Oncogene 22,1198–1205 [DOI] [PubMed] [Google Scholar]
- 44.Philip S., Bulbule A., Kundu G. C. (2001) J. Biol. Chem. 276,44926–44935 [DOI] [PubMed] [Google Scholar]
- 45.Nagase H., Suzuki K., Morodomi T., Enghild J. J., Salvesen G. (1992) Matrix 1, (suppl.) 237–244 [PubMed] [Google Scholar]
- 46.Tuck A. B., Arsenault D. M., O'Malley F. P., Hota C., Ling M. C., Wilson S. M., Chambers A. F. (1999) Oncogene 18,4237–4246 [DOI] [PubMed] [Google Scholar]
- 47.Coppola D., Szabo M., Boulware D., Muraca P., Alsarraj M., Chambers A. F., Yeatman T. J. (2004) Clin. Cancer Res. 10,184–190 [DOI] [PubMed] [Google Scholar]
- 48.Nemoto H., Rittling S. R., Yoshitake H., Furuya K., Amagasa T., Tsuji K., Nifuji A., Denhardt D. T., Noda M. (2001) J. Bone Miner. Res. 16,652–659 [DOI] [PubMed] [Google Scholar]
- 49.Rangaswami H., Kundu G. C. (2007) Oncol. Rep. 18,909–915 [PubMed] [Google Scholar]
- 50.Xuan Y., Lin Z. (2009) J. Cancer Res. Clin. Oncol. 135,235–240 [DOI] [PubMed] [Google Scholar]
- 51.Zhang X., Harrington N., Moraes R. C., Wu M. F., Hilsenbeck S. G., Lewis M. T. (2009) Breast Cancer Res. Treat. 115,505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubo M., Nakamura M., Tasaki A., Yamanaka N., Nakashima H., Nomura M., Kuroki S., Katano M. (2004) Cancer Res. 64,6071–6074 [DOI] [PubMed] [Google Scholar]
- 53.Bar-Eli M. (2007) Pigment Cell Res. 20,341–342 [DOI] [PubMed] [Google Scholar]
- 54.Li X., Deng W., Lobo-Ruppert S. M., Ruppert J. M. (2007) Oncogene 26,4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y., Evers B. M., Zhou B. P. (2009) J. Biol. Chem. 284,640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alonso S. R., Tracey L., Ortiz P., Pérez-Gómez B., Palacios J., Pollán M., Linares J., Serrano S., Sáez-Castillo A. I., Sánchez L., Pajares R., Sánchez-Aguilera A., Artiga M. J., Piris M. A., Rodríguez-Peralto J. L. (2007) Cancer Res. 67,3450–3460 [DOI] [PubMed] [Google Scholar]
- 57.Hedley B. D., Welch D. R., Allan A. L., Al-Katib W., Dales D. W., Postenka C. O., Casey G., Macdonald I. C., Chambers A. F. (2008) Int. J. Cancer 123,526–534 [DOI] [PubMed] [Google Scholar]
- 58.Urquidi V., Sloan D., Kawai K., Agarwal D., Woodman A. C., Tarin D., Goodison S. (2002) Clin. Cancer Res. 8,61–74 [PubMed] [Google Scholar]
- 59.Sharp J. A., Sung V., Slavin J., Thompson E. W., Henderson M. A. (1999) Lab. Invest. 79,869–877 [PubMed] [Google Scholar]
- 60.Ellison G., Klinowska T., Westwood R. F., Docter E., French T., Fox J. C. (2002) Mol. Pathol. 55,294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rae J. M., Ramus S. J., Waltham M., Armes J. E., Campbell I. G., Clarke R., Barndt R. J., Johnson M. D., Thompson E. W. (2004) Clin. Exp. Metastasis 21,543–552 [DOI] [PubMed] [Google Scholar]
- 62.Rae J. M., Creighton C. J., Meck J. M., Haddad B. R., Johnson M. D. (2007) Breast Cancer Res. Treat. 104,13–19 [DOI] [PubMed] [Google Scholar]
- 63.Lacroix M. (2009) Cancer Chemother. Pharmacol. 63,567. [DOI] [PubMed] [Google Scholar]
- 64.Sellappan S., Grijalva R., Zhou X., Yang W., Eli M. B., Mills G. B., Yu D. (2004) Cancer Res. 64,3479–3485 [DOI] [PubMed] [Google Scholar]
- 65.Brown L. F., Papadopoulos-Sergiou A., Berse B., Manseau E. J., Tognazzi K., Perruzzi C. A., Dvorak H. F., Senger D. R. (1994) Am. J. Pathol. 145,610–623 [PMC free article] [PubMed] [Google Scholar]
- 66.Hayashi C., Rittling S., Hayata T., Amagasa T., Denhardt D., Ezura Y., Nakashima K., Noda M. (2007) J. Cell. Biochem. 101,979–986 [DOI] [PubMed] [Google Scholar]
- 67.Wai P. Y., Guo L., Gao C., Mi Z., Guo H., Kuo P. C. (2006) Surgery 140,132–140 [DOI] [PubMed] [Google Scholar]
- 68.Pazolli E., Luo X., Brehm S., Carbery K., Chung J. J., Prior J. L., Doherty J., Demehri S., Salavaggione L., Piwnica-Worms D., Stewart S. A. (2009) Cancer Res. 69,1230–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamataki D., Ulloa F., Tsoni S. V., Mynett A., Briscoe J. (2005) Genes Dev. 19,626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.