Abstract
Enzymes of the AID/APOBEC family, characterized by the targeted deamination of cytosine to generate uracil within DNA, mediate numerous critical immune responses. One family member, activation-induced cytidine deaminase (AID), selectively introduces uracil into antibody variable and switch regions, promoting antibody diversity through somatic hypermutation or class switching. Other family members, including APOBEC3F and APOBEC3G, play an important role in retroviral defense by acting on viral reverse transcripts. These enzymes are distinguished from one another by targeting cytosine within different DNA sequence contexts; however, the reason for these differences is not known. Here, we report the identification of a recognition loop of 9–11 amino acids that contributes significantly to the distinct sequence motifs of individual family members. When this recognition loop is grafted from the donor APOBEC3F or 3G proteins into the acceptor scaffold of AID, the mutational signature of AID changes toward that of the donor proteins. These loop-graft mutants of AID provide useful tools for dissecting the biological impact of DNA sequence preferences upon generation of antibody diversity, and the results have implications for the evolution and specialization of the AID/APOBEC family.
The polynucleotide cytosine deaminases have been identified as key contributors to both the adaptive and innate immune responses to pathogens. This enzyme family includes activation-induced cytidine deaminase (AID),2 which initiates antibody diversification, and the APOBEC3 enzymes, which inhibit retroviral infection (1, 2). Although the enzymes share a common chemical mechanism for catalyzing the deamination of cytosine to generate mutagenic uracil within DNA, they have distinct biological functions based on differences in their expression, localization, and DNA sequence specificity (2, 3).
AID is a B-cell-specific enzyme whose catalytic action on mammalian antibody genes initiates somatic hypermutation and class switch recombination (4, 5). The uracil deamination product is processed by familiar DNA repair proteins, including uracil DNA N-glycosylase (UNG) and mismatch repair enzymes. However, the cellular environment within activated B-cells promotes mutagenic processing by these pathways, resulting in point mutations that ultimately enhance antibody affinity in somatic hypermutation or promote clustered double-strand breaks that alter antibody isotype in class switch recombination (6, 7).
The activity of AID is restricted to relatively small 3-kb regions within the immunoglobulin locus around rearranged variable genes and heavy chain switch regions. Within these regions, the targeting of AID is partially dictated by its DNA sequence preferences. For example, it has been noted that the mutable complementarity determining regions from variable gene segments have an abundance of AGC codons as compared with adjacent framework regions that function in stabilizing antibody structure (8). In vivo, somatic hypermutation is focused on WRCW (W = A/T, R = A/G) motifs in variable and switch regions (9–11). Biochemical assays with heterologously expressed AID have confirmed that this mutational footprint reflects the inherent sequence preferences of AID for deamination within WRC motifs (12–14).
In contrast to AID, members of the APOBEC3 subfamily have evolved to function in the innate immune response (15). In the case of HIV, a significant number of hypermutated viral sequences can be found in infected patients, with viral genomes showing a high frequency of G → A substitutions within two sequence motifs, GA and GG (16). The family members APOBEC3F and APOBEC3G preferentially target TC and CC motifs, respectively, within viral cDNA following reverse transcription, accounting for the observed mutational spectrum in the HIV genome (17–19). APOBEC3F and -3G both contain duplicated deaminase domains, with sequence preference and catalytic activity attributed to the C-terminal domain (20, 21). Controversy exists over the protective mechanisms used by APOBEC3 family members, as antiviral activity can be dissociated from deamination (22). However, sequence specificity of the APOBEC family is thought to be significant for antiviral effects, as almost all of the viral open reading frames in the hypermutated HIV genomes contain mutations that lead to premature termination (18).
Despite the fact that these distinguishing sequence preferences are important for activity, the enzymatic determinants of sequence targeting have not been elucidated. Here, we report that a protein loop in the AID/APOBEC family is a key contributor to the unique sequence preferences of individual family members. Grafting the loop from the antiviral deaminases APOBEC3F or -3G into the AID scaffold alters hot spot selectivity toward that of the two donor enzymes. These constructs reveal the modular nature of substrate recognition and provide biochemical tools to probe the importance of sequence specificity in the physiologic function of this important enzyme family.
EXPERIMENTAL PROCEDURES
Cloning of AID and Loop Variants
A synthetic gene encoding Escherichia coli codon-optimized AID was synthesized by GenScript and cloning oligonucleotides were obtained from Integrated DNA Technologies (supplemental Table S1). The AID construct was cloned downstream of an N-terminal maltose-binding protein (MBP) sequence. Expression plasmids for the loop variants and C-terminal truncations of AID were generated through a multistep PCR-based procedure (supplemental Fig. S1).
Protein Expression and Purification
Expression vectors were transformed into BL21(DE3)-Star E. coli (Novagen) in the presence or absence of a trigger factor expression plasmid (generously provided by LiChung Ma and Gaetano T. Montelione, Rutgers University). For protein production, 600-ml cultures were grown to an A600 of 0.6 at 37 °C. Cultures were shifted to 16 °C for 16 h after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The pelleted cells were resuspended in 50 mm Tris-Cl (pH 7.5), 150 mm NaCl, 10% glycerol (wash buffer) and lysed through a microfluidizer. The soluble fraction, filtered after high-speed centrifugation, was incubated with 3 ml of amylose resin (New England Biolabs) for 2 h at 4 °C. The resin was washed extensively prior to elution with wash buffer plus 10 mm maltose. For small scale analysis, 1 ml of culture was processed with BugBuster reagent (Novagen) into soluble and insoluble fractions.
Deamination Assay on Oligonucleotide Substrates
Unlabeled 60-mer oligonucleotide substrates (S60-XXC) contained a solitary C at position 24 and differed only at the −1 and −2 positions relative to the target cytosine (for full sequences see supplemental Table S1). Variants for XX include all 16 permutations of A, G, T, or 5-methyldeoxycytosine (mC). 20-μl assays were carried out in 20 mm Tris-Cl (pH 8.0), 30 mm NaCl, 1 mm dithiothreitol, 5 mm EDTA. Substrate (1 μm) was incubated with no enzyme or 0.5–3 μg of enzyme and 2.5 units of UNG (New England Biolabs) for 2–12 h at 30 °C, then heated to 95 °C for 20 min. Abasic sites were cleaved by addition of 10 mm magnesium acetate and incubation with human apurinic-apyrimidinic endonuclease 1 (APE), purified as previously described (23), for 2 h at 37 °C, along with RNase A (0.2 μg) to degrade contaminating RNA from the enzyme purification. Samples were run on a 20% acrylamide/Tris-borate-EDTA/urea gel. Gels were stained with SybrGold (Invitrogen) and imaged using UV transillumination. A standard titration curve with a uracil-containing product control (S60-AGU) was run on each gel and used to quantify product formation. Sequence preference profiles were generated by a two-step calculation. First, the product formation was averaged for all sequences that contain the same nucleotide at either the −1 or −2 positions. For example, P60-XAC (the average product formation for the S60-XAC substrates) was calculated by averaging product formation with S60-AAC, -mCAC, -GAC, and -TAC. Next, the probability of deamination for each nucleotide at the −1 or −2 positions was calculated relative to other nucleotide variants. For example, the percent preference for A at −1 was calculated as (P60-XAC/(P60-XAC + P60-XmCC + P60-XGC + P60-XTC)) × 100. The in vivo sequence preference profiles for AID are taken from sequencing data on the ung−/−, msh2−/− mice as previously reported and interpreted (12, 24). The mutational sequence preference profile for APOBEC3F and APOBEC3G are derived from the previously reported mutagenesis patterns on the retrovirus Moloney leukemia virus, which packages APOBEC3 molecules in an analogous manner to HIV (20).
Sequence Preferences Determined by Rifampin Mutagenesis Assay
A synthetic gene for uracil DNA-glycosylase inhibitor was obtained from GenScript and cloned into the SphI/AvrII-digested plasmid pETcoco-2 (Novagen). The plasmid was transformed into BL21(DE3)-pLysS (Novagen) cells followed by pET41 control plasmid or enzyme expression plasmids. A saturated culture derived from a single colony was diluted to A600 of 0.15. After growth for 1 h at 37 °C, the log-phase culture was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. After 4 h, an aliquot was plated on LB-agar containing rifampin (100 μg/ml, Sigma) and LB with selection antibiotics to determine total cell density. The mutagenesis frequencies were calculated by the ratio of rifampin-resistant colonies divided by the total cell population. Single colonies were subjected to colony PCR, and rpoB mutations that conferred rifampin resistance were determined by well established protocols (25).
RESULTS
Identification of a Potential DNA Sequence Recognition Loop
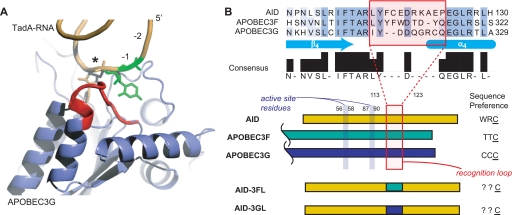
Although recent structures of the C-terminal catalytic domain of APOBEC3G have increased our understanding of the overall enzymatic-fold, there are currently no structures of a polynucleotide cytosine deaminase bound to nucleic acid to elucidate how hot spot sequences are recognized (26, 27). To overcome this limitation, we examined the more distantly related structure of RNA-bound TadA, an adenosine deaminase that generates the universal base inosine at the wobble position of tRNAArg (28). Alignment of the unliganded APOBEC3G structure with TadA suggested that a loop between the β4 strand and the α4 helix is poised to interact with the sequence 5′ to the deamination target (Fig. 1A) (2, 27). Although flanked on either side by highly conserved amino acid residues, this loop itself is poorly conserved between family members, supporting a potential role in specific substrate recognition (Fig. 1B). In prior studies, site-directed mutations have also suggested subtle shifts in specificity that point to this loop. For example, the D311Y mutation of APOBEC3F modestly increased deamination of normally disfavored sequences with A at the −1 position (20). Similarly, the D316R/D317R double mutant of APOBEC3G increased deamination of the off-target cytosines at the −1 or −2 positions in the preferred CCC motif of APOBEC3G (27).
FIGURE 1.
A protein loop in the AID/APOBEC family is poised for potential interactions with target DNA. A, a role for the hot spot recognition loop. Unliganded APOBEC3G (Protein Data Bank code 3E1U) and the RNA-bound TadA (Protein Data Bank code 2B3J) structures were aligned by the Dali server (43). The structure shown is APOBEC3G with the aligned TadA RNA substrate. The structure suggests a role for a loop (red) in recognition of the nucleotides (−1 position in green) upstream of the deamination target (*). B, alignment of AID/APOBEC family members. An area downstream from the catalytic residues is shown in partial sequence alignment. The structural loop noted in A is poorly conserved (red box) and flanked by highly conserved sequences. AID and the C-terminal catalytic domains of APOBEC3F and -3G are shown schematically with their distinct sequence preferences noted. The active site residues are upstream from this loop and AID residue numbering is noted. The loop variants were constructed by replacing the highlighted loop from AID with that from APOBEC3F (AID-3FL) or APOBEC3G (AID-3GL) and their sequence preferences investigated in this work.
To examine the role of this loop in substrate specificity, we aimed to construct AID mutants where the peptide sequence from Leu113 to Pro123 was directly replaced with the corresponding loops from the catalytic domains of APOBEC3F or APOBEC3G (Fig. 1B). These enzymes were selected based on their distinct sequence preferences relative to AID that allow for evaluation of the proposal that loop graft mutants of AID may alter sequence preferences in a predictable manner (19–21, 29, 30).
High Level Expression System for AID
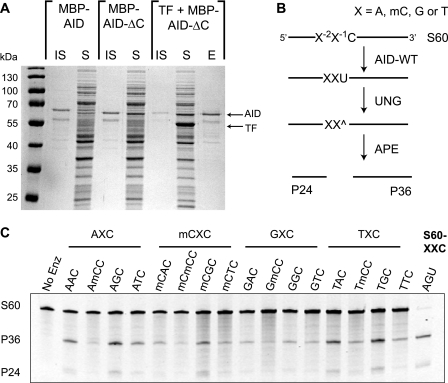
Biochemical characterization of AID has been hampered by difficulties in obtaining large amounts of soluble enzyme from either native B-cell sources or heterologous expression systems (6). To facilitate evaluation of AID and loop graft mutants, we first aimed to develop a facile route to large amounts of soluble, active protein from bacterial cultures. Through screening of multiple fusion constructs, we found that an N-terminal fusion of MBP to AID yielded some encouraging expression of soluble proteins (Fig. 2A). Then, based on the hypothesis that a potentially poorly structured C-terminal region of AID may contribute to its insolubility, we deleted residues 182–198. C-terminal truncation was expected to be non-perturbing based on prior observations that truncation resulted in higher in vivo mutagenesis rates, as well as the lack of conservation of this sequence within the larger APOBEC family (31). Indeed we found that soluble expression increased with MBP-AID-ΔC (hereafter AID-WT) and that the catalytic activity was enhanced about 3-fold over the full-length protein (supplemental Fig. S2). Finally, we found that co-expression of the ribosome-associating chaperone trigger factor produced soluble protein in high yield (∼10 mg/liter) upon purification by amylose resin affinity chromatography (Fig. 2A).
FIGURE 2.
AID-WT targets deamination to WRC hot spot sequences. A, expression of soluble AID-WT. 1-ml cultures from full-length MBP-AID, the C-terminal truncation (MBP-AID-ΔC or AID-WT), and co-expression with the chaperone trigger factor (TF) were separated into soluble (S) and insoluble (IS) fractions demonstrating progressively increasing soluble expression. The elution from the amylose affinity resin is shown (E). B, oligonucleotide deamination assay. An array of substrates (S60-XXC) with a single substrate cytosine in a 60-mer single-strand DNA are incubated with AID-WT and UNG. The substrates include variations where the −1 (X−1) or −2 positions (X−2) are A, G, or T or mC. The resulting abasic sites (^) are cleaved with the endonuclease APE, giving P24 and P36 products. C, AID-WT prefers WRC hot spot sequences. 1 μm substrate was incubated with 1 μg of AID-WT (or no enzyme control) for 3 h. The P24/P36 products generated by UNG and APE treatments were detected on a denaturing gel by staining with SybrGold. S60-AGU serves as a product control for the activity of UNG and APE and as a size standard.
AID-WT Prefers to Deaminate WRC Hot Spot Sequences
To examine the sequence preferences of the new AID-WT construct, we assayed an array of substrates with a single cytosine and variations at the −1 and −2 positions immediately 5′ to the target cytosine (Fig. 2B). The assay is based on the notion that AID-catalyzed deamination generates uracil, which can be excised by UNG. The abasic product is then nicked by APE to cleave the DNA phosphate backbone and generate two product fragments of different lengths. The array includes substrates with an XXC sequence where X = A, G, or T or mC. mC serves as a surrogate for cytosine recognition at these positions, as competing cytosines would be potential sites for deamination, thereby complicating the analysis. In contrast, it has been previously shown that mC is a poor substrate for AID, and if a small amount of deamination were to occur at the mC sites, the product (T) would not be a substrate for UNG (32). The assay therefore cleanly reports on deamination of the single cytosine in the S60-XXC oligonucleotide substrates.
To quantify the sequence preferences of AID-WT, we carried out multiple replicates of the assay under fixed enzyme and substrate conditions where product formation was approximately ≤35% for all substrates. As expected, incubation of AID-WT with the substrate array resulted in preferential deamination of substrates containing A or T at the −2 position, with selection against mC and G (Fig. 2C). At the −1 position, a slight preference of G over A, discrimination against T, and strong selection against mC, was observed. Thus, in vitro sequence specificity of the truncated AID protein with an MBP tag is similar to that previously reported for full-length AID constructs (14, 33).
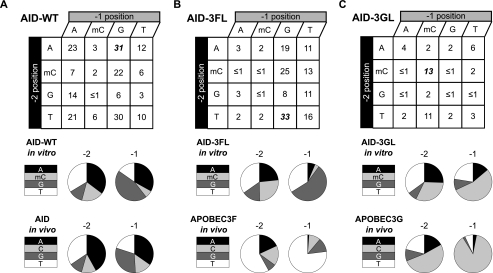
The in vitro sequence preferences of AID-WT also recapitulate the observed hypermutation sequence preferences derived from in vivo data. In vivo sequence preferences of AID were previously obtained through genetic studies by knocking out both the base excision repair enzyme UNG and the mismatch repair enzyme MSH2 with the aim of preventing AID-generated uracil from being processed by downstream repair pathways. Accordingly, the mutational spectrum of the immunoglobulin variable regions of B-cells from ung−/−, msh2−/− mice reflects the in vivo sequence preferences for AID-catalyzed deamination (Fig. 3A) (12, 24). By measuring deamination efficiency of AID-WT with individual members of our substrate array, we constructed an analogous in vitro sequence preference activity profile for AID-WT and compared it with the in vivo profile (Fig. 3A). The two profiles are very similar, with clear preservation of the WRC sequence preference. Of interest, mC appears to be an excellent surrogate for C at the −2 position and a good surrogate at the −1 position, as judged by similar activity profiles derived from in vivo experiments with C and the in vitro system that used mC.
FIGURE 3.
Grafting of a loop from APOBEC enzymes onto AID results in predictable shifts in sequence preference profiles. A, product formation with AID-WT correlates with in vivo sequence preferences. B, loop grafting in AID-3FL, and C, AID-3GL alter sequence preferences. For each condition, 1 μm S60-XXC substrate was subjected to 1 μg of enzyme for 3 h under assay conditions. The products from 3 to 5 replicates of each condition were quantified and the average values for % product formation are shown for AID-WT, AID-3FL, and AID-3GL. The best substrates for each construct are shown in bold italics. The detection limit of the assay is ≤1%. For substrates with 2–5% conversion to product, standard deviation was 1–2%; for 6–10% conversion, standard deviation was 2–3%; for 11–20% conversion, standard deviation was 5–10%; for 21–33% conversion, standard deviation was 6–11%. The data were used to construct the sequence preference profile for AID-WT and the loop graft variants. The in vivo deamination sequence preferences are shown for comparison. These values are derived from previously reported sequences of variable regions of B-cells mutated in ung−/−, msh2−/− mice for AID (12, 24) or from the reported sequencing of hypermutated retroviral genomes for APOBEC3F and APOBEC3G (20).
Construction and Expression of Loop Graft Mutants
Cloning of the loop variants was achieved by overlap extension PCR, leaving N-terminal regions upstream from Leu113 and C-terminal region downstream of Pro123 unchanged from AID-WT and introducing the shorter 10-amino acid loop from APOBEC3F (Leu306 to Gln315) or 9-amino acid loop from APOBEC3G (Ile314 to Gln322) (Fig. 1B). The AID-APOBEC3F loop variant (AID-3FL) and the AID-APOBEC3G loop variant (AID-3GL) were overexpressed as MBP fusion proteins in high yield using the chaperone co-expression system developed for AID-WT. Despite the alteration of a large protein loop, upon overexpression, the proteins were unchanged in their distribution between soluble and insoluble fractions and were obtained in nearly equivalent purity and yield (supplemental Fig. S3).
Loop Grafting Confers APOBEC3F and APOBEC3G Preferences to AID
To examine sequence preferences, we assayed the enzymes for deamination of the S60-XXC substrate array. Qualitative differences between the loop variants are easily observed under high turnover conditions (supplemental Fig. S4). To allow for quantitative comparisons using the substrate array, each of the three enzymes were assayed using a fixed concentration of substrate (1 μm) and enzyme (1 μg) under conditions where product formation was ≤35% to approximate the initial rate conditions (Fig. 3). The reported values in Fig. 3 are averages from three to five replicate measurements.
Loop grafting in AID-3FL and AID-3GL did not significantly compromise enzymatic activity. This conclusion is supported by the observation that AID-WT activity against its best substrate (S60-AGC) is equivalent to the activity of AID-3FL against its best substrate (S60-TGC). This indicates that the specificity and not the overall activity of AID-3FL has been altered relative to AID-WT. Similarly, AID-3GL is 6.5-fold more active with its preferred substrate (S60-mCmCC) as compared with AID-WT with the same substrate, even though the maximal activity of AID-3GL is about one-third of AID-WT with its preferred substrate. Thus, loop grafting in AID-3GL results in a more selective, albeit slightly less active enzyme. Notably, the sequence preference profiles are unlikely to be affected by modest differences in activity, as these comparisons involve the same enzyme acting on different substrates in the S60-XXC array.
As compared with AID-WT, significant changes in sequence preferences for deamination were introduced by the AID-3FL and AID-3GL loop swaps, and these preferences mimic those inferred from hypermutated retroviral genomes (Fig. 3, B and C) (20). For AID-3FL, S60-TGC was the best substrate, accurately reflecting the preference of APOBEC3F for T at the −2 position. The shifts to APOBEC3F-like preferences were also extended to the −1 position. For instance, AID-3FL shows a >2-fold increase in its preference for a −1 T and a >5-fold selection against −1 A relative to AID-WT, which mimics the preferences of APOBEC3F that have been surmised from analyses of hypermutated retroviral genomes (19–21).
The AID-3GL loop swap negated the classic AID substrate preference, in agreement with poor recognition of WRC sequences by APOBEC3G (Fig. 3C) (20, 29, 30). As compared with AID-WT, the overall preference for a −1 mC increased more than 9-fold with AID-3GL, in line with the preference of APOBEC3G. At the −2 position, where selectivity is less stringent with the entire AID/APOBEC family members, a modest ∼1.5-fold increase in deamination of mC containing substrates was observed with AID-3GL relative to AID-WT. Thus, the AID-3GL loop swap recapitulates the two main sequences targeting attributes of APOBEC3G: its preference for the CCC sequence and its discrimination against the WRC sequence preferred by AID-WT.
Loop Grafting Alters Mutational Hot Spot Targeting in E. coli
To further explore the changes in sequence preferences, we examined the mutagenesis patterns using a rifampin-based mutagenesis assay in E. coli (25). Rifampin resistance is conferred by a limited number of C/G → T/A transition mutations in a small portion of the RNA polymerase gene, rpoB. These mutations occur in various trinucleotide sequence contexts. Prior studies have established that each deaminase family member has a distinctive mutational spectrum based on their individual hot spot preferences, which provides a second system to validate the observed shifts in sequence preference (20, 21, 25, 30). The assay takes advantage of the potent protein, uracil DNA-glycosylase inhibitor, to prevent repair of uracil lesions. C/G → T/A transition mutations therefore persist and can be located by sequencing individual rifampin-resistant colonies.
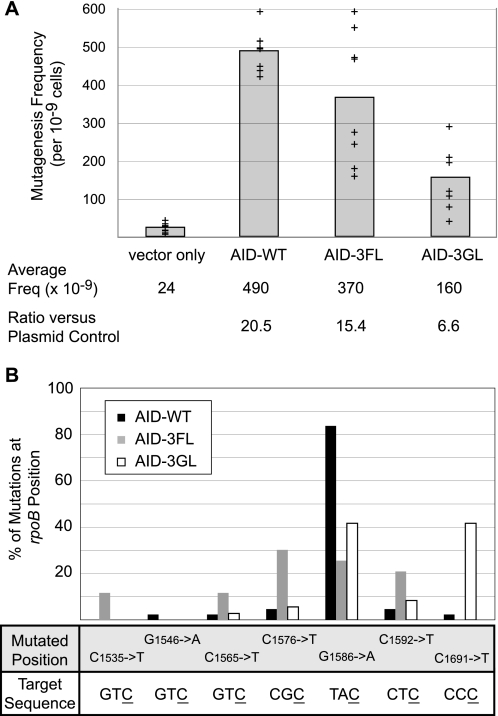
Expression of AID-WT, AID-3FL, or AID-3GL results in a 6–20-fold increase in the frequency of mutations resulting in rifampin resistance in E. coli (Fig. 4A). Sequencing of rpoB from rifampin-resistant clones established that the in vitro sequence preferences extended to the intracellular environment, where potentially competitive DNA binding sequences could have attenuated the expected results. For all resistant clones, only a single mutation was observed within the sequenced portion of rpoB, and all were known mutations that confer rifampin resistance (25). The mutations on the coding strand were exclusively C → T or G → A, and therefore attributable to replication over unrepaired uracil introduced by the deaminase on either the coding or non-coding strands. For AID-WT, 84% of rifampin mutations were G1586 to A transitions, which can result from a non-coding strand C → T mutation within an ATC trinucleotide sequence (Fig. 4B). Notably, this is the only locus within the rpoB gene where a C → T mutation at a canonical WRC hot spot has been shown to result in rifampin resistance in wild-type E. coli. For AID-3FL, a new mutational spectrum was seen, with many of the new deamination sites containing T at the −1 position, which is the same preference observed in the in vivo studies (Fig. 4B). Remarkably, the most preferred site for mutation by the loop variant AID-3FL was C1576 (30% frequency), which has been previously reported as the preferred site of deamination by APOBEC3F with identical frequency (19). For AID-3GL, more than 40% of the mutations occurred at CCC locus C1691, which has been previously established as the preferred site of deamination for APOBEC3G (30, 34). This preference contrasts significantly with AID-WT, where <3% of mutations were observed at this site. Thus, the predicted shift in preferences for both loop-grafted AID variants extend to an in vivo environment.
FIGURE 4.
Loop graft variants alter the mutational targeting conferring resistance to rifampin in E. coli. A, induction of a mutator phenotype in E. coli. Expression of AID or loop variants along with the uracil DNA glycosylase inhibitor leads to an increase in rifampin resistance. The mutagenesis frequency was calculated based on the number of rifampin-resistant colonies per total cells. Shown are the data for eight individual experiments (+) for each expression plasmid. The average mutagenesis frequency is shown with the gray bar graph. The ratio of mutagenesis frequency with AID-WT, AID-3FL, or AID-3GL over the control pET41 plasmid are listed below the plot. B, loop variants have an altered mutational spectrum. The rpoB gene from individual colonies was sequenced to determine the mutation spectrum generated by each enzyme. The total number of sequences obtained were: AID-WT (43), AID-3FL (43), and AID-3GL (36). The mutated position of the rpoB gene is shown, along with the surrounding trinucleotide sequence targeted for deamination with the deaminated cytosine underlined. The plot reports on the frequency of mutation at all detected positions demonstrating the alteration in mutational targeting by AID-3FL and AID-3GL.
DISCUSSION
This work identifies a recognition loop that significantly accounts for the different substrate sequence preferences in the AID/APOBEC family. These studies were facilitated by an improved protein expression system that yielded large amounts of soluble AID, expressed as N-terminal MBP fusion and C-terminal truncation. Importantly, this construct exhibits improved activity relative to the full-length protein and the same hot spot preferences seen in B-cells or with heterologously expressed full-length enzyme (12–14, 35).
The hot spot recognition sequence of AID, and its degree of specificity for this sequence, would be expected to reflect an evolutionary balance between multiple potentially competing requirements. The requirement to generate a broad range of amino acid changes by somatic hypermutation would select for a certain degree of promiscuity in the selectivity of AID. In this regard, deaminated products can be detected for all S60-XXC substrate sequences, which would tend to enhance diversification of target sequences. Class switch recombination has different requirements, where clustering of specific AID target motifs could result in recombinogenic double-strand breaks. High activity of AID on WGC sequences and the prevalence of WGCW in switch regions are also consistent with this requirement.
A significant unanswered question in the understanding of the generation of antibody diversity by AID is the mechanism by which the enzyme is targeted. Targeting occurs at multiple levels. Globally, AID must target the immunoglobulin locus, because aberrant global targeting could result in oncogenic lesions (36, 37). Upon finding the immunoglobulin locus, AID must then be preferentially localized to the variable regions to promote somatic hypermutation and to the heavy chain switch regions to yield class switch recombination. Within these regions, AID further selects its hot spot WRCW sequences, preferentially promoting productive mutations within the antigen complementarity determining regions over structurally essential framework regions.
In exploring these layers of targeting, our work serves as a basis for understanding how AID can achieve sequence level targeting and demonstrates the utility of loop swapping in manipulating sequence preferences. Examination of related enzyme structures led us to postulate that a loop region with poor sequence conservation in the AID/APOBEC family might play a role in DNA sequence recognition. The loop is enriched for aromatic, acidic, and basic side chains, which make it a potential source of base stacking and hydrogen bonding interactions that could contribute to sequence specificity. Flexible loops often play useful roles in the recognition of macromolecules (38). Indeed, antibody molecules themselves exploit the flexibility of protein loops to make diverse antigen recognition pockets (39). As compared with random point mutagenesis, loop grafting has several advantages. Grafting potentially preserves subtle geometric constraints important for binding, accommodates differences in loop sizes, and allows for the directed evaluation of a specific hypothesis, namely that a large part of the individual specificities of these enzymes are encoded in their unique loop architectures.
The discovery that a recognition loop can be grafted from APOBEC family members to AID and confer altered specificity has implications for the evolution of this enzyme family (40). The APOBEC enzymes have undergone rapid gene expansion in higher mammals and exhibit one of the strongest positive selection signals in the human genome, which suggests an essential and growing role for these enzymes in pathogen defense (15, 41). The apparently modular nature of the APOBEC family, with segregation of hot spot recognition from the deaminase catalytic core, would offer an elegant strategy for rapid evolution of sequence specificities that are tailored to their individual niches in the immune response to foreign antigens and retroviruses. To further evaluate the generality of loop swaps in the AID/APOBEC family, it will be interesting to see if reciprocal loop swaps, introducing foreign loops into an APOBEC3 scaffold, will alter deamination specificity. This could potentially be pursued by in vivo analysis of loop graft variants acting on retroviral genomes given that in vitro characterization in the AID/APOBEC family often presents potentially significant biochemical challenges (6).
These loop variants provide potentially useful biochemical tools for addressing the importance of sequence specificity in the individual cellular functions of AID/APOBEC family members. The growing enzyme family has clear evidence of specialization. For instance, APOBEC3G and APOBEC3F are able to counteract HIV, whereas many of the other APOBEC enzymes cannot, and APOBEC1 cannot compensate for AID deficiency in B-cells (3, 42). It is unclear if these restricted activities are related to enzymatic sequence preferences. These catalytically competent variants with altered specificities provide valuable tools to now explore the contribution of sequence specificity to antibody diversity, improper gene targeting, and antiviral defense.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM056834-13 (to J. T. S.) and by the Intramural Research Program of the NIA, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- AID
- activation-induced cytidine deaminase
- UNG
- uracil DNA N-glycosylase
- MBP
- maltose-binding protein
- mC
- 5-methyldeoxycytosine
- APE
- human apurinic-apyrimidinic endonuclease 1
- AID-WT
- activation-induced cytidine deaminase wild type
- AID-3FL
- activation-induced cytidine deaminase-APOBEC3F loop variant
- AID-3GL
- activation-induced cytidine deaminase-APOBEC3G loop variant
- HIV
- human immunodeficiency virus.
REFERENCES
- 1.Muramatsu M., Nagaoka H., Shinkura R., Begum N. A., Honjo T. (2007) Adv. Immunol. 94,1–36 [DOI] [PubMed] [Google Scholar]
- 2.Conticello S. G., Langlois M. A., Yang Z., Neuberger M. S. (2007) Adv. Immunol. 94,37–73 [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg B. R., Papavasiliou F. N. (2007) Adv. Immunol. 94,215–244 [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M., Sankaranand V. S., Anant S., Sugai M., Kinoshita K., Davidson N. O., Honjo T. (1999) J. Biol. Chem. 274,18470–18476 [DOI] [PubMed] [Google Scholar]
- 5.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Cell 102,565–575 [DOI] [PubMed] [Google Scholar]
- 6.Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) Annu. Rev. Immunol. 26,481–511 [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R. T., Datta A., Manis J., Alt F. W. (2007) Adv. Immunol. 94,157–214 [DOI] [PubMed] [Google Scholar]
- 8.Wagner S. D., Milstein C., Neuberger M. S. (1995) Nature 376,732. [DOI] [PubMed] [Google Scholar]
- 9.Rada C., Ehrenstein M. R., Neuberger M. S., Milstein C. (1998) Immunity 9,135–141 [DOI] [PubMed] [Google Scholar]
- 10.Ehrenstein M. R., Neuberger M. S. (1999) EMBO J. 18,3484–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martomo S. A., Yang W. W., Gearhart P. J. (2004) J. Exp. Med. 200,61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larijani M., Frieder D., Basit W., Martin A. (2005) Immunogenetics 56,840–845 [DOI] [PubMed] [Google Scholar]
- 13.Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) J. Biol. Chem. 279,51612–51621 [DOI] [PubMed] [Google Scholar]
- 14.Yu K., Huang F. T., Lieber M. R. (2004) J. Biol. Chem. 279,6496–6500 [DOI] [PubMed] [Google Scholar]
- 15.Sawyer S. L., Emerman M., Malik H. S. (2004) PLoS Biol. 2,E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vartanian J. P., Meyerhans A., Sala M., Wain-Hobson S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91,3092–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418,646–650 [DOI] [PubMed] [Google Scholar]
- 18.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11,435–442 [DOI] [PubMed] [Google Scholar]
- 19.Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. (2004) Curr. Biol. 14,1385–1391 [DOI] [PubMed] [Google Scholar]
- 20.Langlois M. A., Beale R. C., Conticello S. G., Neuberger M. S. (2005) Nucleic Acids Res. 33,1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haché G., Liddament M. T., Harris R. S. (2005) J. Biol. Chem. 280,10920–10924 [DOI] [PubMed] [Google Scholar]
- 22.Newman E. N., Holmes R. K., Craig H. M., Klein K. C., Lingappa J. R., Malim M. H., Sheehy A. M. (2005) Curr. Biol. 15,166–170 [DOI] [PubMed] [Google Scholar]
- 23.Erzberger J. P., Barsky D., Schärer O. D., Colvin M. E., Wilson D. M., 3rd (1998) Nucleic Acids Res. 26,2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada C., Di Noia J. M., Neuberger M. S. (2004) Mol. Cell 16,163–171 [DOI] [PubMed] [Google Scholar]
- 25.Garibyan L., Huang T., Kim M., Wolff E., Nguyen A., Nguyen T., Diep A., Hu K., Iverson A., Yang H., Miller J. H. (2003) DNA Repair 2,593–608 [DOI] [PubMed] [Google Scholar]
- 26.Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452,116–119 [DOI] [PubMed] [Google Scholar]
- 27.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456,121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losey H. C., Ruthenburg A. J., Verdine G. L. (2006) Nat. Struct. Mol. Biol. 13,153–159 [DOI] [PubMed] [Google Scholar]
- 29.Beale R. C., Petersen-Mahrt S. K., Watt I. N., Harris R. S., Rada C., Neuberger M. S. (2004) J. Mol. Biol. 337,585–596 [DOI] [PubMed] [Google Scholar]
- 30.Harris R. S., Petersen-Mahrt S. K., Neuberger M. S. (2002) Mol. Cell 10,1247–1253 [DOI] [PubMed] [Google Scholar]
- 31.Barreto V., Reina-San-Martin B., Ramiro A. R., McBride K. M., Nussenzweig M. C. (2003) Mol. Cell 12,501–508 [DOI] [PubMed] [Google Scholar]
- 32.Larijani M., Frieder D., Sonbuchner T. M., Bransteitter R., Goodman M. F., Bouhassira E. E., Scharff M. D., Martin A. (2005) Mol. Immunol. 42,599–604 [DOI] [PubMed] [Google Scholar]
- 33.Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003) Nature 424,103–107 [DOI] [PubMed] [Google Scholar]
- 34.Jónsson S. R., Haché G., Stenglein M. D., Fahrenkrug S. C., Andrésdóttir V., Harris R. S. (2006) Nucleic Acids Res. 34,5683–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue K., Rada C., Neuberger M. S. (2006) J. Exp. Med. 203,2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M., Duke J. L., Richter D. J., Vinuesa C. G., Goodnow C. C., Kleinstein S. H., Schatz D. G. (2008) Nature 451,841–845 [DOI] [PubMed] [Google Scholar]
- 37.Okazaki I. M., Kotani A., Honjo T. (2007) Adv. Immunol. 94,245–273 [DOI] [PubMed] [Google Scholar]
- 38.Tawfik D. S. (2006) Science 311,475–476 [DOI] [PubMed] [Google Scholar]
- 39.Jones P. T., Dear P. H., Foote J., Neuberger M. S., Winter G. (1986) Nature 321,522–525 [DOI] [PubMed] [Google Scholar]
- 40.Khersonsky O., Roodveldt C., Tawfik D. S. (2006) Curr. Opin. Chem. Biol. 10,498–508 [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Webb D. M. (2004) Hum. Mol. Genet. 13,1785–1791 [DOI] [PubMed] [Google Scholar]
- 42.Fugmann S. D., Rush J. S., Schatz D. G. (2004) Eur. J. Immunol. 34,844–849 [DOI] [PubMed] [Google Scholar]
- 43.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24,2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.