Abstract
The degradation of a selected mRNA species by RNA interference requires a high degree of homology between the short interfering or short hairpin RNA (si or shRNA) and its target. Recent reports have demonstrated that the number and location of nucleotide mismatches affect the activity of si/shRNA. Here, we systematically examined the effect of single nucleotide mutations in all 21 positions of an effective shRNA that targets the gag gene of HIV-1. We found that all mutant shRNAs exerted RNAi activity but were less effective in gene silencing compared to the wild-type gag shRNA. The most pronounced reduction in function was observed with mutations in the central and 5′ regions of the shRNA. Our results demonstrate that optimal gene silencing requires perfect homology between shRNA and the chosen target, but that a variable degree of silencing occurs, depending upon the precise location of nucleotide mismatches.
INTRODUCTION
RNA interference (RNAi) refers to the ability of double-stranded RNA (dsRNA) to induce sequence-specific degradation of homologous mRNA (1). Originally identified in Caenorhabditis elegans, RNAi in mammalian cells can be triggered by the cellular introduction of small interfering RNAs (siRNA) by transfection or electroporation of synthetic short (21–23 nt) RNA duplexes (2–4). Given the limited application of these techniques, a considerable advance in the field is the design of si/shRNA expression cassettes that can be introduced into cells by DNA transfection or retroviral and adenoviral gene transfer (5–11). The resulting ability to stably express si/shRNA in human cells has been utilized to target disease-specific mRNAs, such as those encoding oncogenes or viral proteins (12–15). Thus, RNAi holds the potential of constituting a new form of gene therapy.
Although a critical feature of RNAi has been the presumed sequence specificity of si/shRNA, the degree of homology between a particular siRNA and its target required for optimal RNAi is unclear. Recent reports that have addressed this issue have yielded conflicting results. Using Drosophila melanogaster embryo lysates, Elbashir et al. (16) demonstrated that a single nucleotide mismatch 10 or 11 nt downstream of the target position complementary to the extreme 3′ nucleotide of the 21 nt complementary guide siRNA sequence abolished gene silencing activity. Zeng and Cullen, applying a microRNA backbone to express siRNA targeting luciferase, found that gene silencing was compromised but not eliminated by single mismatches in positions 10, 11 and 12 (17). In contrast, Boutla et al. found that siRNA targeting Notch and hedgehog genes in Drosophila tolerated mismatches even when located in central regions of siRNA (18). Location-specific effects were observed by Amarzguioui et al. who found that gene silencing was severely compromised when mutations were located in the 3′ half of the sense strand but not when located in the 5′ half of a siRNA targeting blood clotting initiator Tissue Factor (19,20). Jackson et al. found that regions of sequence similarity were most important in the center and 3′ end of the siRNA sense strand and that as few as 11–15 continuous homologous nucleotides were sufficient for gene silencing (21). While these investigations have employed synthetic siRNA, little data are available on position-specific effects of nucleotide mismatches in shRNA.
Given that one possible clinical application of RNAi may be in the treatment of chronic viral infections such as HIV-1, a critical issue is the degree of sequence specificity of shRNA required for optimum gene silencing. The nucleotide sequence heterogeneity of HIV-1 among different patients and within the same host is likely to present a major challenge to using RNAi as a therapeutic if gene silencing requires perfect homology between shRNA and targeted regions of the viral genome. To better characterize the effect of sequence variation on the effect of HIV-1-specific RNAi, we first identified a potent shRNA that targeted a conserved region of the gag protein of HIV-1. We then introduced point mutations in all 21 positions of the shRNA. Compared to wild-type gag shRNA, we found reduced antiviral activity in all mutants, especially those containing mutations in the central positions 9–11 of the molecule.
MATERIALS AND METHODS
Construction of wild-type and mutant gag shRNA expression vector
The human U6 promoter was PCR-amplified from genomic DNA isolated from HeLa cells using primers 5′-AAG GTC GGG CAG GAA GA-3′ and 5′-CGG TGT TTC GTC CTT TCC AC-3′. A second round PCR including primers, 5′-GTG ACA GCG GCC GCA AGG TCG GGC AGG AAG A-3′ and 5′-CAA CAG GCG GCC GCA TCG ATG TTA GGA GAT CTA AAA ACT CGA GAT TTC ATC TAG AGG TGT TTC GTC CTT TCC-3′ was performed on the first round PCR product to introduce appropriate restriction sites for further cloning.
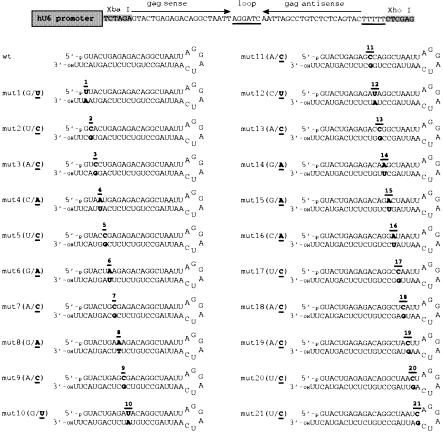
The PCR product was gel-purified and introduced into the NotI sites of the DNA vector pCMV-MCS (Stratagene, Valencia, CA) resulting in vector pCMV-U6-MCS. A neomycin resistance cassette, including the SV40 early promoter and the SV40 polyadenylation signal, was inserted into the BglII and ClaI sites of pCMV-U6-MCS. The U6 driven shRNA expression cassette was subcloned into the NotI sites of pAAV-MCS (Stratagene). Individual oligonucleotides (Qiagen, Valencia, CA) targeting HIV-1NL4.3 gag (position 2066–2086) were annealed and introduced into the XbaI and XhoI sites of pAAV-U6-MCS. The resulting shRNA expression sequence consists of 21-nt gag sense and antisense sequences separated by an HIV-1 sequence-unrelated hexaloop (AGGACT). A five-thymidine pol III termination signal was included at the end of the gag-specific hairpin sequence. HIV-1 gag point mutations were introduced from position 1 to position 21 of the target sequence resulting in a total of 21 different single-mutant and one wild-type shRNAs (Fig. 1). The base pairs G-C and C-G were systematically mutated to base pairs T-A and A-T. In addition, a control vector was generated, which expresses shRNA targeting the luciferase gene.
Figure 1.
Schematic of mutated and wild-type versions of gag shRNA. Each nucleotide position in the shRNA was mutated as indicated in parentheses. The mutants are numbered according to the position of the mutation with respect to the 5′-end of the sense strand.
Cell culture and transient transfections
The human embryonal kidney cell line 293 T (ATCC) was cultured in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Twenty-four hours prior to transfection, 293T cells were trypsinized and plated at 4 × 105 cells per well in 6-well plates in DMEM plus 10% FBS. Cells were co-transfected with 1 µg HIV-1NL4.3 DNA and 1 µg of the various pAAV-U6-shRNA DNA vectors expressing wild-type or mutant gag shRNAs and luc shRNA using 6 µl Lipofectamine 2000 (Gibco-Invitrogen, San Diego, CA) per reaction. Cell-free supernatant was collected 2 days later and HIV-1 p24 antigen levels were quantified by ELISA (Beckman-Coulter, Fullerton, CA) according to the manufacturer’s instructions.
Northern blot
Total RNA was isolated from transfected 293 T cells from a subset of samples using Trizol Reagent (Gibco-Invitrogen) according to the manufacturer’s instructions. Ten micrograms of total cellular RNA were denatured in glyoxal sample loading dye (Ambion, Austin, TX), separated on a 1.2% agarose gel and transferred using the Turboblotter system (Schleicher & Schuell, Dassel, Germany). Ribosomal RNA was used as a loading control. RNA was blotted on a Zeta-Probe GT membrane (Bio-Rad, Hercules, CA) and immobilized by UV crosslinking. Hybridization was carried out overnight at 65°C using Ultrahyb Hybridization Buffer (Ambion, Austin, TX). Full-length HIV-1 RNA was detected with a random-primed 32P-labeled 480-nt gag DNA probe. Membranes were washed at 60°C twice in 2× SSC, 0.1% SDS and 0.2× SSC, 0.1% SDS.
Real-time RT–PCR for intracellular HIV-1 RNA
Extracted RNA was treated with RQ1 DNase I (Promega, Madison, WI) according to the manufacturer’s protocol. One microgram of DNase I-treated RNA was added to a reverse transcription (RT) reaction containing Powerscript Reverse Transcriptase (Clontech, Palo Alto, CA), 1 mM each of dNTP, 1× First-Strand buffer (Clontech), 200 ng random hexamers (Promega) and 10 U RNasin (Promega). Reverse transcription was performed at 42°C for 1 h followed by heat-inactivation of the RT enzyme at 70°C for 15 min. PCR was performed with 2 µl 1/10 diluted cDNA in a final volume of 30 µl and contained 1× Titanium Taq PCR buffer (Clontech), 20 pmol of sense primer Gag-A, 5′-GGA CCA AAG GAA CCC TTT AGA GA-3′, and antisense primer Gag-B, 5′-GGA CCA ACA AGG TTT CTG TCA TC-3′, 1 mM dNTPs, SYBR Green I (1:75000), 10 nM fluorescein, and 1× Titanium Taq polymerase (Clontech). To normalize the samples for absolute RNA amount, a GAPDH–PCR was performed with primers G1, 5′-GAT TTC TCC CCC TTC TGC TGA TG-3′ and G2, 5′-CCT TGG CTG GGG GTG CTA A-3′. An external standard curve was created using a 466-nt gag RNA in vitro transcript generated with the MEGAscript in vitro transcription kit (Ambion). The template for the in vitro transcription was generated by PCR using primers T7-Gag, 5′-TAA TAC GAC TCA CTA TAG GAT GGA TGA CAC ATA ATC C-3′ and Gag-C, 5′-GCT A TG TGC CCT TCT TTG CC-3′. Real-time PCR was carried out in an iCycler (Bio-Rad) using the following thermal cycling profile: 95°C for 1 min, followed by 35 cycles of amplification (95°C 15 s, 63°C 30 s, 68°C 30 s). All samples and in vitro transcripts were run in triplicates.
RESULTS
We have previously demonstrated that a single nucleotide mismatch between a shRNA and its HIV-1 target sequence abrogates gene silencing (22). This observation prompted the current investigation, which sought to systematically determine whether certain regions of a potent HIV-1 gag shRNA are able to tolerate nucleotide mismatches and retain antiviral activity.
The specificity of target recognition and the effects of mutations on siRNA activity remain a controversial subject. For durable shRNA-mediated inhibition of viral replication it would be important to determine the general tolerance of the specific shRNA sequence to single nucleotide mismatches, i.e. polymorphisms. We assessed the specificity of target recognition and the effects of mutations on shRNA function by testing the antiviral activity of a panel of shRNA species that targeted HIV-1 gag. The shRNA sequence was determined by systematic alignments of computer-predicted secondary structures (mfold 2.0) of overlapping 300-nt long sequence stretches of the gag gene. The five lowest free energy structures were analyzed, and a sequence that was conserved among all energy structures was scanned for favorable siRNA target sites. We selected a 21-nt gag region with favorable predicted secondary structure features such as free energy (ΔG) of the target site, total numbers of unpaired nucleotides and major loop size. We introduced a single point mutation at each position of the 21-nt target sequence resulting in 21 different mutant and one wild-type gag shRNA expression vectors (Fig. 1). The introduced point mutations in the forward strand of the oligonucleotides were associated with complementary changes in the reverse sequence to allow continuous base pairing of the shRNA. To simplify analysis, guanosine-cytosine base pairs were mutated to adenosine-thymidine base pairs. Oligonucleotides with the individual gag point mutations were synthesized, annealed and introduced into unique restriction sites of a U6 promoter driven shRNA expression vector. The target sequence and the reverse complementary sequence were separated by a hexaloop and terminated by a five-thymidine stretch (Fig. 1).
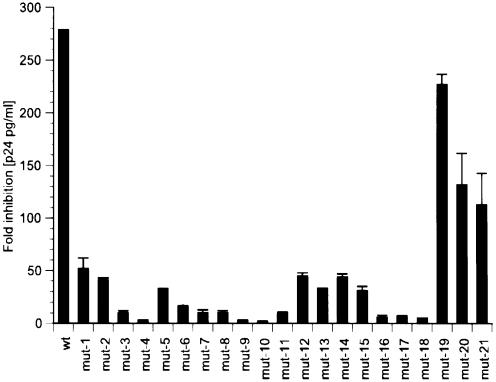
To assess the effect of single mutant gag shRNAs on HIV-1 viral replication, 293 cells were co-transfected with the infectious molecular clone HIV-1NL4.3 and the respective mutant and wild-type gag shRNA expression vectors. HIV-1 p24 antigen level was quantified from cell-free supernatant 48 h post-transfection. A control shRNA expression vector targeting luciferase was included as a negative control. The wild-type gag shRNA was the most effective and reduced HIV-1 p24 antigen expression by 275-fold. All 21 mutant shRNAs exhibited a reduction of antiviral activity compared to wild-type shRNA. The majority of shRNAs (mut-1 to mut-18) demonstrated a low tolerance to mutations (Fig. 2). Mutations at positions 3, 4, 7–11 and 16–18 exhibited severely impaired antiviral activity with only 0.7–3.5% of the antiviral activity of the wild-type shRNA (Fig. 2). Silencing was almost completely abolished with centrally located mutations mut-9 (1.1%), mut-10 (0.7%) and 5′-end located mutation mut-4 (1.1%). Mismatches in the 3′-end of the shRNA (mut-19, -20, -21) were better tolerated and affected gene silencing by 18–60% reduction of antiviral activity. Mutations localized in the 3′ region from the midpoint of the shRNA (mut-12 through mut-15) and 5′-end (mut-1, mut-2) showed 11–19% residual antiviral activity compared to wild-type shRNA.
Figure 2.
The antiviral activity of mutant gag shRNA was assessed by quantifying HIV-1 p24 antigen level in culture supernatant after co- transfection of 293 T cells with HIV-1 and DNA vectors containing the shRNA expression cassette. The inhibitory effect of gag shRNAs on HIV-1 replication is expressed as fold inhibition of p24 production compared to cells transfected with a DNA vector expressing luciferase shRNA. Wild-type shRNA reduced p24 production by 278-fold. All experiments were performed in duplicates with the range indicated.
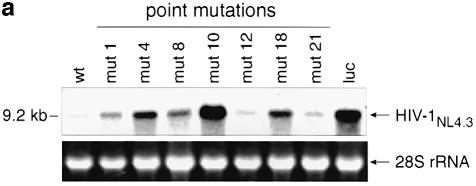
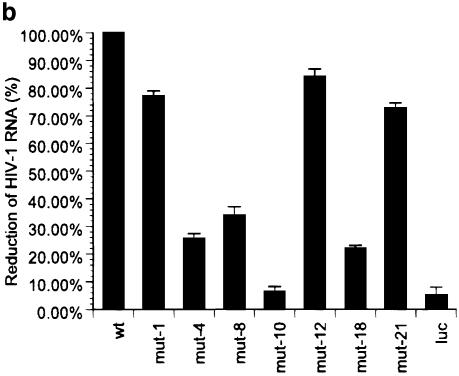
Given that shRNA-mediated gene silencing operates on the mRNA level, we assessed the effect of wild-type and a selected subset of mutant gag shRNAs on the degradation of full-length HIV-1 RNA by northern blot analysis and real-time PCR. The subset of shRNA mutants was selected according to different antiviral activities of representative positions (1, 4, 8, 10, 12, 18, 21) obtained from the HIV-1 p24 antigen data. There was overall concordance between the p24 antigen results and the observed RNA degradation, determined by northern blot and real-time RT–PCR (Fig. 3a and b). The introduction of a central mutation at position 10 (mut-10) led to complete loss of gene silencing with no effect on HIV-1 RNA degradation. A severe loss of antiviral activity was observed with shRNA constructs mut-4 and mut-18 (Fig. 3a and b). Mutations at the extreme end of the target sequence (mut-1 and mut-21) were well tolerated and revealed up to 77% residual antiviral activity compared to wild-type shRNA (Fig. 3b).
Figure 3.
(a) Northern blot analysis of full-length HIV-1 RNA. Degradation of full-length HIV-1 RNA in 293 T cells mediated by a selected panel of DNA vectors expressing wild-type and mutant gag shRNAs. 293 T cells were co-transfected with the indicated vectors and HIV-1NL4.3. A luc shRNA expression vector served as negative control. Ribosomal RNA expression was used as a loading control. (b) Real-time RT–PCR analysis of total HIV-1 RNA degradation by gag shRNAs. A panel of representative gag shRNA expression constructs was selected for the assessment of wild-type and mutant gag-specific shRNA-mediated HIV-1 degradation. Variation of RNA input was normalized by GAPDH real-time RT–PCR. An external standard curve was created with known copy numbers of gag in vitro transcripts. Threshold cycles were converted to absolute copy numbers and the antiviral effect of wild-type gag shRNA was set arbitrarily to 100%. The inhibitory effect of the mutant gag shRNAs is expressed in percentage relative to the wild-type gag shRNA. Values represent averages of three independent experiments with the range indicated.
Finally, we assessed whether variation in the antiviral effect of the mutant shRNA transcripts might be due to G:U wobble or other non-Watson–Crick interactions between the mutant guiding antisense shRNA sequence and the gag target sequence. The introduction of 21 single point mutations resulted in eight gag shRNA constructs containing a G:U wobble at the site of the mutation. The G:U wobble did not seem to have any beneficial effect on gene silencing compared to shRNA mutants, which led to C:U or A:G base pairing. For example, mut-4 with C:U target interactions at the position of the introduced mutation resulted in a higher degree of gene silencing when compared to mut-5 involving G:U base pairing with the gag target sequence. Mut-16 [C:U] and mut-17 [G:U] showed equal levels of antiviral activity as did mut-13 [A:G] when compared with mut-14 [G:U].
DISCUSSION
The use of RNAi as a gene knockout tool and a potential therapeutic modality has been forwarded by the specificity and potency of the resulting gene silencing. Previous work has demonstrated that RNAi can be applied effectively to control the replication of viruses such as HIV-1, HCV and HBV (22–25). Thus far, the sequence specificity of RNAi has been largely demonstrated by creating mutant siRNA with nucleotide mismatches in certain portions of the molecule. We have extended these findings by introducing mutations in every position of a shRNA targeting HIV-1 gag. Our results demonstrate that maximally effective HIV-1-specific RNAi requires perfect homology between shRNA and the chosen target. Variable degrees of gene silencing occur when target and shRNA differ in a single nucleotide, depending on the position of the nucleotide mismatch within the shRNA. Sequence-specific target recognition is severely affected by single mutations at positions 9–11 in the middle of the 21-nt target sequence. Mismatches in the extreme 3′-end of the shRNA sense sequence are tolerated much more so than those in the extreme 5′-end region of the target sequence. The importance of perfect sequence homology in the center of the siRNA molecule and the mRNA target sequence follows from our understanding of siRNA-mediated RNA degradation. It is thought that the cleavage of target RNA proceeds from the middle of the complementary region to the RNA duplex (4). Using D.melanogaster lysates, Elbashir et al. demonstrated that target cleavage occurs 11 or 12 nt downstream of the target position complementary to the extreme 3′-nucleotide of the sequence-complementary 21-nt guide siRNA duplex. In our experiments, mismatches in corresponding positions 10 and 11 relative to the mRNA target sequence compromised RNAi to 1% remaining activity when compared to wild-type shRNA. Although most investigators have found that sequence homology in central regions of siRNA is critical for function, results have varied when sequence mismatches are placed toward the 5′- or 3′-end of the target sequence. The diverse results regarding the tolerance to nucleotide mismatches in both ends of a siRNA might be due to differences in the accessibility to the guide siRNA due to the secondary structure of the individual target mRNA.
A proposed therapeutic application of RNAi is its use in selectively silencing disease-associated RNA transcripts, such as those encoding oncogenes. For example, Brummelkamp et al. used RNAi to selectively degrade oncogenic K-RAS species that differed from wild-type by a single nucleotide (5). Interestingly, the point mutant fell in the middle of the shRNA allowing investigators to demonstrate selective degradation of oncogenic K-RAS with preservation of the wild-type counterpart. Our results indicate that the selective degradation of a mRNA species containing a single point mutation would be best achieved by designing shRNA such that the mutation falls in the center of the molecule. In this manner, the discriminatory effect of shRNA/siRNA would be maximized to allow specific degradation of the targeted nucleotide sequence with minimal unspecific degradation of the mRNA that differs by a single nucleotide mismatch.
A particular challenge of translating RNAi into an antiviral therapeutic derives from the replication kinetics of viruses such as HIV-1 (26). The chronically infected host harbors a heterogeneous swarm of viruses that is unlikely to be effectively targeted by a single shRNA species. Even if a highly conserved region of the virus is targeted, all published reports on HIV-1-specific RNAi demonstrate the reduction but not complete elimination of viral replication. Recent work has demonstrated that residual HIV-1 replication in human cell lines engineered to stably express shRNA targeting HIV-1 leads to the emergence of viral strains with mutations in the shRNA target sequence (22). Thus, a necessary requirement of antiviral RNAi will likely be the need to simultaneously target different regions of the viral genome, not only to increase antiviral potency, but also to decrease the chance of virus escape from RNAi.
In conclusion, our results demonstrate that optimal gene silencing requires perfect homology between shRNA and the chosen target, but that a variable degree of silencing occurs, depending upon the precise location of nucleotide mismatches. These findings should assist investigators in increasing the specificity of shRNA and optimizing RNAi-mediated gene silencing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Brown/Tufts/Lifespan CFAR Retrovirology Core Laboratory for assay support. We acknowledge the AIDS Research and Reference Reagent Program Division of AIDS, NIAID, NIH for supplying H9 cells (contributed by Dr Robert Gallo) and HIVNL4-3 (contributed by Dr Malcolm Martin). This work was supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation, a Daland Fellowship in Clinical Investigation from the American Philosophical Society and a Career Development Grant from NIAID/NIH (B.R.).
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Novina C.D., Murray,M.F., Dykxhoorn,D.M., Beresford,P.J., Riess,J., Lee,S.K., Collman,R.G., Lieberman,J., Shankar,P. and Sharp,P.A. (2002) siRNA-directed inhibition of HIV-1 infection. Nature Med., 8, 681–686. [DOI] [PubMed] [Google Scholar]
- 3.Coburn G.A. and Cullen,B.R. (2002) Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol., 76, 9225–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell, 2, 243–247. [DOI] [PubMed] [Google Scholar]
- 6.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 7.Barton G.M. and Medzhitov,R. (2002) Retroviral delivery of small interfering RNA into primary cells. Proc. Natl Acad. Sci. USA, 99, 14943–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen C., Buck,A.K., Liu,X., Winkler,M. and Reske,S.N. (2003) Gene silencing by adenovirus-delivered siRNA. FEBS Lett., 539, 111–114. [DOI] [PubMed] [Google Scholar]
- 9.Miyagishi M. and Taira,K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 10.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 11.Paddison P.J., Caudy,A.A., Bernstein,E., Hannon,G.J. and Conklin,D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) Effective expression of small interfering RNA in human cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 13.Boden D., Pusch,O., Lee,F., Tucker,L., Shank,P.R. and Ramratnam,B. (2003) Promoter choice affects the potency of HIV-1 specific RNA interference. Nucleic Acids Res., 31, 5033–5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacque J.M., Triques,K. and Stevenson,M. (2002) Modulation of HIV-1 replication by RNA interference. Nature, 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gitlin L., Karelsky,S. and Andino,R. (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature, 418, 430–434. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng Y. and Cullen,B.R. (2003) Sequence requirements for micro RNA processing and function in human cells. RNA, 9, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 19.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson A.L., Bartz,S.R., Schelter,J., Kobayashi,S.V., Burchard,J., Mao,M., Li,B., Cavet,G. and Linsley,P.S. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol., 21, 635–637. [DOI] [PubMed] [Google Scholar]
- 22.Boden D., Pusch,O., Lee,F., Tucker,L. and Ramratnam,B. (2003) Human immunodeficiency virus type 1 escape from RNA interference. J. Virol., 77, 11531–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaffrey A.P., Nakai,H., Pandey,K., Huang,Z., Salazar,F.H., Xu,H., Wieland,S.F., Marion,P.L. and Kay,M.A. (2003) Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol., 21, 639–644. [DOI] [PubMed] [Google Scholar]
- 24.Yokota T., Sakamoto,N., Enomoto,N., Tanabe,Y., Miyagishi,M., Maekawa,S., Yi,L., Kurosaki,M., Taira,K., Watanabe,M. et al. (2003) Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep., 4, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson J.A., Jayasena,S., Khvorova,A., Sabatinos,S., Rodrigue-Gervais,I.G., Arya,S., Sarangi,F., Harris-Brandts,M., Beaulieu,S. and Richardson,C.D. (2003) RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl Acad. Sci. USA, 100, 2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramratnam B., Bonhoeffer,S., Binley,J., Hurley,A., Zhang,L., Mittler,J.E., Markowitz,M., Moore,J.P., Perelson,A.S. and Ho,D.D. (1999) Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet, 354, 1782–1785. [DOI] [PubMed] [Google Scholar]