Abstract
Loss-of-function mutations in the parkin gene (PARK2) and PINK1 gene (PARK6) are associated with autosomal recessive parkinsonism. PINK1 deficiency was recently linked to mitochondrial pathology in human cells and Drosophila melanogaster, which can be rescued by parkin, suggesting that both genes play a role in maintaining mitochondrial integrity. Here we demonstrate that an acute down-regulation of parkin in human SH-SY5Y cells severely affects mitochondrial morphology and function, a phenotype comparable with that induced by PINK1 deficiency. Alterations in both mitochondrial morphology and ATP production caused by either parkin or PINK1 loss of function could be rescued by the mitochondrial fusion proteins Mfn2 and OPA1 or by a dominant negative mutant of the fission protein Drp1. Both parkin and PINK1 were able to suppress mitochondrial fragmentation induced by Drp1. Moreover, in Drp1-deficient cells the parkin/PINK1 knockdown phenotype did not occur, indicating that mitochondrial alterations observed in parkin- or PINK1-deficient cells are associated with an increase in mitochondrial fission. Notably, mitochondrial fragmentation is an early phenomenon upon PINK1/parkin silencing that also occurs in primary mouse neurons and Drosophila S2 cells. We propose that the discrepant findings in adult flies can be explained by the time of phenotype analysis and suggest that in mammals different strategies may have evolved to cope with dysfunctional mitochondria.
Many lines of evidence suggest that mitochondrial dysfunction plays a central role in the pathogenesis of Parkinson disease, starting from the early observation that the complex I inhibitor 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine induced acute and irreversible parkinsonism in young drug addicts (for review, see Refs. 1–3). In support of a crucial role of mitochondria in Parkinson disease, several Parkinson disease-associated gene products directly or indirectly impinge on mitochondrial integrity (for review, see Refs. 4–6). A clear link between Parkinson disease genes and mitochondria has recently emerged from studies on PINK1 (PTEN-induced putative kinase 1), a mitochondrial serine/threonine kinase, and parkin, a cytosolic E3 ubiquitin ligase. Drosophila parkin null mutants displayed reduced life span, male sterility, and locomotor defects due to apoptotic flight muscle degeneration (7). The earliest manifestation of muscle degeneration and defective spermatogenesis was mitochondrial pathology, exemplified by swollen mitochondria and disintegrated cristae. Remarkably, Drosophila PINK1 null mutants shared marked phenotypic similarities with parkin mutants, and parkin could compensate for the PINK1 loss-of-function phenotype but not vice versa, leading to the conclusion that PINK1 and parkin function in a common genetic pathway with parkin acting downstream of PINK1 (8–10). We recently demonstrated that PINK1 deficiency in cultured human cells causes alterations in mitochondrial morphology, which can be rescued by wild type parkin but not by pathogenic parkin mutants (11). We now present evidence that parkin plays an essential role in maintaining mitochondrial integrity. RNAi3-mediated knockdown of parkin increases mitochondrial fragmentation and decreases cellular ATP production. Notably, mitochondrial fragmentation induced by PINK1/parkin deficiency is observed not only in human neuroblastoma cells but also in primary mouse neurons and insect S2 cells. Alterations in mitochondrial morphology are early manifestations of parkin/PINK1 silencing that are not caused by an increase in apoptosis. The mitochondrial phenotype observed in parkin- or PINK1-deficient cells can morphologically and functionally be rescued by the increased expression of a dominant negative mutant of the fission-promoting protein Drp1. Moreover, manifestation of the PINK1/parkin knockdown phenotype is dependent on Drp1 expression, indicating that an acute loss of parkin or PINK1 function increases mitochondrial fission.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were used: anti-parkin rabbit polyclonal antibody (pAb) hP1 (12), anti-parkin mouse monoclonal antibody (mAb) PRK8 (Millipore, Schwalbach, Germany), anti-parkin polyclonal antibody 2132 (Cell Signaling, Danvers, MA), anti-FLAG M2 mAb (Sigma), anti-FLAG M2 horseradish peroxidase mAb (Sigma), anti-β-actin mAb (Sigma), anti-Drp1 mAb (BD Transduction Laboratories), anti-Mfn2 pAb (Sigma), anti-OPA1 pAb (13), anti-PINK1 pAB (Novus Biologicals, Hamburg, Germany), penta-His horseradish peroxidase conjugate mouse IgG (Qiagen, Hilden, Germany), horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibody (Promega, Mannheim, Germany), anti-active caspase-3 pAb (Promega), anti-V5 mAb (Invitrogen), cyanine 3 (Cy3)-conjugated anti-rabbit IgG antibody (Dianova, Hamburg, Germany), anti-neuron specific β III Tubulin rabbit-pAb (Abcam, Cambridge, UK), and CyTM 3-conjugated Affinity Pure Donkey anti-rabbit IgG (heavy and light chain) (Jackson ImmunoResearch, Newmarket, Suffolk, UK). Staurosporine, rotenone, cycloheximide, and carbonyl cyanide 3-chlorophenylhydrazone were purchased from Sigma, complete protease inhibitor mixture was from Roche Applied Science, and 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)), and MitoTracker Red CMXRos was from Invitrogen.
DNA Constructs
The following constructs were described previously: wild type human parkin, W453X, R42P, G430D, Δ1–79 parkin mutant (12, 14, 15), PINK1-V5 and PINK1-G309D-V5 (11), Mfn2-His6, OPA1-MycHis, Drp1-EYFP, Drp1(K38E)-ECFP (16, 17), and Bcl-2-FLAG (18). Mfn2 containing a C-terminal FLAG tag was subcloned into pcDNA3.1/Zeo (+) (Invitrogen). Drp1 was subcloned into the pCMV-Tag 2B (Stratagene, Amsterdam, Netherlands) vector adding an N-terminal FLAG tag. mCherry (19) was subcloned into the pCS2+ vector. For the generation of small interfering RNA (siRNA)-resistant wild type parkin, four silent mutations were introduced into the siRNA target sequence by PCR. The plasmid encoding enhanced yellow fluorescent protein (EYFP) was purchased from Clontech (Mountainview, CA).
Lentiviruses
The sequence of the PINK1 shRNA 5′-GCG GTA ATT GAC TAC AGC AAA-3′, which corresponds to nucleotides 1353–1373 of the PINK1 gene, was cloned into the pLL3.7 vector via the HpaI and XhoI cloning sites. The shRNA is driven by a U6 promotor. The green fluorescent protein portion of the pLL3.7 vector was exchanged by cloning a cytomegalovirus-driven mito-EYFP (pEYFP-mito, Clontech) into the NheI and EcoRI site. As a control virus we used the same virus without the PINK1 shRNA sequence. Lentiviruses were produced in HEK293T cells as described by Consiglio et al. (20). The titer of the viruses ranged from 4 × 108 to 1 × 109 colony-forming units/ml. The infection efficiency of primary neurons was not affected by different titers. shRNAs were designed using the pSico-Oligomaker 1.5 (developed by A. Ventura). Conditional knockdowns were generated by cloning shRNAs into pSico as described previously (21).
Cell Culture, Transfection, and RNA Interference
SH-SY5Y (DSMZ number ACC 209) cells were transfected with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Drosophila S2 cells were cultivated in Schneider's Drosophila medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Sigma) and maintained at 26 °C. Transfection of S2 cells was performed in 12-well plates in serum-free medium with 5 μg of dsRNA. Serum-containing medium was added 45 min after transfection. For RNA interference, SH-SY5Y cells were reverse-transfected with Stealth siRNA (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen). For each target gene at least two different effective siRNAs were used. For RNAi treatment of Drosophila S2 cells, the following clones from the Drosophila Genomics Resource Center (Bloomington, IN) were used: parkin (SD01679) and PINK1 (GH20931). They served as templates for T7-tagged primers, which were designed by a program for the de novo design of long dsRNAs (E-RNAi Webservice, German Cancer Research Center): T7-parkin forward primer, 5′-taa tac gac tca cta tag ggC TGT TGA CAC GCG AGG AGT A-3′, and T7-parkin reverse primer, 5′-taa tac gac tca cta tag ggA TTT TGG ACA GGG CTT TGT G-3′; T7-PINK1 forward primer, 5′- taa tac gac tca cta tag ggG CCA TGT ACA AGG AGA CGG T-3′, and PINK1 reverse primer, 5′-taa tac gac tca cta tag ggA TTG AGT ACG GCA AAC GGA C-3′. After synthesis of T7-cDNA templates by PCR, the Ambion Megascript RNA synthesis kit (Austin, TX) was used to generate dsRNA.
Preparation, Transduction, and Analysis of Primary Mouse Hippocampal Neurons
Hippocampi of C57/BL6 mice from two different litters were prepared at embryonic stage E15.5 and transferred to dissection medium (48.8 ml of Hanks' balanced salt solution (Invitrogen), 500 μl of HEPES (1 m, Invitrogen), 600 μl of MgSO4 (1 m), and 500 μl of penicillin/streptomycin (100×, Invitrogen)). Tissue was washed with prewarmed trypsin 0.05% with EDTA (Invitrogen) 2 times, trypsinated for 15 min at 37 °C, and thereafter washed 3 times in culture medium (48 ml of Neurobasal (Invitrogen), 1 ml of B-27 (Invitrogen), 500 μl of l-glutamine (Invitrogen), and 500 μl of penicillin/streptomycin (100×)). Cells were dispersed with a fire-polished tip of a Pasteur pipette. 40 × 103 cells were cultivated on poly-d-lysine-coated coverslips in 24-well plates. On day 4 cells were infected with lentiviruses. Three days after infection cells were fixed in 4% paraformaldehyde for 10 min. Neurons were detected by immunocytochemistry using anti-neuron-specific β III tubulin rabbit pAB (Abcam) as primary antibody and CyTM 3-conjugated affinity pure donkey anti-rabbit IgG (H+L) as secondary antibody (Jackson ImmunoResearch). Pictures were acquired using confocal microscopy (LSM 510, Carl Zeiss, Göttingen, Germany) to analyze the length of mitochondria in soma and processes. For quantification of mitochondrial lengths, a specified analysis software (Axiovision 4.7, Carl Zeiss) was used. Classification of mitochondria was done as follows: fragmented mitochondria (<0.5 μm), intermediate mitochondria (0.5–5 μm), and tubular mitochondria (>5 μm).
Quantitative RT-PCR
For the analysis of parkin or PINK 1 knockdown efficiencies, quantitative real-time PCR was performed. Total cellular RNA from human SH-SY5Y and Drosophila S2 cells was isolated at the time indicated and treated with DNase I (RNeasy mini kit; Qiagen). cDNA was synthesized from 1 mg of total RNA using a High-Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). For human SH-SY5Y cells, RT-PCR was performed with 2× TaqMan Universal PCR Master Mix and TaqMan Gene expression assay (parkin: Hs00247755_m1, PINK1: Hs02330592_s1; Applied Biosystems). For Drosophila S2 cells RT-PCR was performed with 2× Power SYBR Green PCR Master Mix (Applied Biosystems) and 1 μm concentrations of each primer pair (S2 parkin forward primer, 5′-AGC CTC CAA GCC TCT AAA TG-3′; S2 parkin reverse primer, 5′-CAC GGA CTC TTT CTT CAT CG-3′; S2 PINK1 forward primer, 5′-GCT TTC CCC TAC CCT CCA C-3′; S2 PINK1 reverse primer, 5′-GCA CTACAT TGA CCA CCG ATT-3′; S2 Rp49 forward primer, 5′-CCA AGC ACT TCA TCC GCC ACC-3′; S2 Rp49 reverse primer, 5′-GCG GGT GCG CTT GTT CGA TCC-3′) (22). Quantification was performed with 7500 Fast Real Time PCR System (Applied Biosystems). Triplicates were performed with each primer set for each RNA sample. RNA expression was normalized with respect to an endogenous reference gene; β-actin for SH-SY5Y cells and Rp49 for S2 cells, respectively. Relative expression was calculated for each gene using the delta delta cycle threshold method. For quantification of the knockdown efficiency in primary neuronal cultures, cells were prepared and cultivated as described above and lysed directly on the plate according to the manual of the RNeasy mini kit (Qiagen). cDNA was transcribed using cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed with the TaqMan expression assay (PINK1 mouse: 4m00550827_m1) with β-actin as an internal control.
Western Blotting
SDS-PAGE and Western blotting was described previously (14). Antigens were detected with the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences) or the Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Fluorescent Staining of Mitochondria
SH-SY5Y cells were grown on 15-mm glass coverslips, and S2 cells were grown on coverslips coated with concanavalin A (Sigma). Cells were fluorescently labeled with either 0.1 μm DiOC6(3) or 0.05 μm MitoTracker Red CMXRos in cell culture medium for 15 min. After washing coverslips with medium, living cells were analyzed for mitochondrial morphology by fluorescence microscopy as described previously (11, 13) using a Leica DMRB microscope (Leica, Wetzlar, Germany). Cells were categorized in either two or three classes according to their mitochondrial morphology: tubular, fragmented, or highly connected. Cells displaying an intact network of tubular mitochondria were classified as tubular. When this network was disrupted and mitochondria appeared predominantly spherical or rod-like, they were classified as fragmented. Cells with considerably elongated mitochondria that were more interconnected were classified as highly connected. The mitochondrial morphology of at least 300 cells per plate was determined in a blinded manner, i.e. the researcher was blind to transfection status. Quantifications were based on triplicates of at least three (SH-SY5Y cells) or two (S2 cells) independent experiments.
Measurement of Cellular ATP Levels
Cellular steady state ATP levels were measured using the ATP Bioluminescence assay kit HS II (Roche Applied Science) according to the manufacturer's instructions. SH-SY5Y cells were reversely transfected with the indicated siRNAs and/or DNA constructs. 24 h before harvesting cells, the culture medium was replaced by medium containing 3 mm glucose. Cells were washed twice with phosphate-buffered saline, scraped off the plate, and lysed according to the provided protocol. Bioluminescence of the samples was determined using an LB96V luminometer (Berthold Technologies), analyzed with WinGlow Software (Berthold Technologies), and normalized to total protein levels. Each transfection was performed at least in triplicate, and all experiments were repeated at least three times.
Apoptosis Assays
For detection of apoptotic cells the ApopTag Fluorescein Direct in Situ Apoptosis detection kit (Chemicon, Temecula, CA) was used according to the manufacturer's protocol. Briefly, SH-SY5Y/S2 cells were grown on glass coverslips. At days 1, 2, 3, and 4 after siRNA/dsRNA transfection the positive controls were incubated with staurosporine (1 μm, SH-SY5Y cells) or cycloheximide (10 μm, S2 cells) for 4 and 6 h, respectively. The cells were fixed with 1% paraformaldehyde for 10 min at room temperature, permeabilized with a 2:1 mixture of ethanol and acetic acid for 5 min at −20 °C, and equilibrated with the supplied buffer. Fixed cells were incubated with Working Strength TdT enzyme in a humidified chamber for 1 h at 37 °C in the dark. After washing with Working Strength Stop/Wash buffer (Chemicon, Temecula, CA) for 10 min at room temperature cells were mounted onto glass coverslides using FluorSave Reagent (Calbiochem/Merck KGaA). Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). To detect cells undergoing apoptosis, the number of TdT-mediated dUTP nick end labeling (TUNEL)-positive cells of at least 300 DAPI-stained cells was determined using a Zeiss Axioscope 2 plus microscope (Carl Zeiss). Quantifications were based on at least three independent experiments. Activation of caspase-3 was determined as described previously (18, 23). Briefly, SH-SY5Y cells were grown on glass coverslips. 3 days after transfection cells were incubated with rotenone (10 μm, 3 h) as a positive control. The cells were then fixed, and activated caspase-3 was detected by indirect immunofluorescence. Nuclei were stained with DAPI, and caspase-3-positive cells were quantified as outlined above.
Statistical Analyses
For cell culture experiments, data are expressed as the means ± S.E. Experiments were performed in triplicate and repeated at least three times. Statistical analysis was carried out using analysis of variance; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. For primary mouse hippocampal neurons, data are expressed as the means ± S.E. Statistical analysis of the mitochondrial lengths were carried out using analysis of variance. The statistical analysis of the fragmented and intermediate mitochondria were analyzed via analysis of variance, whereas the p values of tubular mitochondria were calculated via Fisher's Exact Test. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
RESULTS
Down-regulation of Parkin Induces Fragmentation of the Mitochondrial Network
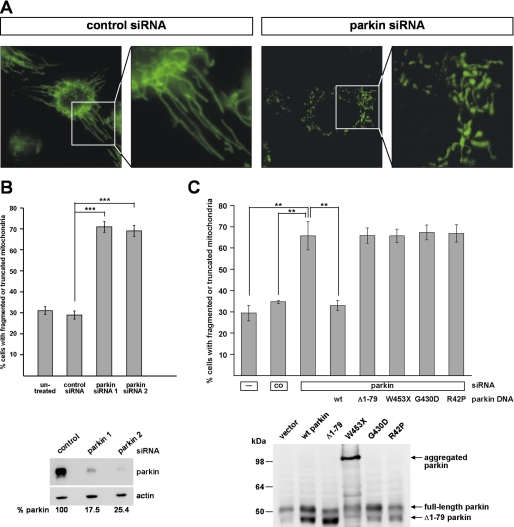
We recently reported that RNAi-mediated down-regulation of PINK1 in cultured human cells results in abnormal mitochondrial morphology, which can be rescued by the enhanced expression of parkin (11). To address a possible role of parkin in mitochondrial integrity, we performed life cell imaging by fluorescence microscopy in human SH-SY5Y cells after siRNA-mediated down-regulation of parkin. Under physiological conditions, the majority of the cells (about 70%) showed a network of tubular mitochondria, which is in line with previous reports (24–26). Upon knockdown of parkin, the percentage of cells with truncated or fragmented mitochondria significantly increased from about 30% in control siRNA-treated cells up to 70% (Fig. 1, A and B). This alteration in mitochondrial morphology also occurred when a second parkin-specific siRNA was used (Fig. 1B) and could be prevented by the co-transfection of siRNA-resistant wild type parkin (Fig. 1C), demonstrating the specificity of the effects observed. The pathogenic parkin mutants W453X, G430D, and R42P did not show rescue activity (Fig. 1C). Notably, the Δ1–79 parkin mutant, which lacks the N-terminal ubiquitin-like domain and is generated in vivo due to the presence of an internal translation initiation site at codon 80 (15), could not compensate for the mitochondrial phenotype observed in parkin-deficient cells. The Δ1–79 mutant has been reported previously to be impaired in its neuroprotective capacity and ubiquitylation activity (14, 27). These results indicate that an acute loss of parkin function significantly affects mitochondrial morphology.
FIGURE 1.
Down-regulation of parkin by RNAi leads to alterations in mitochondrial morphology. A and B, SH-SY5Y cells transfected with control siRNA or siRNA targeting parkin were stained with the fluorescent dye DiOC6(3) to visualize mitochondria and analyzed by fluorescence microscopy. The analysis was performed 3 days after transfection. Cells displaying an intact network of tubular mitochondria were classified as tubular. When this network was disrupted and mitochondria appeared predominantly spherical or rod-like, they were classified as fragmented. B, for quantification, the mitochondrial morphology of at least 300 cells per plate was determined in a blinded manner. Quantifications were based on triplicates of at least three independent experiments. Shown is the percentage of cells with fragmented or truncated mitochondria. Lower panel, the efficiency of parkin knockdown is shown by Western blotting using the monoclonal anti-parkin antibody PRK8. β-Actin was used as a loading control. C, SH-SY5Y cells were transfected with parkin-specific siRNA and either siRNA-resistant wild type (wt) parkin, Δ1–79, W453X, G430D, or R42P mutant parkin. The cells were analyzed by fluorescence microscopy as described under A. Lower panel, expression of parkin or parkin mutants was analyzed by Western blotting using the monoclonal anti-parkin antibody PRK8. Please note that the overexpression of parkin gives rise to two parkin species, full-length parkin of 52 kDa and a smaller parkin species of about 42 kDa, due to the usage of the second translation initiation site (15).
Alterations in Mitochondrial Morphology and Cellular ATP Production Caused by Parkin or PINK1 Deficiency Can Be Rescued by Mfn2, OPA1, or Dominant Negative Drp1
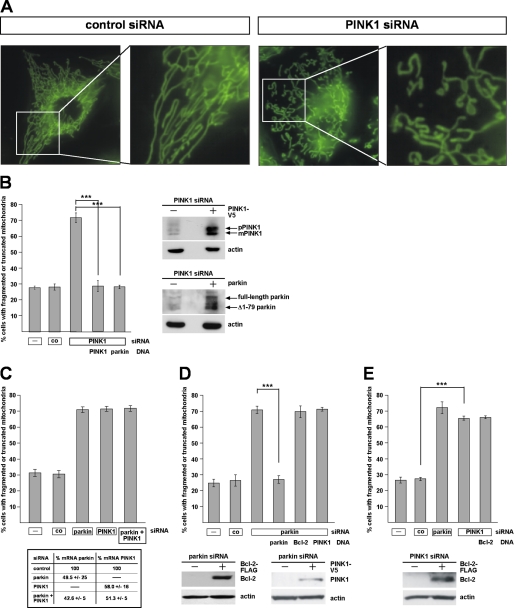
In our previous study we used HeLa cells to assess the effects of PINK1 on mitochondrial morphology. HeLa cells do not express parkin due to the localization of the parkin gene within FRA6E (6q26), a common fragile site of the human genome that is frequently mutated in ovarian tumors (28). To compare the effects of parkin and PINK1 on mitochondria, we treated SH-SY5Y cells with PINK1-specific siRNA. The effects on mitochondrial morphology induced by PINK1 deficiency were comparable with those observed in HeLa cells. Moreover, PINK1-deficient SH-SY5Y displayed qualitative and quantitative alterations similar to parkin-deficient SH-SY5Y cells, i.e. a significant increase in the percentage of cells with truncated or fragmented mitochondria (Fig. 2, A and B). Consistent with the data from Drosophila melanogaster, we observed that parkin rescues the mitochondrial pathology in PINK1-deficient cells (Fig. 2B), whereas enhanced PINK1 expression cannot compensate for the parkin knockdown phenotype (Fig. 2D). Also in line with the fly model, a simultaneous down-regulation of parkin and PINK1 did not enhance alterations in mitochondrial morphology observed in a single parkin or PINK1 knockdown (Fig. 2C). As no anti-PINK1 antibody is available to detect endogenous PINK1 in SH-SY5Y cells (29, 30), we verified the PINK1 knockdown efficiencies by real-time PCR (Fig. 2C, lower panel). Based on the observation that Bcl-2 suppressed the mitochondrial phenotype in PINK1 null Drosophila mutants (9), we tested a possible effect of Bcl-2 in our model. However, overexpression of Bcl-2 had no effect on the mitochondrial morphology in parkin- or PINK1-deficient human cells (Fig. 2, D and E), suggesting that the increase in mitochondrial fragmentation was not a consequence of apoptosis (see also Fig. 6).
FIGURE 2.
PINK1-deficient SH-SY5Y cells show alterations in mitochondrial morphology similar to parkin-deficient cells. A, SH-SY5Y cells transfected with control siRNA or siRNA targeting PINK1 were stained with the fluorescent dye DiOC6(3) to visualize mitochondria and analyzed by fluorescence microscopy as described in Fig. 1. B, down-regulation of PINK1 by RNAi leads to an increase in mitochondrial fragmentation, which can be rescued by parkin. SH-SY5Y cells were transfected with PINK1-specific siRNA and either siRNA-resistant PINK1 or wild type parkin. The cells were analyzed by fluorescence microscopy as described in Fig. 1. Right panel, expression of PINK1 and parkin was analyzed by Western blotting using a monoclonal anti-V5 antibody or the anti-parkin anti-serum hP1. pPINK1, precursor form; mPINK1, mature form. C, simultaneous down-regulation of parkin and PINK1 does not increase mitochondrial fragmentation over the single parkin or PINK1 knockdown. SH-SY5Y cells were transfected with parkin-specific siRNA and/or PINK1-specific siRNA, and the cells were analyzed by fluorescence microscopy as described above. Lower panel, efficiency of PINK1 and/or parkin down-regulation was determined by quantitative RT-PCR as described under “Experimental Procedures.” D and E, anti-apoptotic Bcl-2 has no effect on the mitochondrial morphology in parkin or PINK1 knockdown cells. PINK1 cannot rescue the parkin knockdown phenotype (D). SH-SY5Y cells were transfected with parkin-specific siRNA (D) or PINK1-specific siRNA (E) and a Bcl-2 or PINK1 expression plasmid. The cells were analyzed as described in Fig. 1. Lower panels, expression of Bcl-2 and PINK1 was analyzed by Western blotting. ***, p ≤ 0.001.
FIGURE 6.
The increase in mitochondrial fragmentation observed in parkin- or PINK1-deficient S2 or SH-SY5Y cells is not associated with an increase in apoptosis. A, S2 cells were treated with control dsRNA and parkin-specific or PINK1-specific dsRNA. At days 2, 3, and 4 after treatment, cells were fixed and permeabilized. Apoptotic cells were detected by fluorescently labeling the free 3′-OH ends of DNA strand breaks (TUNEL). As a positive control, cells were treated with cycloheximide (10 μm, 6 h). Shown is the percentage of apoptotic cells, determined by the number of TUNEL-positive cells of at least 300 DAPI-stained cells. Quantifications were based on at least three independent experiments. B, parkin or PINK1 knockdown efficiencies in S2 cells corresponding to the experiments shown in Figs. 5A and 6A. Cells were harvested at days 2, 3, and 4 after treatment. Total cellular RNA was isolated and subjected to quantitative RT-PCR using parkin- and PINK1-specific primers. The amount of RNA of each sample was normalized with respect to the endogenous housekeeping gene Rp49. The efficiencies of the parkin/PINK1 double knockdown are shown in the right panel. C, SH-SY5Y cells were transfected with control siRNA and parkin-specific or PINK1-specific siRNA. At days 1, 2, 3, and 4 after transfection, cells were fixed and permeabilized. Apoptotic cells were detected by the TUNEL assay described in A. As a positive control, cells were treated with staurosporine (1 μm, 4 h). Shown is the percentage of apoptotic cells, determined by the number of TUNEL-positive cells of at least 300 DAPI-stained cells. Quantifications were based on at least three independent experiments. D, quantification of parkin or PINK1 knockdown efficiencies in SH-SY5Y cells corresponding to the experiment shown under C. SH-SY5Y cells were harvested at days 1, 2, 3, and 4 after siRNA transfection. Total cellular RNA was isolated and subjected to quantitative RT-PCR using parkin- and PINK1-specific primers. The amount of mRNA of each sample was normalized with respect to the endogenous housekeeping gene β-actin. E, examples of the direct immunofluorescence analysis described under C. Apoptotic cells (TUNEL-positive) were fluorescein-labeled (green), and nuclei were stained with DAPI (blue). F, in addition to the TUNEL assay, a single cell analysis for activated caspase-3 was performed in SH-SY5Y cells. Two days after transfection with siRNA, SH-SY5Y cells were fixed, permeabilized, and analyzed by indirect immunofluorescence. Activation of caspase-3 was detected using an anti-active caspase-3 antibody. As a positive control, cells were treated with rotenone (10 μm, 3 h). Shown is the percentage of apoptotic cells, determined by the number of activated caspase-3-positive cells of at least 300 DAPI-stained cells. Quantifications were based on triplicates of at least three independent experiments.
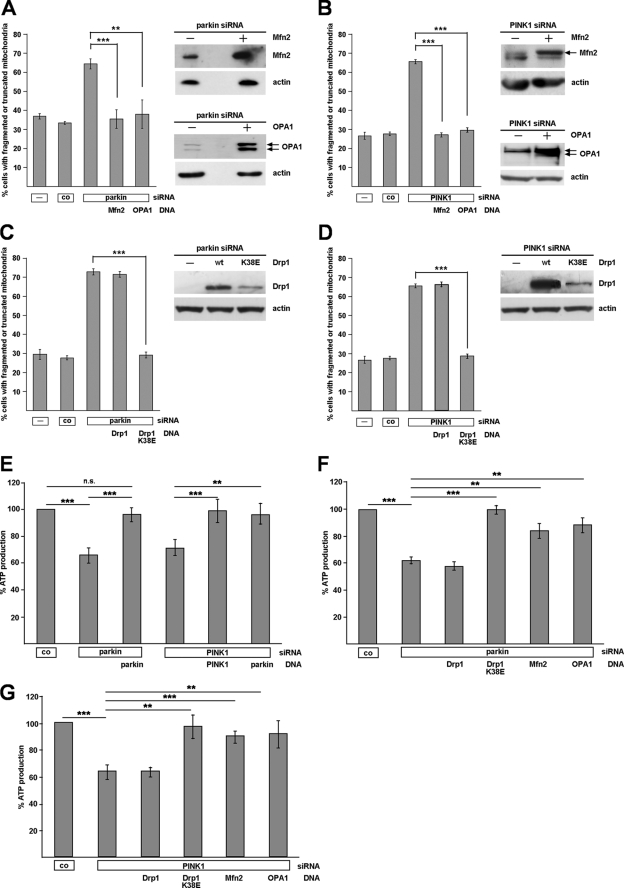
The increased percentage of cells with fragmented mitochondria observed in parkin- or PINK1-deficient cells prompted us to address the question of whether mitochondrial dynamics might be perturbed by a loss of parkin or PINK1 function. Overexpression of Mfn2, which mediates mitochondrial outer membrane fusion, or OPA1, implicated in inner membrane fusion, prevented the changes in mitochondrial morphology induced by parkin or PINK1 knockdown (Fig. 3, A and B, and supplemental Fig. 1). In line with these observations, overexpression of a dominant negative mutant of the fission-promoting GTPase Drp1 (Drp1 K38E) fully reverted the morphological mitochondrial alterations in parkin- or PINK1-deficient cells (Fig. 3, C and D, and supplemental Fig. 1). To test whether manipulating mitochondrial dynamics might have an impact on mitochondrial function in parkin- or PINK1-deficient cells, we determined steady state cellular ATP levels. ATP levels were significantly decreased in parkin-deficient cells (to 66.3 ± 4.7%) and PINK-deficient cells (to 70.9 ± 6.9%; Fig. 3E). Co-transfection of siRNA-resistant parkin or PINK1 prevented ATP depletion in parkin- or PINK1-deficient cells, confirming specificity of the observed effects (Fig. 3E). Consistent with the effect on mitochondrial morphology, parkin could compensate for the impaired ATP production caused by PINK1 silencing (Fig. 3E). Remarkably, increased expression of Drp1 K38E, Mfn2, or OPA1 also prevented the drop in ATP production caused by parkin or PINK1 depletion (Fig. 3, F and G). Collectively, these results indicate that the morphological alterations in parkin- and PINK1-deficient cells are functionally relevant.
FIGURE 3.
Abnormal mitochondrial morphology and function caused by parkin or PINK1 loss of function can be rescued by increasing mitochondrial fusion or decreasing fission. SH-SY5Y cells were cotransfected with siRNA targeting either parkin or PINK1 and the constructs indicated. The cells were analyzed as described under Fig. 1. A–D, Mfn2, OPA1, and dominant negative Drp1 (Drp1 K38E) rescued the mitochondrial phenotype observed in parkin or PINK1 knockdown cells. Shown is the percentage of cells with fragmented or truncated mitochondria. **, p ≤ 0.01; ***, p ≤ 0.001. wt, wild type. Right panel of each set, expression of Mfn2, OPA1, or Drp1 was analyzed by Western blotting. See supplemental Fig. 1 for mitochondrial images. E–G, steady state cellular ATP levels were measured in SH-SY5Y cells transfected with either parkin siRNA or PINK1 siRNA, and the expression plasmids are indicated. The analysis was performed 3 days after transfection.
Manifestation of the Parkin/PINK1 Knockdown Phenotype Is Dependent on Drp1 Expression
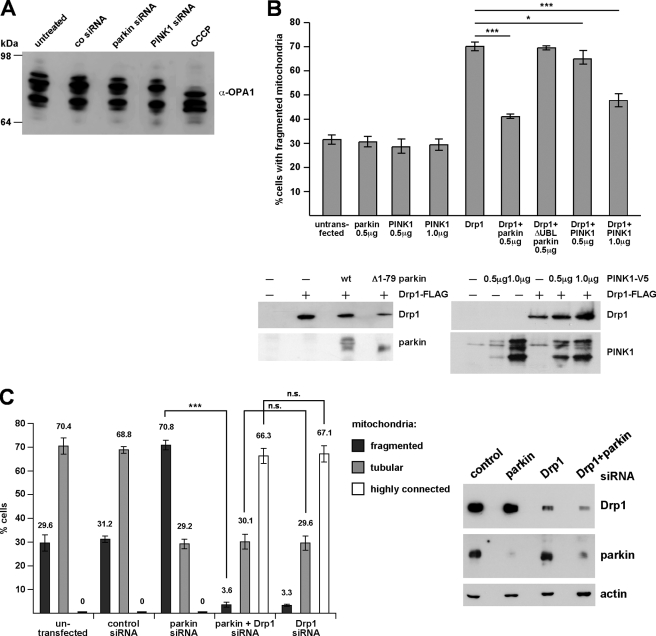
Our observations suggested that a loss of parkin or PINK1 function is associated with an imbalance in the fusion-to-fission rate. However, it remained unclear whether the mitochondrial phenotype was a consequence of reduced fusion or increased fission activity. To address this question experimentally, we first analyzed whether processing of OPA1 is altered under parkin or PINK1 knockdown conditions. The dynamin family GTPase OPA1 is a key player in promoting mitochondrial inner membrane fusion and regulating cristae morphology. Because of extensive alternative splicing and posttranslational proteolytic processing of OPA1, several isoforms are generated in mammalian cells. Recent studies revealed that OPA1 processing is implicated in the regulation of OPA1 function (13, 31, 32). It has been proposed that increased proteolytic processing of OPA1 large to small isoforms in energetically compromised mitochondria reduces the fusion-promoting activity of OPA1 (13). The pattern of endogenous OPA1 isoforms in parkin or PINK1 knockdown cells was analyzed by Western blotting. The mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone induced a decrease in longer isoforms and an increase in shorter OPA1 isoforms; however, no alterations in OPA1 processing were observed upon transient down-regulation of parkin or PINK1 (Fig. 4A). The same results were obtained using stable PINK1 knockdown cells (data not shown). Thus, the effects of parkin or PINK1 down-regulation on mitochondrial morphology are not caused by a decrease in fusion mediated by proteolytically processed OPA1.
FIGURE 4.
A, RNAi-mediated knockdown of parkin or PINK1 does not alter proteolytic processing of OPA1. SH-SY5Y cells transfected with control siRNA or siRNA targeting parkin or PINK1 were analyzed by Western blotting using a polyclonal antibody against OPA1. As a positive control to induce OPA1 processing, cells were treated with carbonyl cyanide 3-chlorophenylhydrazone (CCCP; 20 μm, 30 min). B, parkin and PINK1 (at higher expression levels) can reduce mitochondrial fission induced by Drp1. SH-SY5Y cells were transfected with the constructs indicated. 24 h after transfection mitochondrial morphology of transfected cells (identified by the coexpression of mCherry) was assessed as described in Fig. 1. Lower panel, expression levels of Drp1-FLAG, parkin, and PINK1-V5 in SH-SY5Y cells. 10 μg of protein were loaded per lane. See supplemental Fig. 2 for mitochondrial images. wt, wild type. C, in Drp1-deficient cells the mitochondrial phenotype induced by parkin knockdown does not occur. SH-SY5Y cells were transfected with the siRNAs indicated, and mitochondrial morphology was determined as described in Fig. 1. Right panel, the efficiency of Drp1 and parkin down-regulation by RNAi was shown by Western blotting using a monoclonal anti-Drp1 antibody and the anti-parkin antibody PRK8. β-Actin was used as a loading control. ***, p ≤ 0.001.
An increase in shorter and less tubular mitochondria could also be a consequence of increased mitochondrial fission. To test whether this scenario applies to the effect of parkin and PINK1 on mitochondrial dynamics, we addressed the question of whether Drp1 might be implicated in mediating the parkin or PINK1 knockdown phenotype. First, we analyzed whether parkin or PINK1 might have an impact on mitochondrial fission induced by Drp1. This experiment revealed that the overexpression of neither parkin nor PINK1 induced significant changes in the mitochondrial morphology of transfected SH-SY5Y cells (Fig. 4B and supplemental Fig. 2). However, overexpression of Drp1 increased the percentage of transfected SH-SY5Y cells with fragmented mitochondria from 31.8% under control conditions to 71.0%. Remarkably, coexpression of parkin prevented mitochondrial fragmentation induced by Drp1, whereas Δ1–79 parkin had no effect (Fig. 4B and supplemental Fig. 2). PINK1 was also able to suppress Drp1-induced mitochondrial fragmentation but only at higher expression levels (Fig. 4B and supplemental Fig. 2). Second, we analyzed the consequences of parkin down-regulation in cells deficient in Mfn2 or Drp1. Mitochondrial fragmentation observed in Mfn2 knockdown cells was not influenced by parkin nor by increased parkin expression or by parkin deficiency (supplemental Fig. 3, A and B). In cells treated with Drp1-specific siRNA, the typical increase in cells with fragmented mitochondria upon down-regulation of parkin did not occur (Fig. 4C). Thus, Drp1 expression is necessary to mediate the effects of parkin down-regulation on mitochondrial morphology. Of note, the extent of tubular and highly connected mitochondria, which typically occurs in Drp1-deficient cells, was similar in cells lacking either Drp1 or Drp1 and parkin (Fig. 4C), indicating that mitochondrial fusion is not impaired in parkin-deficient cells. The same results were obtained when we used PINK1 siRNA instead of parkin siRNA (data not shown). These results corroborate that the mitochondrial phenotype induced by parkin or PINK1 deficiency is mediated by increased fission.
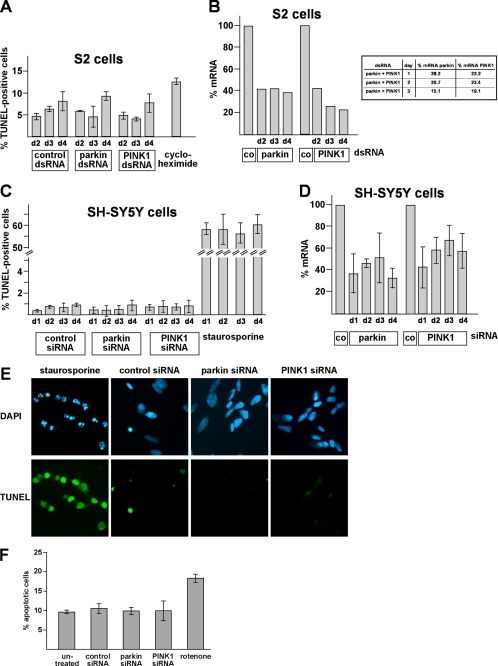
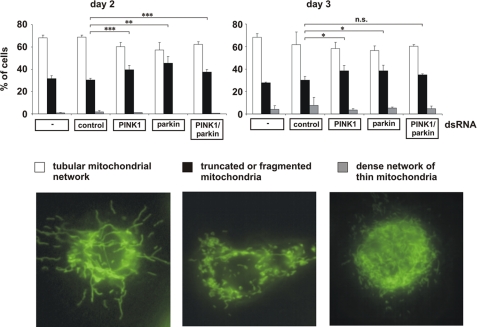
Mitochondrial Fission Is an Early Phenotype of Parkin or PINK1 Deficiency That Also Occurs in Drosophila S2 Cells and Primary Mouse Neurons
It has recently been demonstrated that parkin or PINK1 mutant phenotypes in flies can be rescued by increasing mitochondrial fission or decreasing fusion (33–36). To explain the discrepant results obtained in our model, we performed a comparative analysis of human SH-SY5Y cells and Drosophila S2 cells. For the down-regulation of parkin and PINK1 in S2 cells, we tested 2 and 3 different dsRNAs, respectively, which were all effective as determined by quantitative RT-PCR and showed the same effects on mitochondrial morphology. For the experiments shown in Figs. 5 and 6, we used the most effective dsRNAs, resulting in a 60–70% reduction in parkin- or PINK1-specific mRNA in comparison to control dsRNA-treated S2 cells (Fig. 6B). To monitor alterations in mitochondrial morphology, we performed time course experiments and analyzed the mitochondrial morphology of parkin- or PINK1-deficient S2 cells by fluorescence microscopy 48, 60, and 72 h after dsRNA treatment. We observed the most obvious increase in fragmented mitochondria in parkin- and/or PINK1-deficient S2 cells on day 2 after dsRNA treatment (Fig. 5), explaining why two previous studies analyzing only day 3 or 4 failed to observe mitochondrial fission induced by parkin or PINK1 loss of function (33, 34). Indeed, on day 3 after dsRNA treatment, we observed only minor differences (parkin- or PINK1 knockdown S2 cells) or no significant differences (parkin/PINK1 double knockdown S2 cells) to control cells. We never saw an increase in mitochondrial fusion in parkin- or PINK1 dsRNA-treated S2 cells. However, we noticed a dense network of fine thread-like mitochondria on days 3 and 4 after dsRNA treatment, but these mitochondria also occurred in control dsRNA-treated cells. In comparison to human SH-SY5Y cells, the extent of mitochondrial fragmentation was lower (about 70% of SH-SY5Y cells versus 40–50% of S2 cells), and in contrast to S2 cells, the fragmented state in SH-SY5Y cells persisted for days. As mitochondrial fragmentation can occur as a consequence of apoptosis, we analyzed apoptotic cell death in parkin- or PINK1-deficient SH-SY5Y or S2 cells over a period of 4 days after siRNA treatment. Importantly, mitochondrial fission induced by parkin or PINK1 deficiency was not associated with an increase in apoptosis. At the time we observed an increase in fragmented mitochondria in parkin- or PINK1-depleted SH-SY5Y or S2 cells, there was no activation of apoptosis as determined by a TUNEL assay (Fig. 6, A–D). An additional approach to analyze apoptotic cell death did not reveal SH-SY5Y cells positive for activated caspase-3 upon parkin/PINK1 down-regulation (Fig. 6F). In conclusion, our comparative analysis revealed that mitochondrial fission induced by parkin or PINK1 loss of function occurs both in human SH-SY5Y cells and insect S2 cells; however, in S2 cells this seems to be an early and transient phenotype.
FIGURE 5.
In Drosophila S2 cells, mitochondrial fragmentation is an early phenotype of parkin and/or PINK1 loss of function. S2 cells grown on glass coverslips were treated with control dsRNA and parkin- and/or PINK1-specific dsRNA. 48 h (day 2), 60 h (day 3), and 72 h (day 4, data not shown) after dsRNA treatment, S2 cells were stained with the fluorescent dye DiOC6(3) to visualize mitochondria and analyzed by fluorescence microscopy. Cells were categorized in three classes according to their mitochondrial morphology. For quantification, the mitochondrial morphology of at least 300 cells per plate was determined in a blinded manner. Quantifications were based on triplicates of at least two independent experiments. Shown is the percentage of cells with a tubular mitochondrial network (white columns), fragmented or truncated mitochondria (black columns), or a dense network of thin mitochondria (gray columns). Lower panel, fluorescence microscopy images to illustrate the different categories of mitochondrial morphologies. Efficiencies of parkin and PINK1 down-regulation is shown in Fig. 6B. **, p ≤ 0.01; ***, p ≤ 0.001.
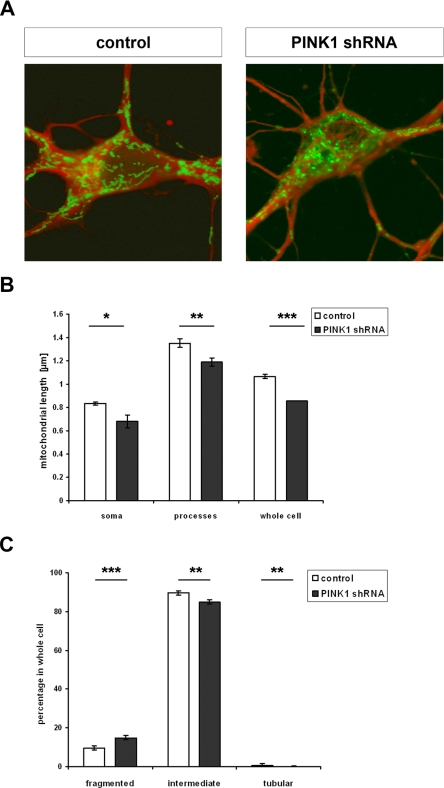
In a next step we included primary mouse hippocampal neurons in our analysis and monitored mitochondrial morphology upon transducing primary neurons by a lentivirus expressing PINK1 shRNA and mito-EYFP (PINK1 shRNA) or mito-EYFP (control, Fig. 7A). 60% of the cells were transduced, and the overall PINK1 knockdown efficiency was 50% (determined by quantitative RT-PCR). First we determined the lengths of mitochondria in primary neurons. To consider possible differences in neuronal soma and processes, we assessed the length of mitochondria in both compartments separately. Primary hippocampal neurons deficient for PINK1 showed a significant decrease in the length of mitochondria both in soma (0.81 μm ± 0.01 in control to 0.68 ± 0.05 μm in PINK1 shRNA, p = 0.014) and processes (1.26 ± 0.04 μm in control to 1.16 ± 0.04 μm in PINK1 shRNA, p = 0.006) (Fig. 7B). Consequently, average mitochondrial length (soma and processes) was significantly decreased in PINK1 knockdown neurons (0.85 ± 0 μm) compared with neurons transduced with the control virus (1.02 ± 0.02 μm; p < 0.001). These differences in mitochondrial length were paralleled by an increase in the percentage of fragmented mitochondria (<0.5 μm; PINK1 shRNA, 15.0 ± 1.12%; control, 9.6 ± 1.03%; p < 0.001) and a decrease in the percentage of intermediate (0.5–5 μm; PINK1 shRNA, 84.9 ± 1.1%; control, 89.7 ± 1.03%; p = 0.002) and tubular mitochondria (>5 μm; PINK1 shRNA, 0.85 ± 0.07%; control, 1.03 ± 1.03%; p = 0.005) in PINK shRNA neurons in comparison to neurons transduced with control virus (Fig. 7C). Thus, our results in primary mouse hippocampal neurons demonstrated a subtle but highly significant mitochondrial phenotype that is consistent with that observed in SH-SY5Y and Drosophila S2 cells.
FIGURE 7.
PINK1-deficient primary mouse hippocampal neurons show a decrease in the length of mitochondria and an increase in mitochondrial fragmentation. A, hippocampal cells of E15.5 C57/BL6 mice were transduced with pLL3.7 + mito-EYFP lentivirus for control or pLL.3.7 + PINK1 shRNA mito-EYFP for down-regulation of PINK1. Mito-EYFP expression was used to determine mitochondrial morphology (green). To visualize neurons, cells were detected with the anti-β III tubulin antibody by immunocytochemistry (red). B, for quantification, the lengths of mitochondria of 40 neurons per group were determined. Shown is the mean mitochondrial length with S.E. in the soma, processes, and in the whole neuron. Down-regulation of PINK1 by shRNA led to significant decrease in mitochondrial length throughout the neuronal cell (soma, p = 0.014; processes, p = 0.006; whole neuron, p < 0.001). C, mitochondria were categorized into fragmented (<0.5 μm), intermediate (0.5–5 μm), and tubular (>5 μm). Shown is the percentage ± S.E. of mitochondria in these categories in whole neurons (n = 40). PINK1 down-regulation via shRNA resulted in a significant increase in fragmented (p < 0.001) mitochondria at the expense of intermediate (p = 0.002) and tubular (p = 0.005) mitochondria.
DISCUSSION
Several studies reported that loss of PINK1 function causes mitochondrial dysfunction (26, 30, 37–47). Parkin has been reported to compensate for PINK1 deficiency in the fly model, and we, therefore, addressed the question of whether parkin itself plays a role in maintaining mitochondrial integrity. We previously could show that parkin is a stress-responsive protein with a wide neuroprotective capacity and that pathogenic mutations and severe proteotoxic stress can induce inactivation of parkin (12, 14, 15, 23). We now present evidence that a loss of parkin function impairs mitochondrial morphology, dynamics, and function. Moreover, the mitochondrial phenotype of parkin-deficient cells is similar to that of PINK1-deficient cells. Parkin- or PINK1-deficient SH-SY5Y cells showed a significant increase in the percentage of cells with truncated or fragmented mitochondria along with a decrease in cellular ATP production. The mitochondrial phenotype could morphologically and functionally be prevented by the enhanced expression of Mfn2, OPA1, or dominant negative Drp1, suggesting that a decrease in mitochondrial fusion or an increase in fission is associated with a loss of parkin or PINK1 function. Several lines of evidence indicated that an increase in mitochondrial fragmentation is responsible for the alterations observed in parkin- or PINK1-deficient cells. First, the mitochondrial phenotype in parkin- or PINK1-deficient cells was not observed in Drp1-deficient cells. Second, parkin as well as PINK1 suppressed mitochondrial fission induced by Drp1. These results are consistent with a recent publication from Sandebring et al. (30) who observed an increase in Drp1-dependent mitochondrial fission in PINK1-deficient human neuroblastoma cells.
A possible role of parkin and PINK1 in modulating mitochondrial morphology/dynamics emerged from recent studies in Drosophila (33–36). In flies, the parkin or PINK1 flight muscle phenotype was suppressed by an increase in mitochondrial fission and a decrease in fusion, leading to the conclusion that the PINK1/parkin pathway promotes mitochondrial fission. Interestingly, we did not observe fragmentation of mitochondria upon overexpression of parkin or PINK1 in human cells; on the contrary, parkin and PINK1 prevented mitochondrial fragmentation induced by Drp1. In line with this observation, the parkin and PINK1 knockdown phenotype in human cells was rescued by increasing mitochondrial fusion and decreasing fission. The parkin/PINK1 studies in flies and mammalian cells appear controversial at first glance. However, our comparative analysis of the consequences of parkin or PINK1 down-regulation in human, mouse, and insect cells revealed that two aspects might be relevant to explain the discrepant findings. First, the time of phenotype analysis seems to be crucial, and second, there could be differences between arthropods and mammals in the regulation of mitochondrial dynamics and/or in the elimination of dysfunctional mitochondria, especially in highly specialized tissues such as flight muscles. Although we looked at an acute manifestation of parkin or PINK1 knockdown in cultured cells, the phenotype observed in adult flies might be influenced by compensatory effects. In support of this scenario, we did observe a significant increase in mitochondrial fragmentation upon parkin or PINK1 knockdown in cultured insect cells, but this was an early and transient phenomenon. Conceptually, flies might try to rescue the parkin/PINK1 null phenotype by activating fusion in an effort to dilute dysfunctional mitochondria. However, in tissues with high energy demands, such as flight muscles, this strategy might not be beneficial in the end, as increased fusion eventually leads to the contamination of the whole mitochondrial network with dysfunctional contents. This might explain why parkin/PINK1 null flies show a phenotype in such tissues that can be rescued by increasing fission, possibly favoring the elimination of dysfunctional mitochondria by mitophagy. In accordance with this concept, a recent study proposed that parkin can promote autophagic clearance of dysfunctional mitochondria (48). Moreover, regulation of the fusion machinery seems to be more complex in mammals compared with insects. Flies do not have the two mitofusins Mfn1 and Mfn2, but have only dmnf/Marf, whereas the expression of Fzo is restricted to the male germ line (49). In addition, regulation of OPA1 function might be different in flies. For example, the presenilin-associated rhomboid-like (PARL) protease, implicated in OPA1 processing, harbors a highly conserved N-terminal regulatory domain in mammals which is not found in insects (24). Phosphorylation of this vertebrate-specific domain inhibits mitochondrial fragmentation, a regulatory mechanism that obviously emerged during vertebrate evolution (24). Consequently, controversial reports on the effects of proteins influencing mitochondrial dynamics may be at least partially attributed to differences in the complex regulation of these proteins.
Our initial observation that PINK1 deficiency causes alterations in mitochondrial morphology in cultured human cells (11) has also been reported by other groups (26, 30, 45, 46), whereas others observed bioenergetic deficits but failed to detect morphological changes (39, 47). This can be explained by the fact that morphological alterations occur early upon parkin/PINK1 down-regulation and are more prominent in a transient knockdown in comparison to a stable knockdown. Accordingly, we observed a decrease in mitochondrial length and connectivity also in primary mouse neurons as an early response to PINK1 down-regulation that can be compensated at later stages, explaining why PINK1 knock-out mice do not show obvious morphological mitochondrial alterations. However, mitochondrial quality control and compensatory mechanisms might not be sufficient to fully restore mitochondrial function, especially in neuronal populations with a low bioenergetic threshold and a high oxidative burden, such as dopaminergic neurons in the substantia nigra.
The next important step will be to address the question of whether parkin/PINK1 play a direct role in the regulation of mitochondrial morphology or dynamics. Recent research revealed that the activity and subcellular localization of Drp1 is regulated by posttranslational modifications, such as phosphorylation, ubiquitylation, and sumoylation (for review, see Ref. 50); therefore, Drp1 would be a prime candidate for such direct regulatory effects. On the other hand, it is also conceivable that parkin or PINK1 exert an indirect effect on mitochondrial morphology by influencing mitochondrial functions, such as complex I activity, or mitochondrial quality control. In this context it will be important to understand the functional interplay between PINK1 and parkin and to focus on compensatory pathways that are induced after PINK1/parkin loss of function.
Supplementary Material
Acknowledgments
We thank Heidi M. McBride for providing the Drp1, Mfn2, and OPA1 plasmids and for helpful discussions, Roger Y. Tsien for providing the mCherry plasmid, and Anita Schlierf, Dominik Paquet, Dr. Richard Page, and Veronika Müller for experimental help. We are grateful to Gerhard Welzl for help with the statistical analysis.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 596 and CEF Macromolecular Complexes), German Ministry for Education and Research (Nationales Genomforschungsnetz plus “Functional Genomics of Parkinson's Disease”), the Helmholtz Alliance Alliance “Mental Health in an Ageing Society,” the Virtual Institute of “Neurodegeneration and Ageing,” the Center for Integrated Protein Science Munich, and the Hans and Ilse Breuer Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- RNAi
- RNA interference
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- DiOC6(3)
- 3,3′-dihexyloxacarbocyanine iodide
- siRNA
- small interfering RNA
- EYFP
- enhanced yellow fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- TUNEL
- TdT-mediated dUTP nick end labeling
- shRNA
- short hairpin RNA
- dsRNA
- double-stranded RNA.
REFERENCES
- 1.Abou-Sleiman P. M., Muqit M. M., Wood N. W. (2006) Nat. Rev. Neurosci. 7,207–219 [DOI] [PubMed] [Google Scholar]
- 2.Lin M. T., Beal M. F. (2006) Nature 443,787–795 [DOI] [PubMed] [Google Scholar]
- 3.Schapira A. H. (2008) Lancet Neurol. 7,97–109 [DOI] [PubMed] [Google Scholar]
- 4.Bogaerts V., Theuns J., van Broeckhoven C. (2008) Genes Brain Behav. 7,129–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodson M. W., Guo M. (2007) Curr. Opin. Neurobiol. 17,331–337 [DOI] [PubMed] [Google Scholar]
- 6.Mandemakers W., Morais V. A., De Strooper B. (2007) J. Cell Sci. 120,1707–1716 [DOI] [PubMed] [Google Scholar]
- 7.Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Nature 441,1162–1166 [DOI] [PubMed] [Google Scholar]
- 9.Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Nature 441,1157–1161 [DOI] [PubMed] [Google Scholar]
- 10.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H. H., Gasser T., Krüger R., Winklhofer K. F., Vogel F., Reichert A. S., Auburger G., Kahle P. J., Schmid B., Haass C. (2007) J. Neurosci. 27,12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winklhofer K. F., Henn I. H., Kay-Jackson P. C., Heller U., Tatzelt J. (2003) J. Biol. Chem. 278,47199–47208 [DOI] [PubMed] [Google Scholar]
- 13.Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M. F., Attardi G., Larsson N. G., Neupert W., Reichert A. S. (2006) J. Biol. Chem. 281,37972–37979 [DOI] [PubMed] [Google Scholar]
- 14.Henn I. H., Bouman L., Schlehe J. S., Schlierf A., Schramm J. E., Wegener E., Nakaso K., Culmsee C., Berninger B., Krappmann D., Tatzelt J., Winklhofer K. F. (2007) J. Neurosci. 27,1868–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henn I. H., Gostner J. M., Lackner P., Tatzelt J., Winklhofer K. F. (2005) J. Neurochem. 92,114–122 [DOI] [PubMed] [Google Scholar]
- 16.Harder Z., Zunino R., McBride H. (2004) Curr. Biol. 14,340–345 [DOI] [PubMed] [Google Scholar]
- 17.Neuspiel M., Zunino R., Gangaraju S., Rippstein P., McBride H. (2005) J. Biol. Chem. 280,25060–25070 [DOI] [PubMed] [Google Scholar]
- 18.Rambold A. S., Miesbauer M., Rapaport D., Bartke T., Baier M., Winklhofer K. F., Tatzelt J. (2006) Mol. Biol. Cell 17,3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22,1567–1572 [DOI] [PubMed] [Google Scholar]
- 20.Consiglio A., Quattrini A., Martino S., Bensadoun J. C., Dolcetta D., Trojani A., Benaglia G., Marchesini S., Cestari V., Oliverio A., Bordignon C., Naldini L. (2001) Nat. Med. 7,310–316 [DOI] [PubMed] [Google Scholar]
- 21.Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y., Nishimura I., Imai Y., Takahashi R., Lu B. (2003) Neuron 37,911–924 [DOI] [PubMed] [Google Scholar]
- 23.Schlehe J. S., Lutz A. K., Pilsl A., Lämmermann K., Grgur K., Henn I. H., Tatzelt J., Winklhofer K. F. (2008) J. Biol. Chem. 283,13771–13779 [DOI] [PubMed] [Google Scholar]
- 24.Jeyaraju D. V., Xu L., Letellier M. C., Bandaru S., Zunino R., Berg E. A., McBride H. M., Pellegrini L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,18562–18567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. (2007) J. Biol. Chem. 282,11521–11529 [DOI] [PubMed] [Google Scholar]
- 26.Weihofen A., Thomas K. J., Ostaszewski B. L., Cookson M. R., Selkoe D. J. (2009) Biochemistry 48,2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darios F., Corti O., Lücking C. B., Hampe C., Muriel M. P., Abbas N., Gu W. J., Hirsch E. C., Rooney T., Ruberg M., Brice A. (2003) Hum. Mol. Genet. 12,517–526 [DOI] [PubMed] [Google Scholar]
- 28.Denison S. R., Wang F., Becker N. A., Schüle B., Kock N., Phillips L. A., Klein C., Smith D. I. (2003) Oncogene 22,8370–8378 [DOI] [PubMed] [Google Scholar]
- 29.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandebring A., Thomas K. J., Beilina A., van der Brug M., Cleland M. M., Ahmad R., Miller D. W., Zambrano I., Cowburn R. F., Behbahani H., Cedazo-Mínguez A., Cookson M. R. (2009) PLoS ONE 4,e5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishihara N., Fujita Y., Oka T., Mihara K. (2006) EMBO J. 25,2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Z., Chen H., Fiket M., Alexander C., Chan D. C. (2007) J. Cell Biol. 178,749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., Lu B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng H., Dodson M. W., Huang H., Guo M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J., Lee G., Chung J. (2009) Biochem. Biophys. Res. Commun. 378,518–523 [DOI] [PubMed] [Google Scholar]
- 37.Gandhi S., Wood-Kaczmar A., Yao Z., Plun-Favreau H., Deas E., Klupsch K., Downward J., Latchman D. S., Tabrizi S. J., Wood N. W., Duchen M. R., Abramov A. Y. (2009) Mol. Cell 33,627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautier C. A., Kitada T., Shen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gegg M. E., Cooper J. M., Schapira A. H., Taanman J. W. (2009) PLoS ONE 4,e4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gispert S., Ricciardi F., Kurz A., Azizov M., Hoepken H. H., Becker D., Voos W., Leuner K., Müller W. E., Kudin A. P., Kunz W. S., Zimmermann A., Roeper J., Wenzel D., Jendrach M., García-Arencíbia M., Fernández-Ruiz J., Huber L., Rohrer H., Barrera M., Reichert A. S., Rüb U., Chen A., Nussbaum R. L., Auburger G. (2009) PLoS ONE 4,e5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunewald A., Gegg M. E., Taanman J. W., King R. H., Kock N., Klein C., Schapira A. H. (2009) Exp. Neurol., in press [DOI] [PubMed] [Google Scholar]
- 42.Hoepken H. H., Gispert S., Morales B., Wingerter O., Del Turco D., Mülsch A., Nussbaum R. L., Müller K., Dröse S., Brandt U., Deller T., Wirth B., Kudin A. P., Kunz W. S., Auburger G. (2007) Neurobiol. Dis. 25,401–411 [DOI] [PubMed] [Google Scholar]
- 43.Liu W., Vives-Bauza C., Acín-Peréz R., Yamamoto A., Tan Y., Li Y., Magrané J., Stavarache M. A., Shaffer S., Chang S., Kaplitt M. G., Huang X. Y., Beal M. F., Manfredi G., Li C. (2009) PLoS ONE 4,e4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marongiu R., Spencer B., Crews L., Adame A., Patrick C., Trejo M., Dallapiccola B., Valente E. M., Masliah E. (2009) J. Neurochem. 108,1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A. Y., Abramov A. S., Miljan E. A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., Downward J., Mansfield L., Jat P., Taylor J., Heales S., Duchen M. R., Latchman D., Tabrizi S. J., Wood N. W. (2008) PLoS ONE 3,e2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dagda R. K., Cherra S. J., 3rd, Kulich S. M., Tandon A., Park D., Chu C. T. (2009) J. Biol. Chem. 284,13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morais V. A., Verstreken P., Roethig A., Smet J., Snellinx A., Vanbrabant M., Haddad D., Frezza C., Mandemakers W., Vogt-Weisenhorn D., Van Coster R., Wurst W., Scorrano L., De Strooper B. (2009) EMBO Mol. Med. 1,99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183,795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwa J. J., Hiller M. A., Fuller M. T., Santel A. (2002) Mech. Dev. 116,213–216 [DOI] [PubMed] [Google Scholar]
- 50.Santel A., Frank S. (2008) IUBMB life 60,448–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.