Abstract
Fibroblasts degrade type I collagen, the major extracellular protein found in mammals, during events ranging from bulk tissue resorption to invasion through the three-dimensional extracellular matrix. Current evidence suggests that type I collagenolysis is mediated by secreted as well as membrane-anchored members of the matrix metalloproteinase (MMP) gene family. However, the roles played by these multiple and possibly redundant, degradative systems during fibroblast-mediated matrix remodeling is undefined. Herein, we use fibroblasts isolated from Mmp13−/−, Mmp8−/−, Mmp2−/−, Mmp9−/−, Mmp14−/− and Mmp16−/− mice to define the functional roles for secreted and membrane-anchored collagenases during collagen-resorptive versus collagen-invasive events. In the presence of a functional plasminogen activator-plasminogen axis, secreted collagenases arm cells with a redundant collagenolytic potential that allows fibroblasts harboring single deficiencies for either MMP-13, MMP-8, MMP-2, or MMP-9 to continue to degrade collagen comparably to wild-type fibroblasts. Likewise, Mmp14−/− or Mmp16−/− fibroblasts retain near-normal collagenolytic activity in the presence of plasminogen via the mobilization of secreted collagenases, but only Mmp14 (MT1-MMP) plays a required role in the collagenolytic processes that support fibroblast invasive activity. Furthermore, by artificially tethering a secreted collagenase to the surface of Mmp14−/− fibroblasts, we demonstrate that localized pericellular collagenolytic activity differentiates the collagen-invasive phenotype from bulk collagen degradation. Hence, whereas secreted collagenases arm fibroblasts with potent matrix-resorptive activity, only MT1-MMP confers the focal collagenolytic activity necessary for supporting the tissue-invasive phenotype.
In the postnatal state, fibroblasts are normally embedded in a self-generated three-dimensional connective tissue matrix composed largely of type I collagen, the major extracellular protein found in mammals (1–3). Type I collagen not only acts as a structural scaffolding for the associated mesenchymal cell populations but also regulates gene expression and cell function through its interactions with collagen binding integrins and discoidin receptors (2, 4). Consistent with the central role that type I collagen plays in defining the structure and function of the extracellular matrix, the triple-helical molecule is resistant to almost all forms of proteolytic attack and can display a decades-long half-life in vivo (4–6). Nonetheless, fibroblasts actively remodel type I collagen during wound healing, inflammation, or neoplastic states (2, 7–13).
To date type I collagenolytic activity is largely confined to a small subset of fewer than 10 proteases belonging to either the cysteine proteinase or matrix metalloproteinase (MMP)2 gene families (4, 14–18). As all collagenases are synthesized as inactive zymogens, complex proteolytic cascades involving serine, cysteine, metallo, and aspartyl proteinases have also been linked to collagen turnover by virtue of their ability to mediate the processing of the pro-collagenases to their active forms (13, 15, 19). After activation, each collagenase can then cleave native collagen within its triple-helical domain, thus precipitating the unwinding or “melting” of the resulting collagen fragments at physiologic temperatures (4, 15). In turn, the denatured products (termed gelatin) are susceptible to further proteolysis by a broader class of “gelatinases” (4, 15). Collagen fragments are then either internalized after binding to specific receptors on the cell surface or degraded to smaller peptides with potent biological activity (20–24).
Previous studies by our group as well as others have identified MMPs as the primary effectors of fibroblast-mediated collagenolysis (20, 25, 26). Interestingly, adult mouse fibroblasts express at least six MMPs that can potentially degrade type I collagen, raising the possibility of multiple compensatory networks that are designed to preserve collagenolytic activity (25). Four of these collagenases belong to the family of secreted MMPs, i.e. MMP-13, MMP-8, MMP-2, and MMP-9, whereas the other two enzymes are members of the membrane-type MMP subgroup, i.e. MMP-14 (MT1-MMP) and MMP-16 (MT3-MMP) (13, 27–29). From a functional perspective, the specific roles that can be assigned to secreted versus membrane-anchored collagenases remain undefined. As such, fibroblasts were isolated from either wild-type mice or mice harboring loss-of-function deletions in each of the major secreted and membrane-anchored collagenolytic genes, and the ability of the cells to degrade type I collagen was assessed. Herein, we demonstrate that fibroblasts mobilize either secreted or membrane-anchored MMPs to effectively degrade type I collagen in qualitatively and quantitatively distinct fashions. However, under conditions where fibroblasts use either secreted and membrane-anchored MMPs to exert quantitatively equivalent collagenolytic activity, only MT1-MMP plays a required role in supporting a collagen-invasive phenotype. These data establish a new paradigm wherein secreted collagenases are functionally limited to bulk collagenolytic processes, whereas MT1-MMP uniquely arms the fibroblast with a focalized degradative activity that mediates subjacent collagenolysis as well as invasion.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Mouse Skin Fibroblasts
Mice with targeted deletions in Mmp14, Mmp2, Mmp9, Mmp8, Mmp13, Mmp16, or Timp2 have been described previously (25, 27, 30–34). Mmp16 (GenBankTM accession number AB021228) was disrupted by replacing sequences from base 814 to base 821 within exon 5 with a LacZ-Neo cassette to generate the Mmp16tmlDgen allele. Frozen embryos were obtained from the Mutant Mouse Regional Resource Center at the University of North Carolina at Chapel Hill. Mmp16 homozygous knock-out mice are maintained on a C57BL/6 background. Mmp16 expression was assessed by RT-PCR using forward primers for mouse Mmp16, 5′-GGAGACAGTTCCCCATTTGA-3′, and reverse primers, 5′-CGTTGGAATGTTCCAGTCCT-3′.
Fibroblasts were isolated from the dorsal skin of 2–8-week-old mice (passage 2–8) and were cultured in Dulbecco's modified Eagle's medium supplemented with either 10% heat-inactivated fetal calf serum (FCS; Atlanta Biologicals) or heat-inactivated mouse serum (from control, Mmp2−/− or Mmp9−/− mice), 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml Fungizone, and 2 mm l-glutamine.
RT-PCR Analysis
Cells were cultured on type I collagen matrix for 2 days in serum-free media in the absence and presence of plasminogen, and total RNA was isolated using TRIzol reagent (Invitrogen). RT-PCR was performed with 1 μg of total RNA and 10 μm concentrations of specific primers (25, 36) using the One-Step RT-PCR System reagent (Invitrogen).
Ecotropic Retrovirus Packaging
Full-length MT1-MMP and deletion mutants (37) were subcloned into the pBMN-Z retroviral vector (Addgene, Cambridge, MA). Retroviral vectors were co-transfected into HEK293 cells with ecotropic packaging plasmids using Lipofectamine 2000 (Invitrogen). Six hours post-transfection, cells were seeded at low density, and retroviral supernatants were collected at 48 h. Supernatants were diluted 1:1 with fibroblast culture medium supplemented with 6 μg/ml Polybrene (Sigma) and added to dermal fibroblast cultures. Cells were infected for 48 h.
Collagen Degradation Assay
Type I collagen degradation was evaluated using a modification of a previously described method (38). In brief, 24-well plates were coated with collagen gel (100 μg/well), and 5 × 104 fibroblasts in 35 μl of medium were seeded in the center of each well and allowed to adhere for 8 h. After washing, 0.5 ml of serum-free medium was added to each well with or without PDGF-BB (10 ng/ml), plasminogen (20 μg/ml), or protease inhibitors as indicated. After 5 days, cells were removed by trypsin/EDTA or detergent lysis, and the remaining collagen film was stained with Coomassie Brilliant Blue. Alternatively, collagen degradation products were quantified by hydroxyproline release into the conditioned medium after an ethanol precipitation step (70% v/v) as described (39). Serum-free media were supplemented with recombinant tissue inhibitor of metalloproteinases-1 or -2 (TIMP-1; 7.5 μg/ml or TIMP-2; 2.5 μg/ml; Fuji Chemical Industries), the synthetic MMP inhibitor, BB-94 (5 μm final concentration in 0.1% DMSO; British Biotechnology, Oxford, UK), the cysteine proteinase inhibitor, E-64d (100 μm; Sigma), the aspartyl proteinase inhibitor, pepstatin A (50 μm; Sigma), or the serine proteinase inhibitors, soybean trypsin inhibitor, SBTI (100 μg/ml), or aprotinin (200 μg/ml; Roche Applied Science).
Invasion Assays
Type I collagen was acid- or pepsin- extracted from rat or mice tail tendons (4, 40). Collagen gels were prepared in 24-mm Transwell dishes (3-μm pore size; Corning, Inc.) at a final concentration of 2.2 mg/ml (40). After gelling (45 min at 37 °C), 1.5–2 × 105 cells in plasminogen-free or plasminogen-supplemented (20 μg/ml) media were added to the upper compartment of the Transwell dishes, and PDGF-BB (10 ng/ml) was added to the lower compartment of the Transwell chambers to initiate fibroblast invasion. Invasive activity was visualized by phase contrast microscopy. The number of invading cells was quantified as the mean ± 1 S.E. of at least three experiments as described (36).
Motility Assay
Fibroblast motility was monitored by culturing 1 × 105 cells atop collagen gels in a 35-μl droplets. After attachment, the loose cells were washed away, and serum-free medium was supplemented with 10% FCS added to initiate a motile response. After 48 h, the cells were stained with toluidine blue. The distance migrated by the advancing front of cells (i.e. 3 or more cells) from the confluent area compared with 0 days was measured in 5 randomly selected fields.
Proliferation
To monitor BrdUrd uptake in proliferating cells, fibroblasts in two-dimensional culture were cultured in 0.5% FCS overnight. Fresh media were then added containing either 0.5% FCS (control), 0.5% FCS supplemented with 10 ng/ml fibroblast growth factor-2, or 10% FCS. After overnight incubation, 10 μm BrdUrd was added for 60 min, after which time the cultures were washed with PBS, fixed, and processed with anti-BrdUrd antibody (Roche Applied Science). The cells were counterstained with propidium iodide (Invitrogen) to determine total cell number. The BrdUrd-positive cells were counted in 10 randomly selected fields and expressed as percent of total cells (mean ± S.E.; n = 3).
Immunofluorescence, Light, and Electron Microscopy
The fluorescence images were captured using Spot digital camera (Diagnostic Instruments, Inc.) through a Leica upright microscope. Collagen cultures were prepared for light microscopy as described (25, 40), and sections (5–7 μm thick) were stained with hematoxylin and eosin. For electron microscopy, gels were fixed in 2% glutaraldehyde, 1.5% paraformaldehyde in 0.1 m sodium cacodylate buffer and processed as described (40, 41).
RESULTS
Pericellular Versus Bulk Collagenolysis by Secreted and Membrane-anchored MMPs
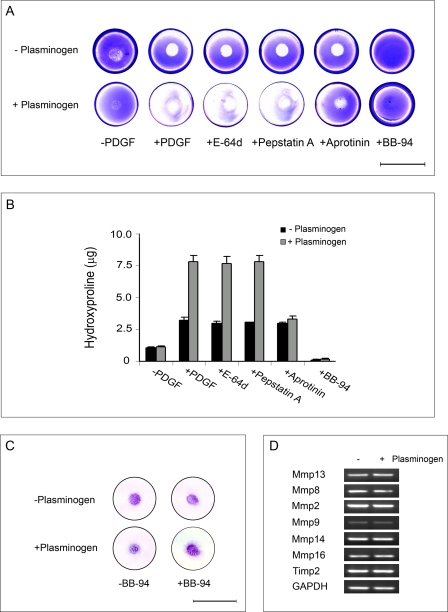
When a concentric island of mouse fibroblasts is established on the surface of a circular, three-dimensional gel of type I collagen and stimulated with PDGF-BB under plasminogen-free conditions, the fibroblasts display increased collagenolytic activity that is confined to the substratum directly subjacent to the area covered by the seeded inoculum of cells (i.e. sites of collagen degradation are visualized as zones of clearing where the underlying gel is no longer observed after staining with Coomassie Blue and de-staining; see Fig. 1A). Although multiple classes of proteolytic enzymes have been linked indirectly to type I collagen degradation (14, 42), fibroblasts stimulated with PDGF-BB in the presence of cysteine (E-64d), aspartyl (pepstatin A), or serine (SBTI or aprotinin) proteinase inhibitors maintain collagenolytic activity as assessed by Coomassie Blue staining or the solubilization of hydroxyproline-containing collagen fragments (Fig. 1, A and B). By contrast, the synthetic MMP inhibitor, BB-94, completely blocks collagenolysis (Fig. 1, A and B).
FIGURE 1.
Profile of plasmin-dependent and independent collagenolytic activity of murine skin fibroblasts. A and B, collagen degradation by mouse skin fibroblasts seeded on a type I collagen gel in the presence or absence of plasminogen as assessed by Coomassie staining (A) or hydroxyproline release (B). In the Coomassie assay fibroblast islands were established in the center of each well, and cells were incubated alone, with PDGF-BB, or with PDGF-BB in the presence of E-64d (100 μm), pepstatin-A (50 μm), aprotinin (20 μg/ml), or BB-94 (5 μm) for 5 days in serum-free media (bar = 20 mm). Hydroxyproline release was monitored under identical conditions without or with PDGF-BB in the absence or presence of inhibitors. Results are shown as the mean ± S.E. of four experiments. C, toluidine blue stain of fibroblast island cultured on collagen gels under serum-free conditions for 5 days in the absence or presence of plasminogen, PDGF-BB, and/or BB-94 (bar = 20 mm). D, RT-PCR analysis of type I collagenases and Timp2 mRNA expression by mouse skin fibroblasts cultured atop type I collagen gels with PDGF-BB for 48 h in the absence or presence of plasminogen. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Although fibroblasts focus collagenolytic activity to the cell-matrix interface under plasminogen-free conditions, these cells can also process plasminogen to plasmin, a potent direct- or indirect- activator of secreted MMP zymogens (43, 44). To determine whether collagen degradation by fibroblasts can be augmented by this serine proteinase axis, PDGF-BB-stimulated cells were cultured atop collagen gels in the presence of plasminogen. Under these conditions plasminogen is processed to plasmin (3.3 ± 0.2 munits/106 cells/24 h) via a plasminogen activator-dependent process (data not shown), which increases collagen degradation 2.5-fold (Fig. 1, A and B). Coincident with the enhancement of collagenolytic activity, the zone of collagen dissolution is extended beyond the confines of the fibroblast island to include the entire surface of the culture dish (Fig. 1, A and B). Although neither E-64d nor pepstatin A affected plasminogen-dependent collagenolysis, the plasmin inhibitor, aprotinin, specifically blocks that portion of the collagen degradation that occurs at sites distant from the seeded cells (herein referred to as “bulk” collagenolysis as opposed to subjacent collagen degradation) while leaving subjacent collagenolysis unaffected (Fig. 1, A and B). Consistent with the conclusion that the enhanced levels of fibroblast-mediated collagen degradation observed in the presence of plasminogen remains dependent on the activity of collagenolytic MMPs, all degradative activity is quenched in the presence of BB-94 (Fig. 1, A and B) without affecting plasmin activity (data not shown). The bulk dissolution of the collagen film observed in the presence of plasminogen does not occur as a consequence of fibroblasts moving from the centrally located inoculation site as the area of collagen film covered by the cells is similar in the absence or presence of either plasminogen or BB-94 (Fig. 1C). Furthermore, the addition of plasminogen does not alter the pattern of Mmp or Timp2 expression (Fig. 1D).
Fibroblast MT1-MMP Activity Defines Plasminogen-independent Collagen Degradation
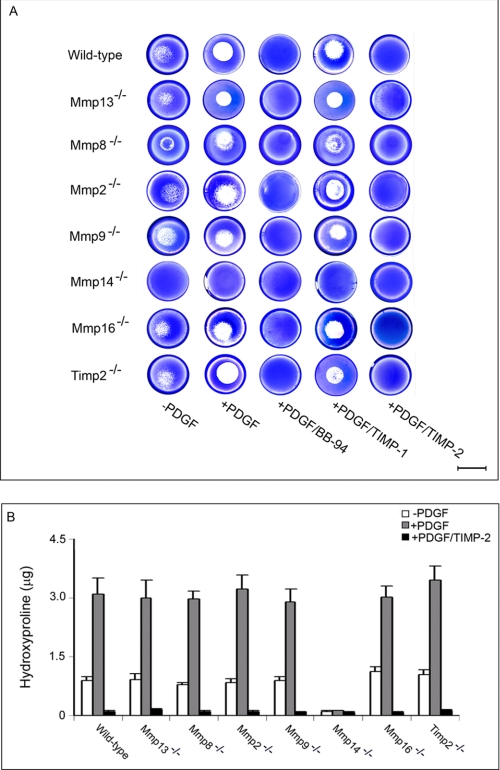
During matrix remodeling events in vivo, fibroblasts express a family of MMP zymogens which potentially participate in collagen degradation by either functioning as direct-acting collagenolysins, promoting MMP activation, or accelerating the further degradation of collagen cleavage fragments (45–49). As previously described (25), fibroblasts stimulated with PDGF-BB atop a collagen substratum express Mmp13, Mmp8, Mmp2, Mmp9, Mmp14, and Mmp16 (Fig. 1D). To identify the individual MMPs, or MMP cascades, that drive the collagenolytic phenotype under plasminogen-free conditions, fibroblasts were isolated from each of the respective null mice, and the ability of resting or PDGF-BB-stimulated cells to degrade type I collagen was assessed. Despite the fact that MMP-13, MMP-8, MMP-2, and MMP-9 can each display collagenolytic activity under cell-free conditions (15, 28, 29), fibroblasts null for any single member of the secreted collagenases degrade collagen comparably to control cells in either the absence or presence of PDGF-BB (Fig. 2, A and B). By contrast, MT1-MMP-deficient fibroblasts display a complete loss of subjacent collagenolytic activity under plasminogen-free conditions (Fig. 2, A and B) while maintaining wild-type levels of Mmp13, Mmp8, Mmp2, Mmp9, and Mmp16 (data not shown). Although MT3-MMP has recently been described as a type I collagenolysin (27), the membrane-anchored enzyme does not play a key role in fibroblast-dependent collagen degradation as the null cells retain full collagenolytic activity (Fig. 2, A and B) without displaying compensatory changes in Mmp14 expression (data not shown). Although endogenously derived TIMP-2 can support MT1-MMP-dependent MMP-2 activation (15, 50, 51), Timp2−/− fibroblasts mount normal collagenolytic responses (Fig. 2, A and B). In each of the null populations studied, save for Mmp14−/− cells, subjacent degradation by fibroblasts remains insensitive to TIMP-1, an endogenous MMP inhibitor that preferentially targets secreted MMPs as well as MT4-MMP and MT6-MMP (52). By contrast, subjacent collagenolysis is blocked completely by BB-94 or TIMP-2, a TIMP family member that inhibits both secreted and membrane-anchored MMPs (Fig. 2, A and B) (25, 52).
FIGURE 2.
MT1-MMP mediates collagen degradation in a plasminogen-independent environment. A and B, collagenolytic activity of primary murine skin fibroblasts isolated from wild-type or Mmp13−/−, Mmp8−/−, Mmp2−/−, Mmp9−/−, Mmp14−/−, Mmp16−/−, or Timp2−/− mice were assessed after a 5-day culture period on type I collagen gels by Coomassie staining (A) or hydroxyproline release (B). In the Coomassie assay, subjacent collagenolysis mediated by fibroblast is limited to MT1-MMP and is blocked by BB-94 (5 μm) or TIMP-2 (2.5 μg/ml) but not by TIMP-1 (7.5 μg/ml) (bar = 10 mm). Hydroxyproline release was monitored under identical conditions without or with PDGF-BB in the absence or presence of TIMP-2. Results are shown as the mean ± S.E. of four experiments.
Plasminogen-dependent Type I Collagen Degradation
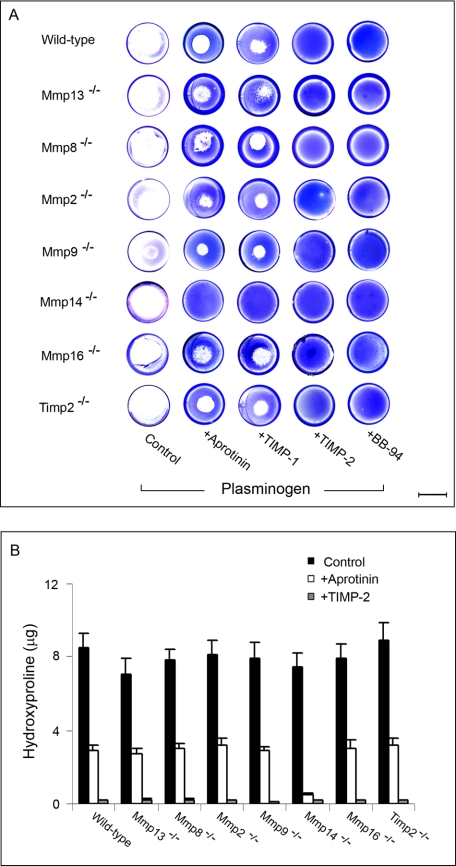
Given the ability of plasmin to augment fibroblast-mediated collagen degradation (see Fig. 1), the collagenolytic potential of each of the MMP-targeted fibroblast populations was reassessed in the presence of exogenous plasminogen. As shown in Fig. 3, type I collagen degradation by wild-type fibroblasts can be divided into two spatial compartments. In the presence of TIMP-1 or aprotinin, collagen degradation is inhibited at sites distant from the fibroblast island seeded in the middle of each well, consistent with the plasmin-dependent activation of secreted collagenases (Fig. 3, A and B). By contrast, proteolysis at the wild-type fibroblast-substratum interface, the site dominated by MT1-MMP-dependent collagenolysis, is not affected by either TIMP-1 or aprotinin and can only be blocked fully by TIMP-2 or BB-94 (Fig. 3A).
FIGURE 3.
Plasmin-dependent collagen degradation. A, collagenolytic activity of PDGF-BB-stimulated wild-type or null fibroblasts seeded atop type I collagen gels was assessed in the presence of plasminogen (20 μg/ml) alone or with plasminogen and either BB-94, TIMP-1, TIMP-2, or aprotinin. Zones of collagenolytic activity are visualized after Coomassie staining after a 5-day culture period (bar = 10 mm). B, hydroxyproline release by PDGF-BB-stimulated wild-type or null fibroblasts cultured atop type I collagen gels was determined in either the absence or presence of aprotinin or TIMP-2. Results are expressed as the mean ± S.E. (n = 4).
In an effort to identify the major secreted collagenase(s) responsible for plasmin-dependent bulk degradation, fibroblasts isolated from Mmp13−/−, Mmp8−/−, Mmp2−/−, Mmp9−/−, or Timp2−/− mice were stimulated with PDGF-BB in the presence of plasminogen, and zones of collagen degradation were visualized by Coomassie Blue staining and quantified as hydroxyproline release (Fig. 3, A and B). Interestingly, each of the null populations degrades the surrounding field of collagen fibrils to a comparable degree via a process sensitive to either aprotinin or TIMP-1 (Fig. 3, A and B). Preliminary studies suggest that MMP-13, MMP-8, and MMP-2 play redundant and compensatory roles in supporting collagenolysis, as bulk collagen degradation is suppressed only after silencing the expression of all three secreted MMPs in tandem (data not shown). Nevertheless, consistent with the ability of MT1-MMP to degrade collagen at the cell-substrate interface, Mmp13-, Mmp8-, Mmp2-, Mmp9-, and Timp2-null fibroblasts continue to express subjacent collagenolytic activity in the presence of aprotinin or TIMP-1 but not TIMP-2 or BB-94 (Fig. 3, A and B).
Mmp14−/− fibroblasts, although unable to mediate subjacent collagenolysis in the absence of plasminogen, express normal levels of Mmp13, Mmp8, Mmp2, Mmp9, and Mmp16 and should retain the ability to activate a complement of their secreted collagenases. Indeed, although MT1-MMP-deficient fibroblasts have previously been characterized as unable to degrade collagen (53), null cells stimulated in the presence of plasminogen degrade the collagen substratum comparably to wild-type cells at sites both subjacent and distant from the fibroblast island (Fig. 3, A and B). As Mmp14−/− fibroblasts are, however, totally reliant on plasmin-activated secreted collagenases for their collagen-degradative activity, collagenolysis is suppressed completely in both subjacent and distant sites by either TIMP-1 or aprotinin (Fig. 3, A and B). Mmp16−/− fibroblasts, by contrast, express a collagenolytic phenotype indistinguishable from wild-type cells in the presence of plasminogen (Fig. 3, A and B). Thus, fibroblasts can mobilize independently operating collagenolytic systems that confer the cells with the ability to use MT1-MMP alone (i.e. in the absence of plasminogen), soluble collagenases alone (i.e. defined by MT1-MMP-deficient fibroblasts stimulated in the presence of plasminogen), or both MT1-MMP and soluble collagenases in tandem (i.e. in wild-type fibroblasts stimulated in the presence of plasminogen).
Fibroblast-derived MMPs and Regulation of the Collagen-invasive Phenotype
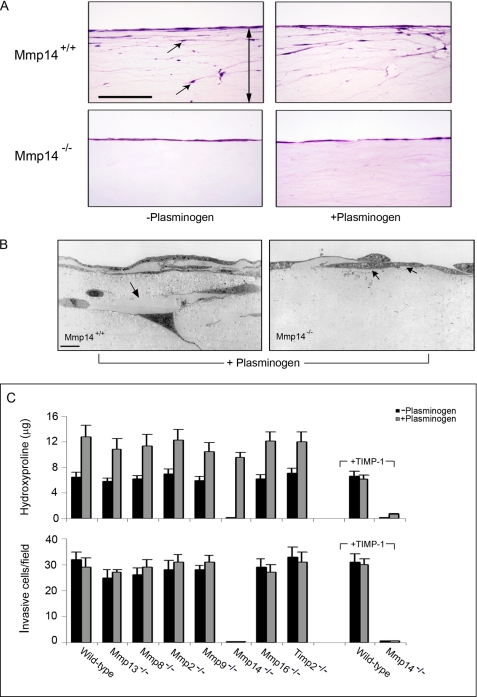
Although fibroblasts mobilize secreted as well as membrane-anchored MMPs to drive type I collagenolytic activity, the ability of these proteolytic systems to support collagen-invasive activity remains controversial (25, 37, 54–57). Hence, wild-type or secreted MMP-, TIMP-2-, or MT3-MMP-deficient fibroblasts were cultured atop three-dimensional type I collagen gels under serum-free conditions in the absence or presence of plasminogen, and invasion was triggered by a chemotactic gradient of PDGF-BB. Under these conditions wild-type fibroblasts display similar, if not identical, invasive activity in the absence or presence of plasminogen despite the fact that collagen degradation is increased ∼2-fold when plasminogen is introduced in the system (Fig. 4, A–C). Although the enhanced collagenolytic activity observed in the presence of plasminogen is suppressed by TIMP-1 (Fig. 4C) or aprotinin (data not shown), invasive activity is unaffected (Fig. 4C). Similarly, each of the fibroblasts deficient for functional MMP-13, MMP-8, MMP-2, MMP-9, TIMP-2, or MT3-MMP displays comparable collagen-invasive activity in the absence or presence of plasminogen (Fig. 4C). By contrast, MT1-MMP-deficient fibroblasts are, as expected, unable to invade collagen gels in the absence of plasminogen (Fig. 4, A–C). Importantly, however, although the collagenolytic activity of Mmp14−/− fibroblasts is increased to nearly wild-type levels in the presence of plasminogen, the null fibroblasts remain unable to express invasive activity as assessed by either light or transmission electron microscopy (Fig. 4, A–C). Hence, although the plasmin-activated, secreted collagenases confer MT1-MMP-deficient fibroblasts with full collagenolytic potential, only MT1-MMP endows fibroblasts with type I collagen-invasive activity.
FIGURE 4.
MT1-MMP regulates collagen-invasive activity. A, Mmp14+/+ or Mmp14−/− fibroblasts were cultured atop type I collagen gels in Transwell culture dishes in serum-free media alone or in serum-free media supplemented with plasminogen (20 μg/ml), and invasion was initiated by the addition of PDGF-BB to the lower chamber of the Transwell dishes. Representative examples of invasion by Mmp14+/+ or Mmp14−/− fibroblasts are shown in hematoxylin and eosin-stained cross-sections (A, bar, 100 μm; the double-headed arrow in the cross-section marks the boundaries of the collagen gel, whereas the black arrows show invasive cells) or assessed by transmission electron microscopy after a 5-day culture period (B). Mmp14+/+ as well as Mmp14−/− fibroblasts are surrounded by zones of collagenolysis in plasminogen-containing medium (black arrows), but in contrast to the wild-type cells, MT1-MMP-deficient−/− fibroblasts fail to invade the underlying matrix (bar, 10 μm). C, in the course of the 5-day culture period, hydroxyproline release and the number of invading cells were quantified in the absence or presence of plasminogen alone or in the absence or presence of plasminogen with TIMP-1. The number of invading cells was determined in 10 randomly selected fields in a single representative experiment of 3 performed, and the supernatants were recovered for hydroxyproline release (mean ± S.E., n = 3).
Tethered Collagenases Drive the Fibroblast Invasion Program
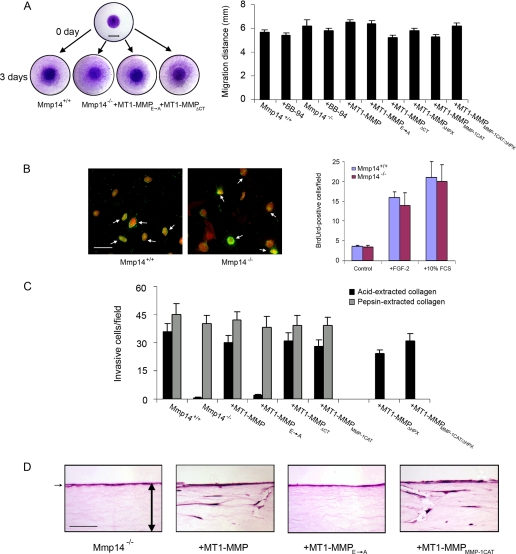
The ability of MT1-MMP to confer pro-invasive activity to fibroblasts is consistent with its pericellular collagenolytic activity, but recent studies suggest that MT1-MMP can exert global effects on cell function independently of its proteolytic potential by triggering a series of signal transduction cascades (51, 58, 59). Hence, to determine whether the loss of invasive activity in MT1-MMP-deficient fibroblasts might be attributed to more generalized defects in cell function, the motile and proliferative activities of the cells were next assessed. Although mouse Mmp14−/− embryonic fibroblasts have been reported to display defects in migration and proliferation under standard, two-dimensional culture conditions (59), wild-type and MT1-MMP-deficient skin fibroblasts migrate at similar rates after stimulation with either fibroblast growth factor-2 or serum (Fig. 5A). Likewise, MT1-MMP-deficient skin fibroblasts proliferate at rates comparable with wild-type fibroblasts (Fig. 5B) and retain full invasive activity in three-dimensional gels of non-cross-linked type I collagen (i.e. pepsin-extracted collagen forms a matrix whose invasion does not require proteolytic activity (25, 54) (Fig. 5C).
FIGURE 5.
Defects in Mmp14−/− dermal fibroblast function are confined to the collagen-invasive phenotype. A, Mmp14+/+, Mmp14−/−, or Mmp14−/− fibroblasts transduced with control or MT1-MMP, MT1-MMPE→A, MT1-MMPΔCT, MT1-MMPΔHPX, MT1-MMPMMP-1CAT, or MT1-MMPMMP-1CAT/ΔHPX retroviral vectors were seeded atop collagen gels as islands in the center of each well and migration monitored in the presence of 10% FCS for 72 h. After 72 h the cells were stained with toluidine blue, and the distance migrated by the advancing front of cells was measured in 5 randomly selected fields (bar = 5 mm). Results are shown for three experiments (mean ± S.E.). B, proliferative activity of Mmp14+/+ or Mmp14−/− skin fibroblasts was measured by BrdUrd uptake in the presence of 0.5% FCS (control), 0.5% FCS, and fibroblast growth factor-2 (10 ng/ml) or 10% FCS as described under “Experimental Procedures.” BrdUrd uptake (green) in cells double-stained with propidium iodide (red) are marked by white arrows (bar, 50 μm) and quantified as the mean ± S.E. in 5 randomly selected fields in a single representative experiment of 3 performed. C and D, invasive activity of Mmp14+/+ or Mmp14−/− skin fibroblasts in acid-extracted, cross-linked type I collagen versus pepsin-extracted, non-cross-linked type I collagen gels. Mmp14+/+, Mmp14−/−, or Mmp14−/− skin fibroblasts transduced with MT1-MMPE→A, MT1-MMPΔCT, MT1-MMPΔHPX, MT1-MMPMMP-1CAT, or MT1-MMPMMP-1CAT/ΔHPX retroviral vectors were seeded atop either acid- or pepsin-extracted collagen gels in the presence of 10% FCS and stimulated with PDGF-BB for a 5-day culture period. The number of invading cells was determined in 10 randomly selected fields in a single representative experiment of 3 performed (C). As shown in hematoxylin and eosin-stained cross-sections, the invasion-null phenotype of Mmp14−/− fibroblasts cultured atop acid-extracted collagen is reversed after transduction with MT1-MMP or MT1-MMPMMP-1CAT but not MT1-MMPE→A retroviral expression vectors (bar = 100 μm). The double-headed arrows mark the boundaries of the collagen gel, whereas the black arrow marks the overlying layer of fibroblasts applied to the surface of the matrix.
Although a required role for MT1-MMP appears restricted to the pro-invasive activity of fibroblasts through native, cross-linked gels of type I collagen, the structural domains that underlie its unique activity relative to the soluble collagenases are largely undefined (37). Despite the fact that MT1-MMP can trigger specific cellular responses via processes independent of the proteinase catalytic activity (51, 59–62), the collagen-invasive activity of Mmp14−/− fibroblasts is not rescued after retroviral transduction with a catalytically inactive form of MT1-MMP harboring an E240A point mutation (MT1-MMPE→A) (Fig. 5, C and D). Furthermore, although the MT1-MMP hemopexin or cytosolic tail domains have been linked to the activation of various signal transduction cascades (51, 59–62), Mmp14-null fibroblasts transfected with MT1-MMP mutants lacking either the hemopexin domain (MT1-MMPΔHPX) or cytosolic tail (MT1-MMPΔCT) display an invasive activity comparable with that observed in wild-type fibroblasts (Fig. 5, C and D). Finally, consistent with the premise that MT1-MMP confers fibroblasts with collagen-invasive activity by displaying pericellular collagenolytic activity alone, the invasion-null phenotype of Mmp14−/− fibroblasts is rescued after transfection with MT1-MMP mutant constructs wherein the wild-type catalytic domain is replaced with that of human MMP-1 (MT1-MMPMMP-1CAT) or an MT1-MMP double-mutant containing the MMP-1 catalytic domain with the MT1-MMP hemopexin domain deleted (MT1-MMPMMP-1CAT/ΔHPX) (Fig. 5, C and D). Given the ability of a membrane-tethered, MMP-1 catalytic domain to support fibroblast invasive activity in the absence of an MT1-MMP hemopexin domain, we conclude that the unique ability of MT1-MMP to confer fibroblasts with collagen pro-invasive activity can be ascribed to the distinct properties of the enzyme as a membrane-anchored type I collagenase.
DISCUSSION
Fibroblasts express multiple proteolytic systems that have been proposed to participate in collagen remodeling events associated with both matrix-resorptive and matrix-invasive states (15, 17, 25, 26, 63–66). Within the mouse MMP family, collagenases include MMP-13, MMP-8, and MMP-2, which all cleave collagen between Gly-775 and Ile-776 in the homodimeric α1 chains and between Gly-775 and Leu-776 in the single α2 chain (67, 68). More recent data suggest that MMP-9 may also express type I collagenolytic activity under physiologic conditions (28). After hydrolysis, the triple-helical fragments unwind to generate gelatin whose exposed motifs are susceptible to further degradation by the collagenases themselves as well as MMP-9, MMP-2, and other MMP family members (15). In addition to the secreted collagenases and gelatinases, MT1-MMP can cleave both collagen at sites identical to the secreted collagenases and degrade gelatin as well (37, 40, 69, 70).
Given the possible collaborations between secreted and membrane-anchored collagenases, we first sought to characterize the major collagen-degradative systems operative in fibroblasts. In the absence of plasminogen, most MMP family members are secreted as inactive zymogens (13, 15, 19). However, a subset of MMPs, including MT1-MMP, undergo intracellular processing by serine proteinases belonging to the proprotein convertase family to generate active proteinases (13, 71–74). In turn, MT1-MMP is able to initiate the processing of either the MMP-2, MMP-13, or MMP-8 zymogens to their active forms (15, 34, 51, 75, 76). However, under plasminogen-free conditions, only MT1-MMP plays a required role in mediating the expression of pericellular collagenolytic activity in fibroblasts (20, 25, 26). Furthermore, the dissolution of collagen to the small Mr peptides detectable by hydroxyproline assay did not require MMP-2 or MMP-9 (77–79). Although MT1-MMP armed fibroblasts with the ability to degrade collagen independently of MMP-2, MMP-9, MMP-13, MMP-8, or TIMP-2, the possibility exists that MT1-MMP could function in concert with other, as yet uncharacterized proteinases. However, purified MT1-MMP cleaves collagen directly (69) and confers collagenolytic-incompetent COS cells with potent degradative activity (37).
Though wild-type fibroblasts are unable to mediate collagenolysis under plasminogen-free conditions by engaging any of the targeted MMPs tested, save for MT1-MMP, the plasminogen activator-plasminogen cascade can directly or indirectly initiate the activation of MMP-13, MMP-8, MMP-2, MMP-9, and MMP-3 (21, 38, 44, 80–82). Consistent with the added participation of the plasmin-activated, secreted collagenases to the pericellular collagenolytic activity conferred by MT1-MMP, degradation extended beyond the borders of the seeded, fibroblast island. These data clearly illustrate the presence of a potent plasmin/secreted collagenase axis, but the individual contributions made by each of the soluble collagenases during collagen fibril dissolution remains to be determined. MMP-13, MMP-8, MMP-2, or MMP-9 can each degrade type I collagen in a TIMP-1-sensitive fashion, but in the presence of plasminogen, Mmp13−/−, Mmp8−/−, Mmp2−/−, or Mmp9−/− fibroblasts were each able to degrade collagen to an equivalent degree. Although the co-activation of multiple secreted collagenases likely compensates for a single deficiency in a given MMP, a fourth secreted collagenase, termed mColA, has been identified in mice and rats (32). The function of mColA in intact fibroblasts has not been defined, but the proteinase expresses only weak collagenolytic activity relative to other collagenases (32), and its expression is restricted to the testes of adult animals (83). Furthermore, mColA-transfected COS cells do not express collagenolytic activity.3 Independent of any specific role for the mouse orthologue, the existence of a redundant set of secreted collagenases is consistent with the fact that neither Mmp13-, Mmp8-, Mmp9-, nor Mmp2-null mice display major defects in type I collagen turnover in the postnatal state (67). Our findings raise the possibility that critical disturbances in bulk remodeling of type I collagen may only be observed under conditions wherein multiple members of the secreted collagenase MMP family are inactivated in tandem.
The ability of fibroblasts to mobilize multiple collagenolytic systems to resorb collagen is a process distinct from that required to allow cells to proteolytically tunnel through the extracellular matrix while maintaining adhesive interactions that support propulsive movement (25, 37, 50, 54). Other reports have implicated multiple MMPs in extracellular matrix-invasive behavior, including secreted collagenases, stromelysin-1, and the gelatinases MMP-2 and MMP-9 (9, 50, 55, 78, 84–87). However, with increasing frequency, these proteases have been found to alter cell function indirectly by cleaving cell adhesion molecules, activating latent growth factors, or initiating signal transduction cascades (13, 88). As previously reported, only MT1-MMP plays a required role in mediating the collagen-invasive activity of fibroblasts (25), even under conditions where secreted collagenase continued to express significant degradative activity. Despite indirect evidence supporting a role for a trimolecular MT1-MMP·TIMP-2·MMP-2 complex or MT1-MMP·MMP-13 axis in collagen-invasive activity (13, 15), fibroblasts require neither MMP-2, TIMP-2, nor MMP-13 to infiltrate collagen barriers. The inability of soluble collagenases to either participate in the invasive process or “rescue” the MT1-MMP-null phenotype may at first glance seem surprising. However, secreted collagenases, although effective in degrading collagen, are unable to restrict their activity to the immediate pericellular environment in a fashion consistent with an invasive phenotype. Secreted collagenases could conceivably participate in tissue-invasive processes by associating with cell surface binding partners (19), but neither latent nor active MMP-13, MMP-8, MMP-2, or MMP-9 was able to rescue the MT1-MMP-null phenotype when expressed in an endogenous fashion by the fibroblast itself.
A priori, the unique ability of MT1-MMP, as opposed to the secreted collagenases, to imbue fibroblasts with collagen-invasive activity is equally consistent with the enzyme's singular presentation as a membrane-tethered collagenase or the possibility that MT1-MMP expresses additional functions distinct from those exhibited by the soluble collagenases (37). In this regard, recent studies indicate that MT1-MMP recognizes a number of membrane-anchored targets whose hydrolysis might potentially alter cell function (13, 51, 58). However, we find that the invasion-null status of MT1-MMP-deficient fibroblasts can be rescued when the catalytic domain of MT1-MMP is replaced with that of human MMP-1. Although similar results have recently been reported using COS cells as a platform for assessing MT1-MMP function (37), COS cells do not express the full complement of secreted collagenases synthesized by stimulated fibroblasts. Whereas the MT1-MMP hemopexin domain might conceivably alter the substrate specificity of the MT1-MMP/MMP-1 chimera (37), MT1-MMP-deficient fibroblasts retain a pro-invasive status even when engineered to express a hemopexin domain-deleted form of membrane-tethered MMP-1. As mice do not express an orthologue of human MMP-1 (32), it seems unlikely that our chimeric constructs fortuitously recapture some unique property of MT1-MMP other than its ability to focus type I collagenolytic activity to the cell surface. Nevertheless, MT1-MMP mutants devoid of catalytic activity can trigger functionally important signal transduction cascades via processes dependent on its short, 20-amino acid-long cytosolic tail (51, 59–62). Indeed, D'Alessio et al. (59) have reported that MT1-MMP regulates the two-dimensional motility and proliferative responses of mouse embryonic fibroblasts via a mechanism dependent on the cytosolic tail of the protease. Using conditions identical to those reported in their study, however, we find that MT1-MMP-deficient dermal fibroblasts obtained from newborn mice display motile and proliferative responses identical to wild-type controls. Furthermore, the motile and proliferative activities of our Mmp14−/− fibroblasts were unaffected after transduction with wild-type MT1-MMP, catalytically inactive MT1-MMP, or cytosolic tail-deleted MT1-MMP. MT1-MMP can clearly initiate signal transduction cascades in a variety of cell populations (51, 59–62), but a central function for these pathways must be considered from the perspective that Mmp14−/− mice develop normally in utero (53). Indeed, these mice only display defects in growth and development in the postnatal state when type I collagen tissue levels begin to increase dramatically, observations consistent with the contention that the loss of pericellular collagenolytic activity underlies the major pathologies that develop in these animals (25, 53, 89, 90). More recently, MT3-MMP has been reported to express type I collagenolytic activity when expressed in an intact cell system (27), but in our hands, the protease express little, if any, collagenolytic activity relative to MT1-MMP (37). In addition, we note that purified MT3-MMP has not been shown to cleave native type I collagen into ¾ and ¼ fragments at 25 °C, although it can hydrolyze the Gly-4—Ile-5 bond in the N-terminal telopeptide of α2(I) chains (91). Independent of these caveats, a precise determination of the role of active MT3-MMP in fibroblasts is further complicated by the fact that the 5′-untranslated region of the Mmp16 mRNA can repress translation in eukaryotic systems (92). Hence, the amount of active MT3-MMP expressed in fibroblasts relative to MT1-MMP awaits the development of specific antibody probes. Nevertheless, our studies demonstrate that MT3-MMP does not play a key role in regulating mouse fibroblast function either in serum-free or serum-containing systems after PDGF-BB stimulation. These results are, in any case, consistent with the more subtle effects observed after Mmp16 versus Mmp14 silencing in vivo (27).
In sum, we have demonstrated that fibroblasts employ both the secreted and membrane-anchored arms of the MMP family in collaborative fashion to effect a collagen-degradative phenotype. Although MT1-MMP collagenolytic activity is confined to the subjacent compartment, the secreted collagenases have an extended range of action that allows them to degrade collagen at sites distant from their point of secretion. While we have used plasmin as a “model” MMP activator, studies performed in plasminogen-deficient mice indicate that other serine proteinases play more important roles in vivo (43, 44). Nevertheless, from a quantitative perspective, secreted collagenases are more likely to play important roles in the bulk resorptive events that distinguish tissue remodeling events such as abscess formation, post-pregnancy uterine involution, post-weaning mammary gland involution, or regression of neovascular beds (82, 93). By contrast, although the MT1-MMP site of action is constrained to the cell surface, the protease resides in a sequestered environment that allows it to remain functional in the presence of plasma antiproteinases that rapidly quench the activity of the secreted collagenases (25). Last, and perhaps most importantly, the ability of MT1-MMP rather than secreted collagenases to serve as a dual-function collagenolysin and pro-invasive factor identifies this alternate class of MMPs as a prototype for sculpting proteolytic activity to the demands of traversing extracellular matrix barriers in the in vivo setting.
Acknowledgments
We thank Robert M. Senior and J. Michael Shipley (Washington University, St. Louis, MO) for providing access to the Mmp9-null mice, S. Itohara (RIKEN Brain Science Institute, Japan) for the Mmp2-null mice, Carlos Lopez-Otin (Universidad de Oviedo, Spain) for access to Mmp8-null mice, and Stephen Krane and Masaki Inada (Harvard) for access to Mmp13-null mice. We acknowledge Galina Gavrilina and the Transgenic Animal Model Core of the University of Michigan Biomedical Research Core Facilities for recovery of B6;129P2-Mmp16tm1Dgen mice from frozen embryos. Core support was provided by National Institutes of Health Grants CA046592, AR048310, and DK034933.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA071699 and CA088308 (to S. J. W.).
F. Sabeh and S. J. Weiss, unpublished observation.
- MMP
- matrix metalloproteinase
- RT
- reverse transcription
- FCS
- fetal calf serum
- PDGF
- platelet-derived growth factor
- BrdUrd
- bromodeoxyuridine.
REFERENCES
- 1.Sweeney S. M., Orgel J. P., Fertala A., McAuliffe J. D., Turner K. R., Di Lullo G. A., Chen S., Antipova O., Perumal S., Ala-Kokko L., Forlino A., Cabral W. A., Barnes A. M., Marini J. C., San Antonio J. D. (2008) J. Biol. Chem. 283,21187–21197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckes B., Zigrino P., Kessler D., Holtkötter O., Shephard P., Mauch C., Krieg T. (2000) Matrix Biol. 19,325–332 [DOI] [PubMed] [Google Scholar]
- 3.Grinnell F. (2003) Trends Cell Biol. 13,264–269 [DOI] [PubMed] [Google Scholar]
- 4.Birkedal-Hansen H. (1987) Methods Enzymol. 144,140–171 [DOI] [PubMed] [Google Scholar]
- 5.Bank R. A., TeKoppele J. M., Oostingh G., Hazleman B. L., Riley G. P. (1999) Ann. Rheum. Dis. 58,35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perumal S., Antipova O., Orgel J. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costea D. E., Kulasekara K., Neppelberg E., Johannessen A. C., Vintermyr O. K. (2006) Am. J. Pathol. 168,1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Wu H., Byrne M., Jeffrey J., Krane S., Jaenisch R. (1995) J. Cell Biol. 130,227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnsen M., Lund L. R., Rømer J., Almholt K., Danø K. (1998) Curr. Opin. Cell Biol. 10,667–671 [DOI] [PubMed] [Google Scholar]
- 10.Singer A. J., Clark R. A. (1999) N. Engl. J. Med. 341,738–746 [DOI] [PubMed] [Google Scholar]
- 11.Arora P. D., Manolson M. F., Downey G. P., Sodek J., McCulloch C. A. (2000) J. Biol. Chem. 275,35432–35441 [DOI] [PubMed] [Google Scholar]
- 12.Crawford Y., Kasman I., Yu L., Zhong C., Wu X., Modrusan Z., Kaminker J., Ferrara N. (2009) Cancer Cell 15,21–34 [DOI] [PubMed] [Google Scholar]
- 13.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8,221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creemers L. B., Hoeben K. A., Jansen D. C., Buttle D. J., Beertsen W., Everts V. (1998) Matrix Biol. 16,575–584 [DOI] [PubMed] [Google Scholar]
- 15.Brinckerhoff C. E., Matrisian L. M. (2002) Nat. Rev. Mol. Cell Biol. 3,207–214 [DOI] [PubMed] [Google Scholar]
- 16.Li Z., Hou W. S., Escalante-Torres C. R., Gelb B. D., Bromme D. (2002) J. Biol. Chem. 277,28669–28676 [DOI] [PubMed] [Google Scholar]
- 17.Ghersi G., Dong H., Goldstein L. A., Yeh Y., Hakkinen L., Larjava H. S., Chen W. T. (2002) J. Biol. Chem. 277,29231–29241 [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Yasuda Y., Li W., Bogyo M., Katz N., Gordon R. E., Fields G. B., Brömme D. (2004) J. Biol. Chem. 279,5470–5479 [DOI] [PubMed] [Google Scholar]
- 19.Ra H. J., Parks W. C. (2007) Matrix Biol. 26,587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen D. H., Engelholm L. H., Ingvarsen S., Hillig T., Wagenaar-Miller R. A., Kjøller L., Gårdsvoll H., Høyer-Hansen G., Holmbeck K., Bugge T. H., Behrendt N. (2007) J. Biol. Chem. 282,27037–27045 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Zhou Z. H., Bugge T. H., Wahl L. M. (2007) J. Immunol. 179,3297–3304 [DOI] [PubMed] [Google Scholar]
- 22.Gaggar A., Jackson P. L., Noerager B. D., O'Reilly P. J., McQuaid D. B., Rowe S. M., Clancy J. P., Blalock J. E. (2008) J. Immunol. 180,5662–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Reilly P. J., Gaggar A., Blalock J. E. (2008) Curr. Opin. Pharmacol. 8,242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin M., Jackson P., Tester A. M., Diaconu E., Overall C. M., Blalock J. E., Pearlman E. (2008) Am. J. Pathol. 173,144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabeh F., Ota I., Holmbeck K., Birkedal-Hansen H., Soloway P., Balbin M., Lopez-Otin C., Shapiro S., Inada M., Krane S., Allen E., Chung D., Weiss S. J. (2004) J. Cell Biol. 167,769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H., Overall C. M., McCulloch C. A., Sodek J. (2006) Mol. Biol. Cell 17,4812–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J., Son M. Y., Yamada S., Szabova L., Kahan S., Chrysovergis K., Wolf L., Surmak A., Holmbeck K. (2008) Dev. Biol. 313,196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bigg H. F., Rowan A. D., Barker M. D., Cawston T. E. (2007) FEBS J. 274,1246–1255 [DOI] [PubMed] [Google Scholar]
- 29.Gioia M., Monaco S., Fasciglione G. F., Coletti A., Modesti A., Marini S., Coletta M. (2007) J. Mol. Biol. 368,1101–1113 [DOI] [PubMed] [Google Scholar]
- 30.Itoh T., Tanioka M., Yoshida H., Yoshioka T., Nishimoto H., Itohara S. (1998) Cancer Res. 58,1048–1051 [PubMed] [Google Scholar]
- 31.Liu Z., Shipley J. M., Vu T. H., Zhou X., Diaz L. A., Werb Z., Senior R. M. (1998) J. Exp. Med. 188,475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balbín M., Fueyo A., Knäuper V., López J. M., Alvarez J., Sánchez L. M., Quesada V., Bordallo J., Murphy G., López-Otín C. (2001) J. Biol. Chem. 276,10253–10262 [DOI] [PubMed] [Google Scholar]
- 33.Inada M., Wang Y., Byrne M. H., Rahman M. U., Miyaura C., López-Otín C., Krane S. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,17192–17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., Juttermann R., Soloway P. D. (2000) J. Biol. Chem. 275,26411–26415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deleted in proof
- 36.Hotary K. B., Yana I., Sabeh F., Li X. Y., Holmbeck K., Birkedal-Hansen H., Allen E. D., Hiraoka N., Weiss S. J. (2002) J. Exp. Med. 195,295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X. Y., Ota I., Yana I., Sabeh F., Weiss S. J. (2008) Mol. Biol. Cell 19,3221–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netzel-Arnett S., Mitola D. J., Yamada S. S., Chrysovergis K., Holmbeck K., Birkedal-Hansen H., Bugge T. H. (2002) J. Biol. Chem. 277,45154–45161 [DOI] [PubMed] [Google Scholar]
- 39.Creemers L. B., Jansen D. C., van Veen-Reurings A., van den Bos T., Everts V. (1997) Biotechniques 22,656–658 [DOI] [PubMed] [Google Scholar]
- 40.Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) J. Cell Biol. 149,1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiraoka N., Allen E., Apel I. J., Gyetko M. R., Weiss S. J. (1998) Cell 95,365–377 [DOI] [PubMed] [Google Scholar]
- 42.Cox S. W., Eley B. M., Kiili M., Asikainen A., Tervahartiala T., Sorsa T. (2006) Oral Dis. 12,34–40 [DOI] [PubMed] [Google Scholar]
- 43.Almholt K., Juncker-Jensen A., Green K. A., Solberg H., Lund L. R., Romer J. (2008) in The Cancer Degradome ( Edwards D., Hoyer-Hansen G., Blasi F., Sloane B. F. eds) pp. 203–222, Springer Science, New York [Google Scholar]
- 44.Green K. A., Almholt K., Ploug M., Rønø B., Castellino F. J., Johnsen M., Bugge T. H., Rømer J., Lund L. R. (2008) J. Invest. Dermatol. 128,2092–2101 [DOI] [PubMed] [Google Scholar]
- 45.Hanemaaijer R., Sorsa T., Konttinen Y. T., Ding Y., Sutinen M., Visser H., van Hinsbergh V. W., Helaakoski T., Kainulainen T., Rönkä H., Tschesche H., Salo T. (1997) J. Biol. Chem. 272,31504–31509 [DOI] [PubMed] [Google Scholar]
- 46.Okada A., Tomasetto C., Lutz Y., Bellocq J. P., Rio M. C., Basset P. (1997) J. Cell Biol. 137,67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohan R., Rinehart W. B., Bargagna-Mohan P., Fini M. E. (1998) J. Biol. Chem. 273,25903–25914 [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto H., Flannery M. L., Kupriyanov S., Pearce J., McKercher S. R., Henkel G. W., Maki R. A., Werb Z., Oshima R. G. (1998) Genes Dev. 12,1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu N., Opalenik S., Liu J., Jansen E. D., Giro M. G., Davidson J. M. (2002) Matrix Biol. 21,149–161 [DOI] [PubMed] [Google Scholar]
- 50.Murphy G., Gavrilovic J. (1999) Curr. Opin. Cell Biol. 11,614–621 [DOI] [PubMed] [Google Scholar]
- 51.Itoh Y., Seiki M. (2006) J. Cell. Physiol. 206,1–8 [DOI] [PubMed] [Google Scholar]
- 52.Nagase H., Visse R., Murphy G. (2006) Cardiovasc. Res. 69,562–573 [DOI] [PubMed] [Google Scholar]
- 53.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) Cell 99,81–92 [DOI] [PubMed] [Google Scholar]
- 54.Sabeh F., Shimizu-Hirota R., Weiss S. J. (2009) J. Cell Biol. 185,11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benbow U., Schoenermark M. P., Mitchell T. I., Rutter J. L., Shimokawa K., Nagase H., Brinckerhoff C. E. (1999) J. Biol. Chem. 274,25371–25378 [DOI] [PubMed] [Google Scholar]
- 56.Jiang X., Dutton C. M., Qi W. N., Block J. A., Garamszegi N., Scully S. P. (2005) J. Cell. Physiol. 202,723–730 [DOI] [PubMed] [Google Scholar]
- 57.Wyatt C. A., Geoghegan J. C., Brinckerhoff C. E. (2005) Cancer Res. 65,11101–11108 [DOI] [PubMed] [Google Scholar]
- 58.Barbolina M. V., Stack M. S. (2008) Semin. Cell Dev. Biol. 19,24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Alessio S., Ferrari G., Cinnante K., Scheerer W., Galloway A. C., Roses D. F., Rozanov D. V., Remacle A. G., Oh E. S., Shiryaev S. A., Strongin A. Y., Pintucci G., Mignatti P. (2008) J. Biol. Chem. 283,87–99 [DOI] [PubMed] [Google Scholar]
- 60.Cao J., Kozarekar P., Pavlaki M., Chiarelli C., Bahou W. F., Zucker S. (2004) J. Biol. Chem. 279,14129–14139 [DOI] [PubMed] [Google Scholar]
- 61.Anilkumar N., Uekita T., Couchman J. R., Nagase H., Seiki M., Itoh Y. (2005) FASEB J. 19,1326–1328 [DOI] [PubMed] [Google Scholar]
- 62.Nyalendo C., Beaulieu E., Sartelet H., Michaud M., Fontaine N., Gingras D., Béliveau R. (2008) Carcinogenesis 29,1655–1664 [DOI] [PubMed] [Google Scholar]
- 63.Tu C., Ortega-Cava C. F., Chen G., Fernandes N. D., Cavallo-Medved D., Sloane B. F., Band V., Band H. (2008) Cancer Res. 68,9147–9156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S., Chow L. H., Pickering J. G. (2000) J. Biol. Chem. 275,35384–35392 [DOI] [PubMed] [Google Scholar]
- 65.Rünger T. M., Quintanilla-Dieck M. J., Bhawan J. (2007) J. Invest. Dermatol. 127,293–297 [DOI] [PubMed] [Google Scholar]
- 66.Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J. F., Harrington K., Sahai E. (2007) Nat. Cell Biol. 9,1392–1400 [DOI] [PubMed] [Google Scholar]
- 67.Krane S. M., Inada M. (2008) Bone 43,7–18 [DOI] [PubMed] [Google Scholar]
- 68.Beare A. H., O'Kane S., Krane S. M., Ferguson M. W. (2003) J. Invest. Dermatol. 120,153–163 [DOI] [PubMed] [Google Scholar]
- 69.Ohuchi E., Imai K., Fujii Y., Sato H., Seiki M., Okada Y. (1997) J. Biol. Chem. 272,2446–2451 [DOI] [PubMed] [Google Scholar]
- 70.Ellerbroek S. M., Wu Y. I., Overall C. M., Stack M. S. (2001) J. Biol. Chem. 276,24833–24842 [DOI] [PubMed] [Google Scholar]
- 71.Pei D., Weiss S. J. (1995) Nature 375,244–247 [DOI] [PubMed] [Google Scholar]
- 72.Yana I., Weiss S. J. (2000) Mol. Biol. Cell 11,2387–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato H., Kinoshita T., Takino T., Nakayama K., Seiki M. (1996) FEBS Lett. 393,101–104 [DOI] [PubMed] [Google Scholar]
- 74.Pei D., Weiss S. J. (1996) J. Biol. Chem. 271,9135–9140 [DOI] [PubMed] [Google Scholar]
- 75.Holopainen J. M., Moilanen J. A., Sorsa T., Kivelä-Rajamäki M., Tervahartiala T., Vesaluoma M. H., Tervo T. M. (2003) Invest. Ophthalmol. Vis. Sci. 44,2550–2556 [DOI] [PubMed] [Google Scholar]
- 76.Knäuper V., Bailey L., Worley J. R., Soloway P., Patterson M. L., Murphy G. (2002) FEBS Lett. 532,127–130 [DOI] [PubMed] [Google Scholar]
- 77.Kerkvliet E. H., Docherty A. J., Beertsen W., Everts V. (1999) Matrix Biol. 18,373–380 [DOI] [PubMed] [Google Scholar]
- 78.Ramos-DeSimone N., Hahn-Dantona E., Sipley J., Nagase H., French D. L., Quigley J. P. (1999) J. Biol. Chem. 274,13066–13076 [DOI] [PubMed] [Google Scholar]
- 79.Atkinson S. J., Patterson M. L., Butler M. J., Murphy G. (2001) FEBS Lett. 491,222–226 [DOI] [PubMed] [Google Scholar]
- 80.Carmeliet P., Collen D. (1998) Thromb. Res. 91,255–285 [DOI] [PubMed] [Google Scholar]
- 81.Milner J. M., Elliott S. F., Cawston T. E. (2001) Arthritis Rheum. 44,2084–2096 [DOI] [PubMed] [Google Scholar]
- 82.Saunders W. B., Bayless K. J., Davis G. E. (2005) J. Cell Sci. 118,2325–2340 [DOI] [PubMed] [Google Scholar]
- 83.Nuttall R. K., Sampieri C. L., Pennington C. J., Gill S. E., Schultz G. A., Edwards D. R. (2004) FEBS Lett. 563,129–134 [DOI] [PubMed] [Google Scholar]
- 84.Kim J., Yu W., Kovalski K., Ossowski L. (1998) Cell 94,353–362 [DOI] [PubMed] [Google Scholar]
- 85.Vu T. H., Werb Z. (2000) Genes Dev. 14,2123–2133 [DOI] [PubMed] [Google Scholar]
- 86.Yu Q., Stamenkovic I. (1999) Genes Dev. 13,35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blackburn J. S., Rhodes C. H., Coon C. I., Brinckerhoff C. E. (2007) Cancer Res. 67,10849–10858 [DOI] [PubMed] [Google Scholar]
- 88.Rowe R. G., Weiss S. J. (2008) Trends Cell Biol. 18,560–574 [DOI] [PubMed] [Google Scholar]
- 89.Zhou Z., Apte S. S., Soininen R., Cao R., Baaklini G. Y., Rauser R. W., Wang J., Cao Y., Tryggvason K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oblander S. A., Zhou Z., Gálvez B. G., Starcher B., Shannon J. M., Durbeej M., Arroyo A. G., Tryggvason K., Apte S. S. (2005) Dev. Biol. 277,255–269 [DOI] [PubMed] [Google Scholar]
- 91.Shimada T., Nakamura H., Ohuchi E., Fujii Y., Murakami Y., Sato H., Seiki M., Okada Y. (1999) Eur. J. Biochem. 262,907–914 [DOI] [PubMed] [Google Scholar]
- 92.Morris M. J., Basu S. (2009) Biochemistry 48,5313–5319 [DOI] [PubMed] [Google Scholar]
- 93.Davis G. E., Saunders W. B. (2006) J. Investig. Dermatol. Symp. Proc. 11,44–56 [DOI] [PubMed] [Google Scholar]