Abstract
Mitosis is an orchestration of dynamic interactions between spindle microtubules and chromosomes, which is mediated by protein structures that include the kinetochores, and other protein complexes present on chromosomes. PinX1 is a potent telomerase inhibitor in interphase; however, its function in mitosis is not well documented. Here we show that PinX1 is essential for faithful chromosome segregation. Deconvolution microscopic analyses show that PinX1 localizes to nucleoli and telomeres in interphase and relocates to the periphery of chromosomes and the outer plate of the kinetochores in mitosis. Our deletion analyses mapped the kinetochore localization domain of PinX1 to the central region and its chromosome periphery localization domain to the C terminus. Interestingly, the kinetochore localization of PinX1 is dependent on Hec1 and CENP-E. Our biochemical characterization revealed that PinX1 is a novel microtubule-binding protein. Our real time imaging analyses show that suppression of PinX1 by small interference RNA abrogates faithful chromosome segregation and results in anaphase chromatid bridges in mitosis and micronuclei in interphase, suggesting an essential role of PinX1 in chromosome stability. Taken together, the results indicate that PinX1 plays an important role in faithful chromosome segregation in mitosis.
During mitosis, chromosome movements are orchestrated by the interactions between spindle microtubules and chromosomes. Studies over the last 2 decades have described the kinetochore as the major site where microtubule-chromosome attachment occurs (1). Electron microscopy has revealed that the kinetochore is composed of four layers as follows: an inner plate, an interzone, an outer plate, and an outermost fibrous corona (2). The outer plate and fibrous corona layers are thought to be the main microtubule-binding sites (1), and it is known that several protein complexes harboring microtubule binding ability are located in these layers (3–7). Meanwhile, through recruiting several microtubule-dependent motor proteins, the kinetochores generate tension and power chromosome movements in mitosis (6, 8). Advancements in genomics and proteomics have enabled the identification of additional kinetochore components that are important in governing faithful chromosome segregation (9, 10).
PinX1 is a 328-amino acid protein that was originally identified as a Pin2/TRF1 interacting protein in a yeast two-hybrid screen. PinX1 binds to Pin2/TRF1 through its C-terminal 142–254 amino acids. Overexpression of PinX1 or its telomerase inhibitory domain suppresses telomerase activity, causes telomere shortening, and induces cells into crisis, whereas depletion of PinX1 increases telomerase activity and elongates telomeres (11). Moreover, PinX1 can directly interact with the human telomerase RNA-binding domain of human telomerase reverse transcriptase as well as human telomerase RNA subunit in vitro (12), suggesting that it acts as an endogenous telomerase inhibitor. Yeast PinX1 inhibits telomerase by sequestering its catalytic subunit in an inactive complex lacking telomerase RNA in nucleoli (13). It has been reported that yeast PinX1 is also involved in rRNA and small nucleolar RNA maturation (14). The rat homolog of PinX1 also localizes to nucleoli in interphase and regulates telomere length (15). In human cells, it is reported that PinX1 has an effect on mediating human telomerase reverse transcriptase nucleolar localization (16). Collectively, these studies demonstrate that the functions of PinX1 in cell growth regulation are well conserved during evolution. Indeed, loss of heterozygosity of PinX1 occurs at a high frequency in many human cancers (17), and animal studies showed that depletion of endogenous PinX1 promotes tumorigenicity in nude mice (11).
As described above, the localization of PinX1 in interphase and its role in regulating telomere length have been well investigated. However, it has remained elusive as to whether PinX1 plays any role in mitosis and what happens if PinX1 is deficient. In this study, we have demonstrated that PinX1 is localized to the outer plate of kinetochores during mitosis. PinX1 is essential for spindle stability because depletion of PinX1 in HeLa cells destabilizes kinetochore microtubules and results in lagging chromosomes. Importantly, PinX1 interacts with microtubules. Our functional analyses show that PinX1 plays an important role in governing chromosome segregation and genomic stability.
EXPERIMENTAL PROCEDURES
Cell Culture and Synchronization
HeLa cells (American Type Culture Collection, Manassas, VA) were maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 8% CO2. Cells were synchronized at G1/S with 5 mm thymidine for 12–16 h and then washed with phosphate-buffered saline five times and cultured in thymidine-free medium for 10 h.
Plasmid Construction
The cDNA of PinX1 (NM_017884) was kindly provided by Dr. Kunping Lu (Harvard University). To generate green fluorescent protein (GFP)3 -tagged and bacterial expression constructs of PinX1 and deletion mutants, PCR-amplified cDNAs were cloned into pEGFP-C1 (Clontech) and pGEX-5X-3 (Amersham Biosciences) vectors by EcoRI and SalI.
Antibodies and RNA Interference
The following antibodies were used: anti-PinX1 mouse serum (Abnova), anti-BubR1 monoclonal antibody (Millipore), anti-Hec1 (9G3) monoclonal antibody (Abcam), anti-Cenp-F rabbit antibody (Novus Biologicals), human anti-centromere antibody (ACA), anti-tubulin antibody DM1A (Sigma), anti-CENP-E rabbit antibody (2). Two sets of siRNAs targeting to different regions of PinX1 were purchased from Qiagen. siRNAs of CENP-E, Hec1, CENP-F, and BubR1 were obtained as described previously (6, 40). All the siRNAs were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen). Transfection efficiency was assessed by fluorescence microscopy to follow the uptake of fluorescein isothiocyanate-labeled oligonucleotide. Transfection efficiency was typically 87 ± 5%.
The efficiency of siRNA-mediated repression of target proteins was assessed by Western blotting analyses using stepwise diluted starting material for reference. Typically, we achieved an 8.3-fold suppression of PinX1 protein using siRNA oligonucleotides targeted to two different regions of PinX1 mRNA.
Immunofluorescence Microscopy
Cells were seeded onto sterile, acid-treated 12-mm coverslips in 24-well plates (Corning Glass). The next day, the cells were transfected with 1 μl of Lipofectamine 2000 pre-mixed with plasmids or siRNAs described above. If not specified, 48 h after transfection, cells were rinsed with PHEM buffer (100 mm PIPES, 20 mm HEPES, pH 6.9, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol) and permeabilized for 1 min with PHEM plus 0.1% Triton X-100 before fixation in freshly prepared 3.7% formaldehyde for 5 min. After rinsing three times in phosphate-buffered saline, cells were blocked with PBST (0.05% Tween 20 in phosphate-buffered saline) with 1% bovine serum albumin (Sigma), followed by incubation with various primary antibodies in a humidified chamber for 1 h. After three washes in PBST, primary antibodies were visualized by fluorescein isothiocyanate or rhodamine-conjugated goat anti-mouse or rabbit IgG. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Slides were examined under a DeltaVision deconvolution microscopy (Applied Precision Inc.) as specified below. The confocal images in Fig. 1C were collected on a Leica SP5 confocal microscope as described previously (18).
FIGURE 1.
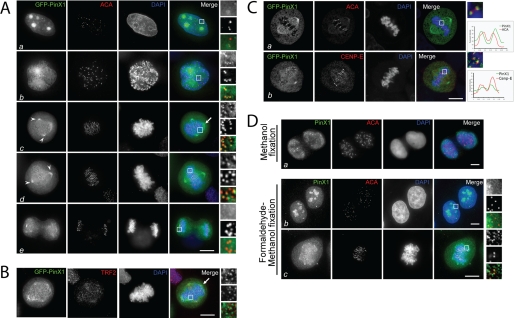
PinX1 localizes to chromosome periphery and outer plate of kinetochore in mitosis. A, subcellular distribution of GFP-PinX1 in HeLa cells. This montage represents optical images collected from PinX1-transfected HeLa cells triply stained for GFP-PinX1 (green), ACA (red), and DAPI (blue). Merged and enlarged images are also shown on the right. In interphase (panel a), it is readily apparent that PinX1 is concentrated in the nucleolus, appearing as four bright spots. No co-distribution of PinX1 with ACA was observed. A merged image shows the PinX1 staining is concentrated in the nucleolus. In prophase (panel b), the centrosomal and kinetochore labeling of PinX1 becomes apparent, which is clearly seen in the enlargement as PinX1 labeling is superimposed onto that of ACA. In prometaphase (panel c), PinX1 labeling remains at the kinetochore in addition to spindle poles (arrowheads) and chromosome periphery (arrow). In metaphase (panel d), PinX1 labeling begins dissociating from the kinetochore, which is apparent as less PinX1 is superimposed onto that of ACA (boxed area). The association of PinX1 to the spindle poles remains unchanged. In anaphase (panel e), PinX1 labeling on the mitotic spindle is decreased. Bar, 10 μm. B, this montage represents optical images collected from PinX1-transfected HeLa cells triply stained for GFP-PinX1 (green), TRF2 (red), and DAPI (blue). The arrow indicates the remnant chromosome peripheral localization of GFP-PinX1 in these pre-extracted cells. Bar, 10 μm. C, this montage represents optical images collected from PinX1-transfected HeLa cells triply stained for GFP-PinX1 (green), CENP-E/ACA (red), and DAPI (blue). GFP-PinX1-expressing metaphase cells were stained with ACA or anti-CENP-E antibody and examined by confocal microscopy to specify the kinetochore localization of PinX1. Linear scans of fluorescent intensities across kinetochore pairs are shown. GFP-PinX1 is shown in green, ACA or CENP-E in red, and DNA in blue. Bar, 10 μm. D, comparison of different fixatives for imaging endogenous PinX1 protein. HeLa cells were fixed either in cold methanol only or first in formaldehyde and then in cold methanol. These cells were subsequently stained with anti-PinX1 mouse serum. The enlargements show kinetochore localization of endogenous PinX1 in mitosis. PinX1 is presented in green, ACA in red, and DNA in blue. Bars, 10 μm.
For time-lapse microscopy, cells were cultured in a glass-bottom culture dish (MatTek, MA) with Leibovitz's L-15 medium (Invitrogen) at 37 °C and examined with a DeltaVision DVI microscopy system. Images were acquired at 2-min intervals and presented in Photoshop.
Fluorescence Intensity Quantification and Kinetochore Distance Measurement
The fluorescence intensity of kinetochore protein labeling was measured using a Zeiss LSM 510 NLO confocal microscope scan head mounted transversely on an Axiovert 200 inverted microscope with a ×100 1.3 numerical aperture PlanApo objective. The images from double labeling were collected using a dichroic filter set with Zeiss LSM 5 image processing software. The distance between sister kinetochores marked with ACA was measured as the distance between the peak fluorescence of the same focal plane as described previously (3).
Quantification of the level of kinetochore-associated protein was conducted as described by Johnson et al. (19). In brief, the average pixel intensities from at least 50 kinetochore pairs from five cells were measured, and background pixel intensities were subtracted. The pixel intensities at each kinetochore pair were then normalized against ACA pixel values to account for any variations in staining or image acquisition. The values of specific siRNA-treated cells were then plotted as a percentage of the values obtained from cells transfected with a control siRNA duplex.
Deconvolution Microscopy and Kinetochore-bound Microtubule Examination
Deconvolution images were collected using a DeltaVision wide field deconvolution microscope system built on an Olympus IX-70 inverted microscope base as described previously (3). For imaging, a ×100 1.35 NA lens was used, and optical sections were taken at intervals of 0.25 μm. Images were processed using DeltaVision Softworx software. Images for display were generated by projecting the sum of the optical sections using the maximum intensity method.
To quantify the role of PinX1 in connecting kinetochores with spindle microtubules, the number of end-on kinetochore microtubules from PinX1-suppressed cell was counted using the projection images constructed from the stack of 0.25-μm optical sections. We measured kinetochore-captured microtubule numbers in 15 cells from three preparations in control, PinX1-suppressed, and Hec1-suppressed cells.
RESULTS
PinX1 Is Localized to the Outer Plate of Kinetochores
PinX1 has been previously identified as a Pin2/TRF1 interacting protein and characterized as a natural telomerase inhibitor (11). Several studies have shown that exogenous PinX1 localizes to nucleoli and telomeres in interphase (11, 16). However, the distribution of PinX1 in mitotic cells has not been well documented.
To determine the precise localization of PinX1 throughout the cell cycle, we adopted a pre-extraction procedure that allows better labeling of kinetochore protein while preserving fine cyto-structure of mitotic cells (2, 20). To this end, HeLa cells were transiently transfected to express GFP-PinX1 followed by the pre-extraction prior to fixation. As shown in Fig. 1A, pre-extracted GFP-PinX1-expressing HeLa cells were stained using the human CREST ACA that reacts primarily with CENP-B, followed by a rhodamine-conjugated goat anti-human secondary antibody to identify the actual centromere (Fig. 1A, red). In the interphase cell shown in Fig. 1A, panel a, GFP-PinX1 staining appears as four bright spots in the nucleus, which is reminiscent of nucleoli. The merged image from three channels demonstrates that PinX1 is located to the nucleolus, which is consistent with previous reports (11). In the prophase cell, PinX1 was released from the nucleoli and translocated to the kinetochores (Fig. 1A, panel b). The merged image with magnification demonstrates that PinX1 is located at the centromere outer domain relative to CENP-B. From prometaphase to metaphase, GFP-PinX1 remains localized to the kinetochores (Fig. 1A, panels c and d). In addition, GFP-PinX1 distributes to the spindle poles. During the metaphase-anaphase transition, PinX1 gradually dissociates from the kinetochores and diffuses to the entire chromosomes in telophase (Fig. 1A, panel e).
It is noteworthy that we found a fraction of GFP-PinX1 occasionally localizes to the chromosome periphery region in some but not all prometaphase and metaphase cells (Fig. 1, A, panel c, and B, arrows). We reason that the pre-extraction strategy employed may remove GFP-PinX1 from the chromosome periphery. To validate our hypothesis, we performed immunocytochemical staining on GFP-PinX1-expressing cells without pre-extraction and compared it with that of pre-extracted cells. As shown in supplemental Fig. S1, strong chromosome peripheral localization of PinX1 was readily apparent in nonextracted cells. However, the kinetochore localization is less apparent due mainly to the overwhelming labeling on the chromosome periphery.
To characterize the spatial order of PinX1 distribution at the kinetochore relative to other well characterized kinetochore proteins such as CENP-E and CENP-B, we conducted confocal microscopic analyses of GFP-PinX1-expressing cells doubly stained with CENP-E antibody and ACA, respectively. From the magnified merged image, it is readily apparent that the PinX1 localization is exterior to the ACA (Fig. 1C, panel a) but relatively interior to CENP-E (Fig. 1C, panel b). Thus, we conclude that PinX1 is a kinetochore outer plate protein.
To verify if the localization of exogenously expressed GFP-PinX1 is representative of the endogenous PinX1, we stained HeLa cells with PinX1 mouse antibody. This mouse antibody can recognize PinX1 in methanol-fixed cells but not in formaldehyde-fixed cells. In the methanol-fixed cells, however, the nucleolar structure was not well preserved, and we failed to detect PinX1 in the nucleoli (Fig. 1D, panel a). We reasoned that chemical cross-linking perhaps masked the epitope for mouse PinX1 antibody. To balance the subcellular structure preservation and antigen accessibility for the PinX1 antibody, we devised a protocol combining a brief 3-min formaldehyde fixation followed by a 5-min cold methanol treatment to expose the epitope for anti-PinX1 antibody. As shown in Fig. 1D, panel b, the nucleolus localization of endogenous PinX1 in interphase is readily apparent. In addition, endogenous PinX1 is clearly seen at the chromosome periphery and the kinetochore (Fig. 1D, panel c, and magnified images). Based on the identical pattern of endogenous and exogenous PinX1 distribution, we conclude that PinX1 is a nucleolar protein in interphase and becomes associated with the mitotic apparatus, including the spindle pole, chromosome periphery, and the kinetochore in mitosis.
Kinetochore Localization of PinX1 Is Mediated by Its Central Region
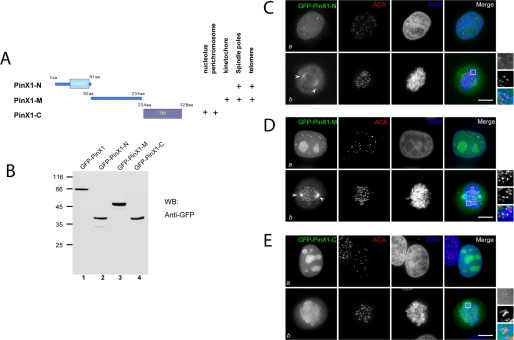
The spatiotemporal dynamics of PinX1 prompted us to identify the structural determinants responsible for PinX1 subcellular localization. To this end, we generated three deletion mutants based on the structural feature of PinX1. These mutants include the N-terminal PinX1 (PinX1-N; 1–91 amino acids), the central domain (PinX1-M; 92–254 amino acids), and C-terminal tail (PinX1-C; 254–328 amino acids) as illustrated in Fig. 2A. These three deletion mutants were tagged with GFP and expressed in HeLa cells as correct sizes based on the Western blotting analysis with a GFP antibody (Fig. 2B).
FIGURE 2.
Characterization of PinX1 structure-localization relationship. A, schematic illustration of PinX1 functional domains and summary of PinX1 structure-localization relationship. aa, amino acids. B, validation of exogenous expression of GFP-PinX1 and its deletion mutants. WB, Western blot; GFP, green fluorescent protein. C, this montage represents optical images collected from PinX1-N transfected HeLa cells triply stained for GFP-PinX1-N (green), ACA (red), and DAPI (blue). In interphase (panel a), it is readily apparent that GFP-PinX1-N appears as six tiny spots in the nucleus. No co-distribution of PinX1 with ACA is observed. In prometaphase (panel b), GFP-PinX1-N labeling appears at the spindle poles (arrowheads). Bar, 10 μm. D, this montage represents optical images collected from PinX1-M transfected HeLa cells triply stained for GFP-PinX1-M (green), ACA (red), and DAPI (blue). In interphase (panel a), it is readily apparent that PinX1 is concentrated in the nucleus, appearing as 7–8 bright spots. No co-distribution of PinX1 with ACA was observed. In prometaphase (panel b), GFP-PinX1-M labeling appears at the spindle poles (arrows) in addition to kinetochore localization. Bar, 10 μm. E, this montage represents optical images collected from PinX1-C-transfected HeLa cells triply stained for GFP-PinX1-C (green), ACA (red), and DAPI (blue). In interphase (panel a), it is readily apparent that PinX1-C is concentrated in the nucleolus, appearing as 5–6 bright spots. In general, PinX1-C and ACA are not co-localized (panel a). In prometaphase (panel b), GFP-PinX1-C labeling appears at perichromosomal regions. Bar, 10 μm.
We then assessed their subcellular distribution in HeLa cells transiently transfected to express GFP-PinX1 deletion mutants. After fixation, the kinetochores and DNA were counter-stained with ACA and DAPI, respectively. As shown in Fig. 2C, PinX1-N, which contains the potential RNA-binding G-patch domain, was dispersed almost evenly in the nucleoplasm with a few bright spots in interphase. In mitosis, PinX1-N has light deposition on the spindle pole localization (Fig. 2C, panel b). Surprisingly, PinX1-M, which contains no obvious structural domain, is localized to telomere-like structures and nucleoli in interphase (Fig. 2D, panel a). In mitosis, PinX1-M is distributed to kinetochores and spindle poles (Fig. 2D, panel b). PinX1-C, containing the telomerase inhibitory domain (11), mainly resides in interphase nucleoli. Interestingly, PinX1-C is localized to the chromosome periphery in mitosis (Fig. 2E, panel b). In sum, the pattern of deletion mutant localization suggests the structural determinants and reflects the dynamic subcellular localization profile of full-length GFP-PinX1 shown in Fig. 1A.
Kinetochore Localization of PinX1 Depends on Hec1 and CENP-E
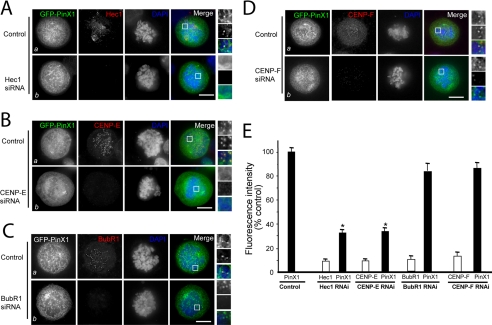
Previous studies revealed that assembly of the kinetochore machine involves several interactive but parallel pathways (6, 21–23). Recent studies reveal that Nuf2 forms a stable and evolutionarily conserved complex with Hec1 and specifies the localization of CENP-E to the kinetochore (6, 24–26). To examine if the Hec1 and CENP-E pathway is responsible for the assembly of PinX1 to the kinetochore, we employed the small interference RNA (siRNA) approach to suppress Hec1 and CENP-E protein expression and assessed their influence on PinX1 localization to the kinetochore. Typically, we achieved 87–93% suppression of Hec1, BubR1, and CENP-E as judged by Western blotting analyses (6). In control cultures, Hec1 localized with PinX1 at the prometaphase kinetochores (Fig. 3A, Control panels). In cells in which Hec1 had been suppressed, the levels of kinetochore-bound PinX1 appeared reduced (Fig. 3A, Hec1 siRNA panels). Quantitation of normalized pixel intensities showed that, when Hec1 was reduced to <10% of its control value, PinX1 levels were reduced to 33 ± 3% of the control (Fig. 3E). However, the association of PinX1 with chromosome periphery was not altered by the Hec1 depletion. We conclude that the kinetochore localization of PinX1 is dependent on Hec1.
FIGURE 3.
Kinetochore localization of PinX1 requires Hec1 and CENP-E. A, Hec1 determines the kinetochore localization of PinX1. Aliquots of HeLa cells were transfected with oligonucleotides (control and siRNA for Hec1) and GFP-PinX1 for 48 h, followed by fixation and immunocytochemical staining as described under “Experimental Procedures.” Optical images were collected from HeLa cells transfected with control siRNA (Control panel) and Hec1 siRNA (Hec1 siRNA panel). Suppression of Hec1 eliminates the kinetochore localization of PinX1, although its perichromosomal distribution is not altered. Scale bars, 10 μm. B, CENP-E determines the kinetochore localization of PinX1. Aliquots of HeLa cells were transfected with oligonucleotides (control and siRNA for CENP-E) and GFP-PinX1 for 48 h, followed by fixation and immunocytochemical staining as described above. Optical images were collected from HeLa cells transfected with control siRNA (Control panel) and CENP-E siRNA (CENP-E siRNA panel). Suppression of Hec1 eliminates the kinetochore localization of PinX1, although its perichromosomal distribution is not altered. Scale bars, 10 μm. C, kinetochore localization of PinX1 is independent of BubR1 in the kinetochore. Aliquots of HeLa cells were transfected with oligonucleotides (control and siRNA for BubR1) and GFP-PinX1 for 48 h, followed by fixation and immunocytochemical staining as described above. Optical images were collected from HeLa cells transfected with control siRNA (Control panel) and BubR1 siRNA (BubR1 siRNA panel). Suppression of BubR1 does not alter the kinetochore localization of PinX1 and PinX1 associated with perichromosome regions. Scale bars, 10 μm. D, kinetochore localization of PinX1 is independent of Cenp-F in the kinetochore. Aliquots of HeLa cells were transfected with oligonucleotides (control and siRNA for Cenp-F) and GFP-PinX1 for 48 h, followed by fixation and immunocytochemical staining as described above. Optical images were collected from HeLa cells transfected with control siRNA (Control panel) and Cenp-F siRNA (Cenp-F siRNA panel). Suppression of Cenp-F does not alter the kinetochore localization of PinX1 and PinX1 associated with perichromosome regions. Scale bars, 10 μm. E, quantitation of Hec1, CENP-E, BubR1, and Cenp-F levels at kinetochores of control and siRNA-treated cells. The pixel intensities of Hec1, CENP-E, Hec1, and Cenp-F (normalized to the ACA signal) in control (closed bars) and Hec1-repressed, CENP-E-repressed, BubR1-repressed, and Cenp-F-repressed cells were measured. Values represent the means ± S.E. of at least 100 kinetochores in 11 different cells. The intensities of target proteins are expressed as open bars, although PinX1 intensities are marked by closed bars. *, p < 0.01 compared with that of the control.
We next examined the requirement of CENP-E for PinX1 assembly to the kinetochore. In control cultures, CENP-E localized with PinX1 at the prometaphase kinetochores (Fig. 3B, Control panels). However, in cells in which CENP-E had been suppressed, the levels of kinetochore-bound PinX1 appeared reduced (Fig. 3B, CENP-E siRNA panels). Quantitation of normalized pixel intensities showed that, when CENP-E was reduced to <10% of its control value, PinX1 levels were reduced to ∼37% (Fig. 3E). Because CENP-E interacts with CENP-F and BubR1 (3, 27), we then tested the requirement of CENP-F and BubR1 for kinetochore localization of PinX1. In control cultures, BubR1 localized with PinX1 at the prometaphase kinetochores (Fig. 3C, Control panels). Interestingly, in cells in which BubR1 had been suppressed, the levels of kinetochore-bound PinX1 appeared unaltered (Fig. 3C, BubR1 siRNA panels). Quantitation of normalized pixel intensities showed that, when BubR1 was reduced to <10% of its control value, PinX1 levels were reduced to ∼83% (Fig. 3E). Similarly, suppression of CENP-F did not alter the localization of PinX1 to the kinetochore (Fig. 3D, CENP-F siRNA panels). We have also examined the effect of repressing PinX1 on the kinetochore localization of Hec1, CENP-E, BubR1, and CENP-F (supplemental Fig. S3). However, quantitation of normalized pixel intensities showed that, when PinX1 was reduced to <10% of its control value, the levels of Hec1, CENP-E, BubR1, and CENP-F were slightly reduced to ∼87–91% of their respective control values (data not shown). Thus, we conclude that PinX1 localization to the kinetochore depends on CENP-E, but not BubR1 and CENP-F.
Depletion of PinX1 Causes Unstable Spindle Microtubules and Lagging Chromosomes
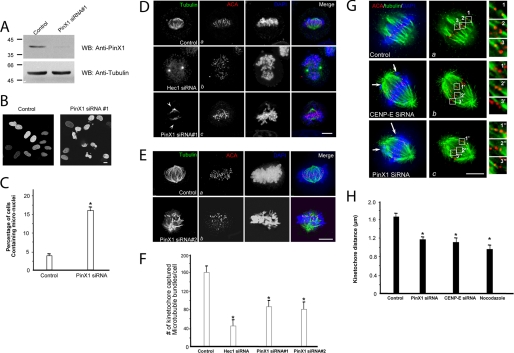
Given its kinetochore localization and dependence on Hec1 and CENP-E, we next examined the role of PinX1 in mitosis. To this end, we introduced siRNA oligonucleotide duplexes to PinX1 by transfection into HeLa cells. Trial experiments revealed that treatment of HeLa cells with 50 nm siRNA for 48 h produced optimal suppression of the target proteins. As shown in Fig. 4A, Western blotting with anti-PinX1 antibody revealed that the siRNA oligonucleotide caused an 8.3-fold suppression of the PinX1 protein level. This suppression was relatively specific, as it did not alter the levels of other proteins such as tubulin and nucleolin (Fig. 4A and data not shown). Surprisingly, our brief examination of DNA staining revealed that the number of cells containing micro-nuclei was readily apparent (Fig. 4B, PinX1 siRNA panel). We surveyed ∼150 interphase cells, positively transfected with PinX1 siRNA, through the stacks of focal planes of the nucleus. As shown in Fig. 4C, a summary from three different experiments shows that the depletion of PinX1 results in significant increases in micronuclei-containing cells (15.7 ± 1.3%) compared with the control group (3.8 ± 0.2%; p < 0.01).
FIGURE 4.
PinX1 is required for faithful chromosome segregation and spindle stability. A, efficiency of siRNA treatments in HeLa cells. Aliquots of HeLa cells were transfected with 50 nm siRNA oligonucleotide duplexes for PinX1 and its control (scrambled oligonucleotide) for 48 h and subjected to SDS-PAGE and immunoblotting. Upper panel, PinX1; lower panel, tubulin. WB, Western blot. B, suppression of PinX1 results in micro-nuclei phenotype. HeLa cells were transfected with siRNA oligonucleotides for PinX1 and examined for nuclear morphology. C, average of 200 cells from three separate experiments was counted for HeLa cells treated with control or PinX1 siRNA oligonucleotides, respectively. Error bars represent S.E.; n = 3 preparations, *, p < 0.01 compared with that of the control. D and E, suppression of PinX1 results in destabilization of spindle microtubules. HeLa cells were transfected with siRNA oligonucleotides (D, oligonucleotide 1; E, oligonucleotide 2) for PinX1 and examined for spindle stability in response to cold treatment. Cold-treated control, Hec1-, or PinX1-depleted cells were stained with tubulin (green) and ACA (red). The kinetochore microtubules became unstable due to cold treatment in the absence of Hec1 or PinX1 (D, panel c, and E, panel b). Bar, 10 μm. Note that the two siRNA oligonucleotides targeting to different regions of PinX1 result in similar repression of PinX1 protein level and subsequent phenotype of chromosome congression errors. F, quantitation of kinetochore-captured microtubule bundles. To quantify the number of kinetochore-captured microtubule bundles as a function of kinetochore protein, the number of end-on kinetochore microtubule bundles per cell from the projection images constructed from the stack of 0.25-μm optical sections was counted. In the control scramble oligonucleotide-transfected cells, the number of kinetochore-captured microtubule bundles is 159 ± 17/cell. The kinetochore-captured microtubule bundle number in Hec1-repressed cells is 47 ± 7/cell, and the number in PinX1-repressed cell is 83 ± 13/cell. Error bars represent S.E.; n = 15 cells, *, p < 0.01 compared with that of the control. G, suppression of PinX1 results in a loss in tension across the sister kinetochore. HeLa cells were transfected with siRNA oligonucleotides for PinX1 and examined for distance across the sister kinetochore. The distance between kinetochore pairs in CENP-E (1′, 2′, and 3′) or PinX1 (1″, 2″, and 3″-depleted cells was measured and compared with that of control cells (1, 2, and 3). Bar, 10 μm. H, quantitation of sister kinetochore distance marked by ACA staining. Kinetochore distance is measured between kinetochores that are marked by ACA staining and localized in the same focal plane. An aliquot of cells was exposed to 100 ng/ml nocodazole to depolymerize all kinetochore microtubules as described under “Experimental Procedures.” The value from siRNA-treated samples was calculated from >100 kinetochores selected from at least 10 different cells. *, p < 0.01 compared with that of the control.
The micronuclei phenotype suggests a possibility of aberrant chromosome segregation in the prior round of mitosis. PinX1 kinetochore localization depends on the Hec1 and CENP-E, two kinetochore proteins essential for spindle microtubule attachment to the kinetochore. If PinX1 functions in spindle microtubule-kinetochore association, suppression of PinX1 could result in an unstable spindle microtubule attachment. To test this hypothesis, aliquots of HeLa cells were transfected with siRNA oligonucleotides to suppress Hec1 and PinX1 followed by thymidine synchronization and release into metaphase (10 h after thymidine wash-out). After an incubation of the coverslips on ice to depolymerize non-kinetochore microtubules, the transfected and synchronized HeLa cells were pre-extracted prior to formaldehyde fixation. The cells were counter-stained with anti-tubulin, anti-ACA antibody, and DAPI. As shown in Fig. 4D, panel a, in control cells, even though the aster microtubules were depolymerized, the spindle microtubules were stable under cold treatment. Conversely, the spindle microtubules were depolymerized in Hec1-depleted cells, consistent with the essential role of Hec1 in spindle microtubule-kinetochore attachment (5). Surprisingly, the spindle microtubules were partially depolymerized in the PinX1-depleted cells (Fig. 4D, panel c), suggesting that PinX1 may function in stabilizing spindle microtubules in mitosis. Using an siRNA targeted to a different region of PinX1 mRNA, we obtained essentially the same phenotype (Fig. 4E, panel b).
To quantify the effect of PinX1 in stabilizing kinetochore microtubules, we counted the number of end-on kinetochore microtubule bundles per cell from the projection images constructed from the stack of 0.25-μm optical sections collected by deconvolution microscopy as described previously (3). As summarized in Fig. 4F from analyses of 15 cells, the number of kinetochore-captured microtubule bundles is 159 ± 17 per cell in control. Such a number is greatly reduced in Hec1-repressed cells (47 ± 7 per cell). The number in PinX1-repressed cell is 83 ± 13 per cell (siRNA 1; n = 15; p < 0.01 compared with that of control) and 78 ± 15 per cell (siRNA 1; n = 15; p < 0.01 compared with that of control). Thus, our quantitative analyses indicate that PinX1 is essential for a stable kinetochore-microtubule connection.
Our previous studies established that CENP-E is essential for stabilizing kinetochore-microtubule attachments (3, 6). Our finding that the CENP-E is required for PinX1 maintenance at kinetochores prompted us to test whether depletion of PinX1 could affect the functional activity of microtubule capturing. The distance between sister kinetochores marked by ACA has been used as an accurate reporter for judging the tension developed across the kinetochore pair (3, 6). In this case, the shortened distance often reflects aberrant microtubule attachment to the kinetochore, in which less tension is developed across the sister kinetochore. Therefore, we measured ACA distance in >100 kinetochore pairs in which both kinetochores were in the same focal plane, in each siRNA-treated cell, including CENP-E, PinX1, and control cells. Nocodazole-treated cells were used as a negative control in which kinetochore pairs were presumably under no tension.
As shown in Fig. 4, G and H, depletion of CENP-E or PinX1 resulted in errors in chromosome alignment at the equator (panels b and c; arrows). Control kinetochores exhibited a separation of 1.65 ± 0.12 μm (Fig. 4G, 1–3), whereas the distance between kinetochores was 1.18 ± 0.13 μm in PinX1-depleted cells (Fig. 4G, 1″ 2″, and 3″; *, p < 0.01 compared with that of the control) and 1.13 ± 0.15 μm in CENP-E-depleted cells (Fig. 4G, 1′, 2′, and 3′; *, p < 0.01 compared with that of the control), comparable with 0.93 ± 0.11 μm in nocodazole-treated cells, indicating that PinX1 functions in stabilizing microtubule-kinetochore association. These data demonstrate that loss of PinX1 is responsible for defects of chromosome congression and aberrant attachment of chromosome to spindle microtubules.
PinX1 Is a Novel Microtubule-binding Protein
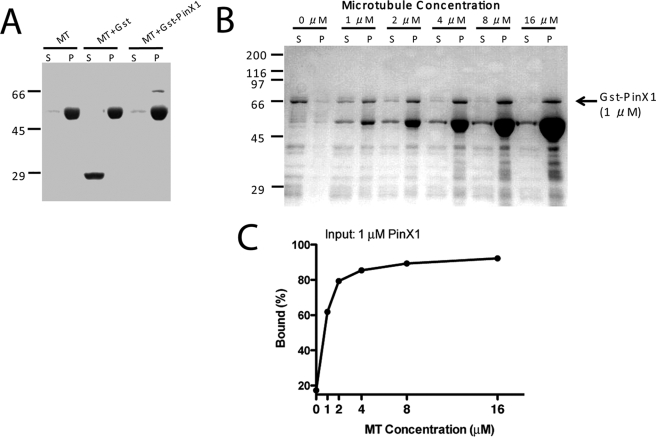
Repression of PinX1 resulting in aberrant chromosome segregation prompted us to examine whether PinX1 exhibits any microtubule binding activity. To this end, we carried out a co-sedimentation assay using recombinant PinX1 protein incubated with pre-formed microtubules. The Coomassie Blue-stained gel in Fig. 5A shows that recombinant GST-PinX1 but not GST co-sedimentated with taxol-stabilized microtubules. No pelleting of PinX1 was observed in the absence of microtubules, suggesting that PinX1 bears microtubule binding activity. To validate this activity, we incubated GST-PinX1 with an increased concentration of taxol-stabilized microtubules followed by co-sedimentation. As shown in Fig. 5B, PinX1 binds to microtubule in a dose-dependent manner. No pelleting of either protein was observed in the absence of microtubules. Quantifying the fraction of PinX1 pelleted with microtubule indicates an apparent dissociation constant of Kd ∼0.83 ± 0.18 μm (Fig. 5C). Therefore, we conclude that PinX1 binds to microtubules directly with a relatively low affinity.
FIGURE 5.
PinX1 is a novel microtubule-binding protein. A, microtubule (MT) co-sedimentation analyses of recombinant GST-PinX1 and GST proteins. B, microtubule co-sedimentation analyses of GST-PinX1 in a dose-responsive manner. C, graph showing the microtubule binding activity of GST-PinX1. The average of two experiments is plotted based on Western blotting of PinX1. S, supernatant; P, pellet.
PinX1 Is Essential for Faithful Chromosome Segregation
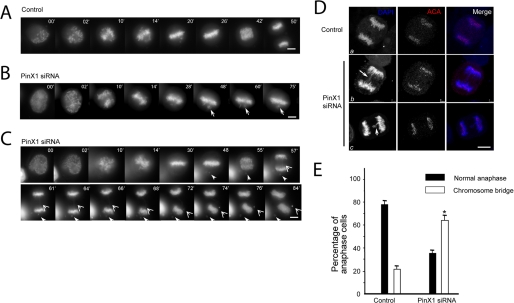
Given the finding that PinX1 binds to microtubules in vitro, we speculated that repression of PinX1 would cause chromosome segregation error. To examine precisely the chromosome movements in cells depleted of PinX1 or transfected by scramble oligonucleotides, GFP-H2B was co-transfected with siRNAs to visualize, in real time, chromosomes in the absence of PinX1 upon nuclear envelope breakdown (NEB). As shown in Fig. 6A, in scramble oligonucleotide-transfected cells, all chromosomes achieve metaphase alignment 26 min after NEB. The anaphase onset was observed 40 min after NEB. The complete segregation of sister chromatids was achieved 50 min after NEB in these scramble-transfected cells. Two examples of PinX1 depleted cells are shown in Fig. 6, B and C. One cell failed to achieve chromosome alignment even at 75 min after NEB, which is similar to nucleolin-depleted cells (Fig. 6B). The other cell (Fig. 6C) did not achieve anaphase onset, in the presence of a lagging chromosome, until 55 min after NEB, which is about 38% longer than that of control cell. Although a majority of sister chromatids separated at the anaphase onset, careful examination revealed a chromatid bridge (Fig. 6C, open arrowheads) in PinX1-suppressed cells. The chromatid bridge was eventually broken and formed a micro-nucleus in the PinX1-depleted telophase cell. We surveyed ∼10 cells from each group. The average time from NEB to anaphase onset is 42.6 ± 2.7 min in control cells, and the length is PinX1-repressed cells is 64.8 ± 8.6 min (p < 0.01 compared with that of control). Thus, we conclude that PinX1 is essential for faithful chromosome segregation in mitosis.
FIGURE 6.
PinX1 governs faithful chromosome segregation. A, real time imaging of chromosome movements in HeLa cells transfected with scramble siRNA oligonucleotides. Chromosomes were marked by GFP-H2B. Bar, 10 μm. B and C, repression of PinX1 by siRNA treatment resulted in chromosome instability phenotype. Chromosome segregation errors are apparent in the absence of PinX1; closed arrows indicate lagging chromosomes during alignment; open arrows indicate chromosome bridges in anaphase. Bars, 10 μm. D, depletion of PinX1 impairs spindle checkpoint by promoting a premature anaphase (panels b and c). HeLa cells were transfected with PinX1 siRNA oligonucleotide and control oligonucleotide for 48 h followed by fixation and indirect immunofluorescence staining. This set of optical images was collected from an anaphase HeLa cell doubly stained for human ACA (red), DAPI (DNA, blue), and their merged images. As shown in panels b and c, PinX1-depleted cell entered in anaphase with a chromatin bridge (panels b and c, arrow). Most interestingly, ACA staining (panel c, arrowhead) verifies that this pair of chromatids failed to separate as the cell entered anaphase, leading to occurrence of aneuploidy. Bars, 10 μm. E, depletion of PinX1 by siRNA effects mitotic defects in chromosome segregation. HeLa cells were transfected with PinX1 siRNA oligonucleotide and control oligonucleotide for 48 h followed by fixation and DNA staining. Cells were then examined under a fluorescence microscopy to score the sister chromatid bridge phenotype. An average of 200 cells from three separate experiments was counted; *, p < 0.01 compared with that of the control.
An unstable kinetochore-microtubule attachment results in aberrant chromosome segregation, although perturbation of kinetochore structure due to PinX1 repression also leads to premature anaphase phenotype (Fig. 6C). To explore whether depletion of PinX1 causes any chromosome segregation defects in HeLa cells, we collected control oligonucleotide-treated and siRNA-treated cells, stained with DAPI and ACA. Control anaphase cells treated with scramble oligonucleotide exhibited a total separation of two sets of sister chromatids (Fig. 6D, panel a). However, cells treated with PinX1 siRNA display a typical chromosome bridge phenotype, in which the majority of sister chromatids is separated with one or more sister chromatids entangled (Fig. 6D, panels b and c, arrows). The failure in separating equal sister chromatids prior to entry into anaphase resulted in unequal distribution of chromatids, which leads to the micro-nuclei phenotype seen in Fig. 4B. We surveyed ∼130 mitotic cells from three different experiments, in which both sets of sister chromatids were in the same focal plane, from PinX1 siRNA oligonucleotide-treated and control oligonucleotide-treated cells. We counted the number of cells displaying a typical chromosome bridge phenotype and expressed it as the percentage of the total cell population that is in mitosis. As shown in Fig. 6E, a summary from three different experiments shows that the depletion of PinX1 resulted in significant increases in cells bearing chromatid bridges (65.7 ± 5.7%; p < 0.01 compared with that of control of 20.3 ± 3.6%). These data demonstrate that loss of PinX1 is also responsible for defects of chromosome segregation associated with premature entry into anaphase.
DISCUSSION
Aneuploidy, a loss or gain of chromosomes, a major form of chromosome instability commonly associated with cancer formation and progression, is thought to arise from aberrant mitotic chromosome segregation. Chromosome segregation in mitosis is orchestrated by the interactions of kinetochores with spindle microtubules and is monitored by the spindle assembly checkpoint proteins (28). In this study, we have demonstrated that PinX1 localizes to the chromosome periphery during mitosis. In addition, we revealed the kinetochore localization of PinX1, which persists from prophase to metaphase, suggesting that PinX1 may be involved in regulating chromosome movements. Suppression of the expression of PinX1 by siRNA in HeLa cells results in depletion of PinX1 during mitosis, destabilization of kinetochore microtubules, and chromosome instability.
Besides its kinetochore and chromosome periphery localization, exogenously expressed GFP-PinX1 was readily apparent on the spindle poles. However, the spindle pole-associated endogenous PinX1 was less prominent in our experimental condition. It is possible that the PinX1 epitope recognized by our antibody is somehow masked at the spindle pole. The apparent spindle pole-associated localization of GFP-PinX1 could also be due to the fact that overexpression of GFP-PinX1 saturates its binding sites at the kinetochore and chromosome periphery leading to accumulation at the poles. Nevertheless, it is worth noting that TRF1, a binding partner of PinX1, is redistributed to the spindle poles in mitosis (29). However, bipolar spindle integrity was not altered in PinX1-depleted cells, suggesting the mitotic defects seen in the PinX1 suppression are not associated with its spindle localization. Thus, we reason that the function of PinX1 in mitosis is mainly tied to its localization on the kinetochores.
It was surprising that PinX1 bears microtubule binding activity. However, our attempt to map the microtubule binding activity using PinX1 deletion mutants was unsuccessful. It is possible that deletion disrupted the secondary structure necessary for the microtubule binding. Given the importance of PinX1 in mitotic chromosome segregation, it would be of great importance to characterize its structure-function relationship. Chromosome instability phenotype seen in PinX1-repressed cells is consistent with the fact that depletion of PinX1 also increases tumorigenicity in nude mice (11).
Recent studies have revealed that other structures on the chromosomes facilitate faithful chromosome segregation (30, 31). Proteomic analysis of mitotic chromosomes identified hundreds of chromosome periphery proteins in addition to the kinetochore proteins (32). The large number of proteins observed in perichromosomal regions during mitosis implies that they play functional roles in this process (33). In fact, Heald et al. (34) have elegantly demonstrated that bipolar spindles can be assembled spontaneously around artificial chromosomes using Xenopus egg extracts, suggesting a role of chromosomes, perhaps chromosome periphery proteins, in microtubule nucleation and spindle assembly. Subsequent investigation identified a Rae1-containing ribonucleoprotein complex that is involved in spindle assembly and particularly in microtubule nucleation and stabilization (35). Several studies demonstrate that Ran-GTP gradients generated by the chromosomal Ran-guanine nucleotide exchange factor RCC1 are essential for chromosome-mediated bipolar spindle assembly (36, 37). More recently, the Ran-importin-β complex was reported to be involved in chromosome loading of human chromokinesin Kid (38). Chromokinesins generate polar ejection forces on chromosomes to push them toward the microtubule plus-end and thus the equatorial plane (39). Considering the chromosome congression defect observed in PinX1-depleted cells, it will be worth verifying whether PinX1 is involved in chromosome loading of chromokinesins or the generation of Ran-GTP gradients.
Taken together, our findings demonstrate a critical role of PinX1 in accurate chromosome segregation. The fact that elimination of PinX1 disrupts kinetochore microtubule-kinetochore association and induces chromosome cross-bridges in HeLa cells prematurely exited from anaphase demonstrates the importance of PinX1-microtubule interaction in facilitating chromosome segregation and the maintenance of genomic stability in mitosis.
Acknowledgments
We thank Dr. Kunping Lu, Dr. Nancy Maizel, and Dr. Guowei Fang for reagents. The facilities used were supported in part by National Institutes of Health Grant G-12-RR03034 from NCRR/RCM1.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-56292, CA89019, CA92080, and CA118948. This work was also supported by Chinese Academy of Science Grants KSCX1-YW-R65 and KSCX2-YW-H10, Chinese 973 Projects 2002CB713700, 2010CB912103, 2007CB914503, and 2006CB943600, Chinese 863 Project 2006AA02A247, Chinese Natural Science Foundation Grants 30270654, 30070349, 90508002, and 30121001, China National Key Projects for Infectious Disease Grant 2008ZX10002-021, American Cancer Society Grant RPG-99-173-01, a Georgia Cancer Coalition Breast Cancer research grant, and an Atlanta Clinical and Translational Science Award Chemical Biology grant (to X. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GFP
- green fluorescent protein
- ACA
- anti-centromere antibody
- DAPI
- 4′,6-diamidino-2-phenylindole
- siRNA
- small interference RNA
- PIPES
- 1,4-piperazinediethanesulfonic acid
- NEB
- nuclear envelope breakdown
- GST
- glutathione S-transferase.
REFERENCES
- 1.Rieder C. L. (1982) Int. Rev. Cytol. 79,1–58 [DOI] [PubMed] [Google Scholar]
- 2.Yao X., Anderson K. L., Cleveland D. W. (1997) J. Cell Biol. 139,435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) Nat. Cell Biol. 2,484–491 [DOI] [PubMed] [Google Scholar]
- 4.Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006) Cell 127,983–997 [DOI] [PubMed] [Google Scholar]
- 5.DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. (2006) Cell 127,969–982 [DOI] [PubMed] [Google Scholar]
- 6.Liu D., Ding X., Du J., Cai X., Huang Y., Ward T., Shaw A., Yang Y., Hu R., Jin C., Yao X. (2007) J. Biol. Chem. 282,21415–21424 [DOI] [PubMed] [Google Scholar]
- 7.Wei R. R., Al-Bassam J., Harrison S. C. (2007) Nat. Struct. Mol. Biol. 14,54–59 [DOI] [PubMed] [Google Scholar]
- 8.Vergnolle M. A., Taylor S. S. (2007) Curr. Biol. 17,1173–1179 [DOI] [PubMed] [Google Scholar]
- 9.Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8,458–469 [DOI] [PubMed] [Google Scholar]
- 10.Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007) Dev. Cell 12,17–30 [DOI] [PubMed] [Google Scholar]
- 11.Zhou X. Z., Lu K. P. (2001) Cell 107,347–359 [DOI] [PubMed] [Google Scholar]
- 12.Banik S. S., Counter C. M. (2004) J. Biol. Chem. 279,51745–51748 [DOI] [PubMed] [Google Scholar]
- 13.Lin J., Blackburn E. H. (2004) Genes Dev. 18,387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guglielmi B., Werner M. (2002) J. Biol. Chem. 277,35712–35719 [DOI] [PubMed] [Google Scholar]
- 15.Oh B. K., Yoon S. M., Lee C. H., Park Y. N. (2007) Gene 400,35–43 [DOI] [PubMed] [Google Scholar]
- 16.Lin J., Jin R., Zhang B., Yang P. X., Chen H., Bai Y. X., Xie Y., Huang C., Huang J. (2007) Biochem. Biophys. Res. Commun. 353,946–952 [DOI] [PubMed] [Google Scholar]
- 17.Wright K., Wilson P. J., Kerr J., Do K., Hurst T., Khoo S. K., Ward B., Chenevix-Trench G. (1998) Oncogene 17,1185–1188 [DOI] [PubMed] [Google Scholar]
- 18.Yuan K., Hu H., Guo Z., Fu G., Shaw A. P., Hu R., Yao X. (2007) J. Biol. Chem. 282,27414–27423 [DOI] [PubMed] [Google Scholar]
- 19.Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S. (2004) J. Cell Sci. 117,1577–1589 [DOI] [PubMed] [Google Scholar]
- 20.Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) J. Biol. Chem. 279,20049–20057 [DOI] [PubMed] [Google Scholar]
- 21.Chan G. K., Jablonski S. A., Starr D. A., Goldberg M. L., Yen T. J. (2000) Nat. Cell Biol. 2,944–947 [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A. W., Teng M., Cleveland D. W., Goldberg M. L., Niu L., Yao X. (2004) J. Biol. Chem. 279,54590–54598 [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., Cui T., Hu R., Dou Z., Zhu J., Xie W., Rao Z., Ding X., Yao X. (2008) J. Biol. Chem. 283,26726–26736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigge P. A., Kilmartin J. V. (2001) J. Cell Biol. 152,349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleland M. L., Gardner R. D., Kallio M. J., Daum J. R., Gorbsky G. J., Burke D. J., Stukenberg P. T. (2003) Genes Dev. 17,101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCleland M. L., Kallio M. J., Barrett-Wilt G. A., Kestner C. A., Shabanowitz J., Hunt D. F., Gorbsky G. J., Stukenberg P. T. (2004) Curr. Biol. 14,131–137 [DOI] [PubMed] [Google Scholar]
- 27.Mao Y., Desai A., Cleveland D. W. (2005) J. Cell Biol. 170,873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musacchio A., Salmon E. D. (2007) Nat. Rev. Mol. Cell Biol. 8,379–393 [DOI] [PubMed] [Google Scholar]
- 29.Nakamura M., Zhou X. Z., Kishi S., Kosugi I., Tsutsui Y., Lu K. P. (2001) Curr. Biol. 11,1512–1516 [DOI] [PubMed] [Google Scholar]
- 30.Ma N., Matsunaga S., Takata H., Ono-Maniwa R., Uchiyama S., Fukui K. (2007) J. Cell Sci. 120,2091–2105 [DOI] [PubMed] [Google Scholar]
- 31.Sakita-Suto S., Kanda A., Suzuki F., Sato S., Takata T., Tatsuka M. (2007) Mol. Biol. Cell, 18,1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchiyama S., Kobayashi S., Takata H., Ishihara T., Hori N., Higashi T., Hayashihara K., Sone T., Higo D., Nirasawa T., Takao T., Matsunaga S., Fukui K. (2005) J. Biol. Chem. 280,16994–17004 [DOI] [PubMed] [Google Scholar]
- 33.Van Hooser A. A., Yuh P., Heald R. (2005) Chromosoma 114,377–388 [DOI] [PubMed] [Google Scholar]
- 34.Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. (1996) Nature 382,420–425 [DOI] [PubMed] [Google Scholar]
- 35.Blower M. D., Nachury M., Heald R., Weis K. (2005) Cell 121,223–234 [DOI] [PubMed] [Google Scholar]
- 36.Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. (2001) Cell 104,95–106 [DOI] [PubMed] [Google Scholar]
- 37.Kalab P., Weis K., Heald R. (2002) Science 295,2452–2456 [DOI] [PubMed] [Google Scholar]
- 38.Tahara K., Takagi M., Ohsugi M., Sone T., Nishiumi F., Maeshima K., Horiuchi Y., Tokai-Nishizumi N., Imamoto F., Yamamoto T., Kose S., Imamoto N. (2008) J. Cell Biol. 180,493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazumdar M., Misteli T. (2005) Trends Cell Biol. 15,349–355 [DOI] [PubMed] [Google Scholar]
- 40.Dou Z., Ding X., Zereshki A., Zhang Y., Zhang J., Wang F., Sun J., Huang H., Yao X. (2004) FEBS Lett. 572,51–56 [DOI] [PubMed] [Google Scholar]