Abstract
Charcot-Marie-Tooth (CMT) is the most frequent inherited neuromuscular disorder, affecting 1 person in 2500. CMT1A, the most common form of CMT, is usually caused by a duplication of chromosome 17p11.2, containing the PMP22 (peripheral myelin protein-22) gene; overexpression of PMP22 in Schwann cells (SC) is believed to cause demyelination, although the underlying pathogenetic mechanisms remain unclear. Here we report an abnormally high basal concentration of intracellular calcium ([Ca2+]i) in SC from CMT1A rats. By the use of specific pharmacological inhibitors and through down-regulation of expression by small interfering RNA, we demonstrate that the high [Ca2+]i is caused by a PMP22-related overexpression of the P2X7 purinoceptor/channel leading to influx of extracellular Ca2+ into CMT1A SC. Correction of the altered [Ca2+]i in CMT1A SC by small interfering RNA or with pharmacological inhibitors of P2X7 restores functional parameters of SC (migration and release of ciliary neurotrophic factor), which are typically defective in CMT1A SC. More significantly, stable down-regulation of the expression of P2X7 restores myelination in co-cultures of CMT1A SC with dorsal root ganglion sensory neurons. These results establish a pathogenetic link between high [Ca2+]i and impaired SC function in CMT1A and identify overexpression of P2X7 as the molecular mechanism underlying both abnormalities. The development of P2X7 inhibitors is expected to provide a new therapeutic strategy for treatment of CMT1A neuropathy.
Charcot-Marie-Tooth disease type 1 (CMT1)3 is a progressive hereditary motor and sensory neuropathy, characterized by distal muscle wasting and weakness, foot deformities, and severe slowing of nerve conduction, because of progressive demyelination (1). With a prevalence of 1 case in 2500, CMT1 is the most common hereditary neurologic disorder, and in the majority of cases (CMT1A) the disease is associated with a duplication on chromosome 17p11.2 of the gene for PMP22 (peripheral myelin protein 22) (2). PMP22 is a 22-kDa glycoprotein mainly expressed by myelinating Schwann cells (SC) and localized in compact myelin (3). The transgenic rat model of CMT1A, obtained by overexpression of PMP22 (4), confirms a role of PMP22 in the pathogenesis of CMT1A. Both PMP22 overexpression because of gene duplication and point mutations of PMP22 are associated with a CMT1A phenotype.
The biochemical mechanisms correlating PMP22 dysfunction with demyelination are still unclear. Some reports indicate that a perturbed homeostasis of the intracellular Ca2+ concentration ([Ca2+]i) might be causally involved in the demyelination process. Conditions inducing an increased [Ca2+]i in SC impair cell differentiation and myelination (5, 6), similarly to what occurs in CMT1A. Incubation of intact rat nerves with Ca2+ and ionophores causes a progressive demyelination, spreading from the paranodes and invading regions of formerly compact myelin, which is dependent upon a rise in the [Ca2+]i of SC (5).
Additional evidence for the detrimental effect of a [Ca2+]i elevation on myelin production by SC comes from application of ATP to murine SC monocultures, inducing an immediate and large increase in the [Ca2+]i. As a result of ATP treatment, maturation and differentiation of SC, as well as expression of the myelin basic protein and production of compact myelin, are completely prevented (6). Taken together, the above observations indicate that abnormally elevated Ca2+ levels are causally related to impairment of myelin production by SC.
In this study, we addressed the possible correlation between PMP22 overexpression and alteration of the [Ca2+]i homeostasis in SC from a rat model of CMT1A. We recorded higher levels of basal [Ca2+]i in affected than in control cells, and we identified the mechanisms responsible for the perturbation of the [Ca2+]i levels in CMT1A SC. Experiments with pharmacological inhibitors and with small interfering RNA (siRNA) unequivocally demonstrated a correlation in CMT1A SC between overexpression of the purinergic receptor P2X7 and influx of extracellular [Ca2+]i across this plasma membrane receptor/channel. In addition, correction of the abnormally elevated [Ca2+]i levels by the use of a P2X7 antagonist or through down-regulation of the expression of P2X7 by transfection with siRNA or with short hairpin RNA-expressing plasmid (shRNA) restored the normal phenotype in CMT1A SC. These findings suggest that CMT1A should be considered as a “calcium disease.” Identification of P2X7 activation as the pathogenetic mechanism underlying demyelination may provide the rationale for a new therapeutic strategy for CMT1A, a disease with no currently available treatment.
EXPERIMENTAL PROCEDURES
Antibodies
Commercially available monoclonal antibodies against myelin basic protein (MBP) (MAB386, Millipore, Milano, Italy), myelin protein zero (MPZ) (P07 extracellular domain, Astexx Ltd. & Co. KEG, Graz, Austria), L1 (MAB5272, Millipore), glial fibrillary acidic protein (GFAP) (G3893, Sigma), and β-tubulin (clone TUB2.1) (T4026, Sigma) and polyclonal antibodies against the transcription factor Krox-20 (Egr2) (PRB-236P, Covance, Princeton, NJ), P2X7 (Calbiochem), and total neurofilament (N4142, Sigma) were used in these experiments. Polyclonal antibody against Gas3/PMP22 was kindly provided by Dr. C. Brancolini (Dipartimento di Scienze e Tecnologie Biomediche, Sezione di Biologia and MATI Center of Excellence, Università di Udine, Italy). Horseradish peroxidase-conjugated secondary antibodies anti-mouse or -rabbit IgG (GE Healthcare) were used in Western blot analyses; ALEXA488 anti-mouse IgG and ALEXA594 anti-rabbit IgG (Molecular Probes Inc., Eugene, OR) antibodies were used for immunofluorescence reactions.
Animal Model
Transgenic Sprague-Dawley rats overexpressing PMP22 (CMT1A rats) were genotyped by PCR (4). Heterozygous (+/−) animals and normal age-matched littermates were used for experiments. Rearing conditions were consistent with the guidelines of the Italian Health Ministry relating to the use and storage of transgenic organisms.
Cell Cultures
Primary SC cultures were isolated from sciatic nerves of adult and newborn CMT1A rats, as described previously (7, 8). Control SC cultures were obtained from wild type rats with the corresponding genetic background. SC from adult animals were grown for 4 days in DMEM/F-12 (Invitrogen) containing 10% fetal calf serum, penicillin, streptomycin, and 10−5 m cytosine arabinoside (Sigma). SC cultures were then processed for molecular, biochemical, and functional assays (migration and release of ciliary neurotrophic factor) 24 h later or after 3 more weeks in culture (5- and 25-day-old cultures, respectively). Unless otherwise indicated, all experiments were performed on 5-day-old cultures from adult CMT1A or from wild type rats.
Characterization of these cells was performed by Western blot and immunofluorescence (see below). SC from newborn rats were used for molecular, biochemical, and myelination experiments. Primary cultures of dorsal root ganglion (DRG) sensory neurons were established from 15-day-old wild type rat embryos. After extraction under sterile conditions, 35–40 DRG were removed from each embryo, incubated for 30 min with 0.25% trypsin in Hanks' solution, and minced to obtain a suspension of DRG cells. Cells were washed and resuspended in neurobasal medium (Invitrogen) supplemented with 15% newborn calf serum, ascorbic acid (100 μg/ml final concentration), and nerve growth factor (NGF) at 5 ng/ml final concentration. This suspension was plated on collagen-coated ACLAR dishes at a density of 15 × 104 cells/dish. After 48 h, cells were treated with 5-fluoro-2′-deoxyuridine medium (neurobasal medium, 15% newborn calf serum, NGF, ascorbic acid, and 10−5 m 5-fluoro-2′-deoxyuridine and uridine) (Invitrogen) for two 48-h cycles to eliminate fibroblasts. For myelination, 3 × 105 primary SC from newborn rats were seeded onto DRG neurons (see below).
Schwann Cells Characterization
Characterization of CMT1A and wild type SC by real time PCR analyses for PMP22, MPZ, MBP, L1, Krox-20, and GFAP was performed as described below.
Western Blot
SC lysates from wild type and CMT1A rats were prepared in 100 μl of the lysis buffer specific for the protein analyzed (myelin proteins buffer: 95 mm NaCl, 25 mm Tris-HCl, pH 7.4, 10 mm EDTA, 2% SDS, 0.01% protease inhibitor mixture; transcription factor (Krox-20) buffer: 20 mm HEPES, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA, 2 mm Na3VO4, 1 mm sodium pyrophosphate, 1 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride; cytoskeleton proteins BUST buffer: 0.5% SDS, 8 m urea, 2% β-mercaptoethanol, 0.01% protease inhibitor mixture, 0.1 m Tris-HCl, pH 6.8). Total protein content was determined by the protein detection kit (Bio-Rad), and equal amounts of proteins (20 μg) from normal and transgenic animals were loaded onto a 10% polyacrylamide gel and separated by SDS-PAGE. Proteins were transferred to 0.2-μm pore size nitrocellulose membranes (Amersham Biosciences) and incubated with the specific primary antibodies against PMP22, MBP, MPZ, L1, KROX-20, or GFAP for 18 h and with the antibody against β-tubulin for 1 h. Following ECL detection (Amersham Biosciences), band intensity was measured with the Gel Doc 1000 imaging system (Bio-Rad). The expression of each protein was normalized on β-tubulin.
Fluorimetric Determination of the [Ca2+]i
Adherent (on 20-mm coverslips) wild type and CMT1A SC were incubated with 10 μm Fura-2AM in complete medium for 45 min at 37 °C and then washed with Hanks' balanced salt solution (HBSS). The coverslips were mounted on the stage of an inverted microscope (Zeiss IM35, Germany), and [Ca2+]i calibrations were performed as described previously (9). Wild type and CMT1A SC were also seeded in 96-well plates (1.5 × 104 cells/well) incubated with 10 μm Fluo-3AM in complete medium for 45 min at 37 °C and then washed with HBSS. Ca2+ measurements upon addition of 1 mm ATP were performed in 96-well plates, and fluorescence (excitation, 485 nm; emission, 520 nm) was measured every 3 s with a fluorescence plate reader (Fluostar Optima; BMG Labtechnologies GmbH, Offenburg, Germany), as described (10). The intensity of emitted light was plotted as a function of time.
Determination of Intracellular ATP Levels
Wild type and CMT1A SC were cultured in 60 × 10-mm dishes; the medium was removed, and adherent cells were rinsed twice with 1 ml of HBSS. Cells were then scraped in 340 μl of water, and 10 μl of 50% trichloroacetic acid were added to 300 μl of the cell extract, whereas the rest of the lysate was used for protein content determination (11). Acid excess was removed with diethyl ether, and high pressure liquid chromatography analysis of cell nucleotides was performed as described (12).
Mitochondrial Membrane Potential in Intact Cells
For measurements of mitochondrial membrane potential, cells were washed and incubated in HBSS containing 10 mm HEPES in the presence of 10 nm tetramethylrhodamine methyl ester at 37 °C for 1 h. Cell fluorescence images were acquired on a Leica TCS SL confocal microscope, equipped with a HCX PL APO CS 63.0 × 1.40 oil objective, at 30-s intervals. The difference between the level of tetramethylrhodamine methyl ester fluorescence before and after addition of the uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (2 μm) was calculated for each cell and was considered as a measure of the ΔΨm (13).
Determination of Intracellular Inositol 1,4,5-Trisphosphate (IP3) and Cyclic ADP-ribose (cADPR) Levels
Wild type and CMT1A SC were cultured in 60 × 10-mm dishes; the medium was removed, and cells were rinsed twice with 1 ml of HBSS. Cells were then scraped in 300 μl of water; 20 μl of perchloric acid (9 m) was added to 250 μl of the cell extract, and the rest of the cell lysate was used for protein content determination (11). Intracellular IP3 levels were measured on the neutralized cell extracts with a specific radioimmunoassay (Amersham Biosciences), as described (10). The cADPR content was measured on the neutralized cell extracts by a highly sensitive enzymatic cycling assay (10, 14).
Assays of ADP-ribosyl Cyclase Activity
Ectocellular ADP-ribosyl cyclase was assayed on intact, adherent wild type and CMT1A SC. Cells were incubated in HBSS at 37 °C in the presence of 0.1 mm NAD+. Aliquots (300 μl) were withdrawn at various times, and 20 μl of 9 m perchloric acid were added to each aliquot; the cADPR concentrations were measured by the enzymatic cycling assay (14, 15).
Real Time Reverse Transcription-PCR
Total RNA was extracted from SC using the RNeasy micro kit (Qiagen, Milan, Italy) according to the manufacturer's instructions and performing the on-column DNase treatment to remove trace amounts of genomic DNA; total RNA was reverse-transcribed into cDNA using the iScriptTM cDNA synthesis kit (Bio-Rad). The cDNA was used as template for real time PCR analysis; reactions were performed in an iCycler iQ5 real time PCR detection system (Bio-Rad). The rat specific primers were designed by using Beacon Designer 2.0 software (Bio-Rad), and their sequences were as follows: P2X7 forward 5′-CTGGAATTGCAGGTGGATATGAGG-3′ and reverse 5′-AAGATGTGGTTGTGAAGGTGTAGG-3′; PMP22 forward 5′-AATAATCCGCTGCCCGAATCAATG-3′ and reverse 5′-CTCCGCCTTCAGGGTCAAGTG-3′; Krox-20 forward 5′-CTCACGCCACTCTCCACCATC-3′ and reverse 5′-CCTCACCGCCTCCACTTGC-3′; MPZ forward 5′-CTGTTGCTGCTGTTGCTCTTCTAC-3′ and reverse 5′-TTGGTGCTTCGGCTGTGGTC-3′; MBP forward 5′-GCACAGAGACACGGGCATCC-3′ and reverse 5′-CGGGCATGAGAAGGCAGAGG-3′; L1 forward 5′-TCTTGCTCCTCATCCTGCTCATC-3′ and reverse 5′-CACTGTACTCGCCGAAGGTCTC-3′; neural cellular adhesion molecules forward 5′-ACCATACTCCAGCACAGCACAG-3′ and reverse 5′-AGCGACTTCCACTCAGCCTTG-3′; GFAP forward 5′-GAAGGTTGAGTCGCTGGAGGAG-3′ and reverse 5′-CGCTGTGAGGTCTGGCTTGG-3′; and p75 forward 5′-CCAGCAGACCCATACGCAGAC-3′ and reverse 5′-GCCAGATGTCGCCAGGTATCC-3′. Each sample was assayed in triplicate in a 25-μl amplification reaction, containing 30 ng of cDNA, primers mixture (0.4 μm each of sense and antisense primers), and 12.5 μl of 2× iQ SYBR Green Supermix Sample (Bio-Rad). The amplification program included 40 cycles of two steps, each including heating to 95 and 60 °C, respectively. Fluorescent products were detected at the last step of each cycle. To verify the purity of the products, a melting curve was produced after each run. Values were normalized to GAPDH, and β-actin mRNA expression was measured using the following primers: GAPDH forward 5′-ATGATTCTACCCACGGCAAG-3′ and reverse 5′-CTGGAAGATGGTGATGGGTT-3′; and β-actin forward 5′-GGGAAATCGTGCGTGACATT-3′ and reverse 5′-GCGGCAGTGGCCATCTC-3′. Statistical analysis of the quantitative real time PCR was obtained using the iQ5 Optical System Software version 1.0 (Bio-Rad) based on the 2−ΔΔCt method, which calculated relative changes in gene expression of the target normalized to GAPDH and β-actin. The quantitative PCR efficiencies were determined by series of 5-fold dilutions for each experiment and each gene and were always between 95 and 102%. Data are presented as means ± S.D.
Small Interfering Oligonucleotide Transfection
Transfection of SC was performed using the Nucleofector system (Amaxa GmbH, Cologne, Germany). Preliminary experiments were carried out with pmaxGFP to select the cell concentration and the program yielding the highest percentage of cell transfection, which was monitored by measuring green fluorescent protein-positive cells. Moreover, viability of SC after transfection was estimated measuring propidium iodide-positive cells by flow cytometry. Thus, the following protocol (giving a transfection efficiency of up to 60% and a cell viability of ∼80–90%) was chosen as optimal. SC were transfected with 2 μg of pmaxGFP, with 2 μm P2X7 or PMP22 targeting duplex small interference RNA (siRNA1 and siRNA2) and with 2 μm negative control (scramble) using the rat neuron Nucleofector kit according to the manufacturer's instructions (Nucleofector program A-033). The P2X7 targeting and negative control siRNA were StealthTM siRNA obtained from Invitrogen (siRNA1, P2X7-RSS310827, 5′-GAUACUUGAAGCCACUGUACUGCCC-3′; siRNA2, P2X7-RSS310829, 5′-AGACAGUUCCAAGAAGUCCGUCUGG-3′); the PMP22-targeting siRNA were ON-TARGETplusTM siRNA obtained from Dharmacon (Chicago, IL). Immediately after transfection, cells were resuspended in 0.5 ml of DMEM/F-12 supplemented with 10% fetal calf serum, penicillin (50 units/ml), and streptomycin (50 μg/ml) and incubated in a humidified 5% CO2 atmosphere at 37 °C for 24 h. Untransfected (UT) SC were treated identically using an equal volume of nucleofection buffer in place of siRNA oligos. After 24 h, green fluorescent protein-positive cells were evaluated by using a FACS-Canto flow cytometer (BD Biosciences), and data, expressed as percentage of propidium iodide-negative (live) cells, were analyzed by using DIVA software. Twenty four and 48 h, 1 week, and 10 days after transfection, RNA was extracted, and real time PCR analyses were performed as described previously; migration and CNTF release were tested on transfected and untransfected cells (see below).
Overexpression of P2X7
The plasmid pcDNA3 containing the full-length rat P2X7 cDNA was kindly provided by Prof. F. Di Virgilio (Dept. of Experimental and Diagnostic Medicine, University of Ferrara, Italy). Wild type SC were transfected using the Nucleofector system, as described above, in the presence of 3 μg of plasmid encoding for the rat P2X7 or in the presence of the empty plasmid (NC). The [Ca2+]i and migration were evaluated 24 h after transfection.
shRNA Plasmid Stable Transfection
Stable transfection with shRNA was performed using the SureSilencingTM shRNA plasmid kit for rat P2X7 (KR44893N SABiosciences, Frederick, MD). The most efficient target sequence for RNA interference was selected among four sequences. The final selected target sequence for rat P2X7 corresponded to coding region 826–846 (5′-CCAGGAAATCGGAGAGAACTT-3′). The shRNA plasmid targeting P2X7 (shRNA-P2X7) or the negative control shRNA plasmid was transfected, as described above, into SC cells from newborn rats. After 48 h, the transfected cells were subcultured at an appropriate density in fresh DMEM/F-12 containing 0.2 mg/ml G418 (Calbiochem). G418-resistant cell populations were established within 10 days. RNA was extracted, and real time PCR analyses were performed as described above; myelination was tested on transfected cells co-cultured with normal sensory neurons (see below).
Migration Assay
To test the ability of CMT1A SC to migrate after the P2X7 receptor siRNA-specific suppression or inhibition by the use of P2X7 receptor inhibitor KN-62, we followed a published protocol (8, 16). In P2X7 down-regulation experiments by specific siRNA, SC were electroporated in the absence of siRNA oligos UT or transfected with P2X7-targeting StealthTM siRNA (Invitrogen). In P2X7 overexpression experiments, SC were electroporated in the presence of the empty plasmid (NC) or of the pcDNA3 containing the full-length rat P2X7 cDNA. After transfection, cells were immediately plated on poly-l-lysine-coated 12-mm coverslips at a density of 1 × 105 cells/coverslip in a 40-μl drop. After 4 h, 200 μl of basal medium (DMEM/F-12 containing 10% fetal calf serum) were added, and cells were cultured for 24 h. The medium was then replaced with basal medium supplemented with 8 μm forskolin (FSK), which mimics the presence of the axon in primary cultures of SC (17).
In KN-62 inhibition experiments, SC were plated as described above; after 24 h, the medium was supplemented with 8 μm FSK in the presence of 100 nm KN-62. Twenty four h later, a cell-free area was generated by gently scratching the cell monolayer with a sterile yellow Gilson pipette tip, thus resulting in the formation of an ∼1-mm-wide gap. Immediately after scratching (time zero, T0), the medium was replaced, and the first microscopic image of the cells was acquired. Twelve hours later (T1), the same SC cultures were photographed again, and images were saved. The extent of cell migration was assessed by subtracting the average gap width at T1 from the average gap width at T0. SC migration was monitored for a maximum of 12 h to avoid interference in the evaluation because of SC proliferation (16). Assessment of migration distance was performed on 10 SC cultures for each condition, using the Image Pro-Plus Software (Immagini e Computer, Rho, Milan, Italy).
Enzyme-linked Immunosorbent Assay Quantification of CNTF
CMT1A and wild type SC were electroporated without siRNA UT or transfected with P2X7-targeting StealthTM siRNA (Invitrogen), and 1 × 106 SC were immediately plated on 60-mm poly-l-lysine-coated dishes. Moreover, 1 × 106 SC/dish were also plated to be treated with 100 nm KN-62. In both experimental settings, SC cultures were supplemented with 8 μm FSK, and 48 h later, the culture medium was collected, and the cells were mechanically disrupted in PBS containing a protease inhibitor mixture (P8340, Sigma). The cell protein concentration was measured (11), and CNTF content in the culture medium was determined using a Rat CNTF DuoSet kit (R & D Systems), according to the manufacturer's instructions.
Myelination Studies
Primary SC from newborn rats (wild type and CMT1A) transfected with the shRNA-P2X7 plasmid or with the negative control shRNA plasmid were seeded (3 × 105 cells/dish) onto DRG neurons in neurobasal medium, supplemented with 15% newborn calf serum, 2 mm glutamine, 5 ng/ml NGF, and 100 μg/ml ascorbic acid.
The myelination medium was changed three times every week, and the cultures were allowed to myelinate for up to 4 weeks and used for light microscopy morphological and morphometric analyses and molecular studies.
Light Microscopy, Immunocytochemistry, and Morphometric Analyses
For immunocytochemistry, rat DRG-SC co-cultures on ACLAR dishes were fixed for 30 min in 4% paraformaldehyde in PBS, permeabilized for 5 min in cold methanol at −20 °C, and incubated for 1 h in blocking buffer (10% normal goat serum in PBS). Incubation with the primary antibodies was performed for 18 h at 4 °C with a monoclonal antibody against MBP (1:100) and a polyclonal antibody against neurofilaments (1:100) in blocking buffer. After washing, cells were incubated with the secondary goat anti-mouse ALEXA594-IgG and goat anti-rabbit ALEXA488-IgG antibodies at a 1:250 dilution for 1 h at 25 °C. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole, and ACLAR dishes were sealed on slides. Images were taken with an Olympus PROVIS AX60 microscope, connected to an Olympus DP70 digital camera.
For morphometric analysis, DRG-SC co-cultures were washed in PBS, fixed overnight in Trump's fixative at 4 °C, post-fixed the following day in 2% osmium tetroxide, and stained with 1% Sudan black in 70% ethanol. Each culture was analyzed for the presence of myelin and for the morphology of the observed fibers. The morphometric study was performed directly on the cultures after fixation, staining, and mounting on slides. It is essential to use a systematic sampling technique because of the following: (i) the culture dish does not provide a completely uniform physical environment; (ii) myelinated fibers are not randomly distributed throughout the dish; and (iii) the differences between control and dys/demyelinating cultures are sometimes so obvious as to compromise a blinded study. A 1-cm diameter circle was superimposed on the culture, with the center of the circle overlying the center of the culture. From each culture 250–300 images were acquired and stored using the ×40 objective. Morphometric evaluation of the digitized images was performed using the Image Pro-Plus Software (Immagini e Computer, Rho, Milan, Italy). A grid mask consisting of 20 circles (50-μm diameter) able to completely cover the culture image below was superimposed to the image itself, and only the myelin segments crossing the circles were counted. For each culture, the total number of myelin segments present in all the images stored was scored, and the density of myelinated fibers was obtained dividing this number by the total area considered for each culture. Each condition was performed in triplicate or quadruplicate, and the data presented are the mean ± S.E. of three independent experiments. To validate this morphometric technique, preliminary experiments were performed on organotypic DRG cultures from 15-day-old CMT1A or wild type embryos. The morphometric evaluation of the myelination rate showed that the number of myelinated segments/mm2 was significantly higher in wild type compared with CMT1A cultures. Indeed, wild type cultures (n = 5) ranged from 0.00040 to 0.00094 myelinated segments/mm2 (mean ± S.E., 0.000728 ± 0.00009), whereas CMT1A cultures (n = 9) ranged from 0.00005 to 0.00036 myelinated segments/mm2 (mean ± S.E., 0.0002089 ± 0.00003). These numbers are ∼1 log higher than those measured in DRG-SC co-cultures (see “Results” and Fig. 7C), which requires separation and isolation of SC and neurons prior to the onset of the co-culture.
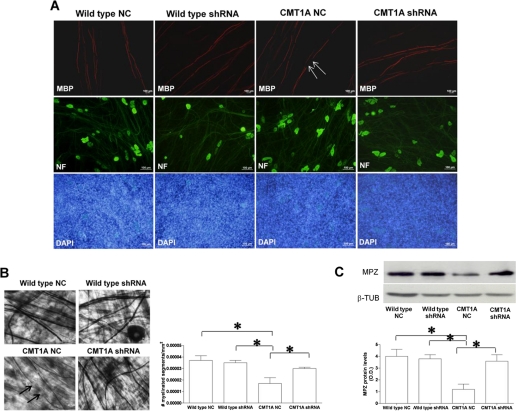
FIGURE 7.
Down-regulation of P2X7 receptor expression in CMT1A SC improves myelination. Wild type and CMT1A SC from newborn rats were electroporated in the presence of negative control shRNA or in the presence of a shRNA plasmid targeting P2X7 (wild type shRNA and CMT1A shRNA). SC were then seeded onto DRG neurons and allowed to myelinate for up to 4 weeks. A, immunocytochemistry for MBP on DRG/SC co-cultures. Axons are stained with an anti-neurofilament antibody, and nuclei of SC are stained with 4′,6-diamidino-2-phenylindole (DAPI). Co-cultures with NC-transfected CMT1A SC show axonal segments devoid of myelin (arrows), along with normally myelinated internodes. Co-cultures with shRNA-P2X7-transfected SC show a higher extent of myelination. B, light microscopy morphometric analysis of myelination in DRG/SC co-cultures. Upper panels show co-cultures stained with Sudan black. Arrows indicate demyelinated internodes. The histograms show the density of myelin segments in the various co-cultures. Data are mean numbers of myelinated segments/mm2 ± S.E. in 250–300 fields of four independent cultures for each condition. C, Western blot analysis of MPZ levels in lysates of the various co-cultures. The histograms show the densitometric quantification of band intensity: values shown are the mean ± S.E. of three independent experiments. β-TUB, β-tubulin.
Immunoblot Analysis
The amount of myelin formation was also evaluated by studying MPZ expression. Briefly, DRG-SC co-cultures were rinsed with PBS and scraped in myelin proteins extraction buffer, and proteins were then separated on 15% SDS gels. Western blot analysis and band intensity quantification were performed as described above.
Statistical Analyses
All parameters were tested by paired t test or by a one-way analysis of variance. p values < 0.05 were considered significant.
RESULTS
Characterization of SC Cultures
The phenotypic characterization of 5-day-old SC cultures from CMT1A and wild type rats revealed expression of PMP22, MPZ, and MBP, which are up-regulated during terminal SC differentiation and myelination (18), together with L1 and GFAP, which should only be expressed by nonmyelinating SC and immature SC precursors (18, 19) (Fig. 1). Co-expression of myelin proteins and early SC markers in CMT1A SC has been already reported (20). We found that CMT1A SC express higher levels of PMP22, MPZ, Krox-20, L1, and GFAP compared with wild type ones (Fig. 1), in line with previously published observations (20).
FIGURE 1.
Characterization of SC cultures. Five-day-old cultures of wild type and CMT1A SC were subjected to the following: A, RNA extraction and real time PCR analysis of gene expression relative to the proteins shown in A; expression in CMT1A SC is shown relative to that of the same gene in wild type SC (n = 3); B, Western blot analysis for the indicated proteins, as described under “Experimental Procedures”; histograms show the densitometric analysis of the corresponding bands, normalized to β-tubulin, and are the mean ± S.E. of three different experiments.
[Ca2+]i Is Increased in CMT1A SC
Previous observations suggested that abnormally elevated Ca2+ levels lead to impairment of myelin production by SC (5, 6). Thus, we compared the basal [Ca2+]i in SC isolated from the rat model of CMT1A (CMT1A SC) and from wild type animals (wild type SC). The [Ca2+]i recorded in SC from CMT1A rats (131 ± 9 nm, n = 10) was significantly higher (p < 0.001), compared with the value observed in SC from wild type rats (57 ± 4 nm, n = 13) (Table 1). An increase of the basal [Ca2+]i could be due, in principle, to either one or to a combination of the following mechanisms: (i) impaired ability of the cells to extrude Ca2+ from the cytoplasm through ATP-dependent pumps on the plasma membrane and/or on the endoplasmic reticulum (ER); (ii) increased release of Ca2+ from intracellular Ca2+ stores; and (iii) increased influx of extracellular Ca2+ (21–24).
TABLE 1.
Intracellular Ca2+ concentration in SC from CMT1A and wild type rats
[Ca2+]i measurements, in the absence or presence of TG, were performed on Fura-2-loaded wild type and CMT1A SC, as described by Ref. 9. Results are the mean ± S.D. of at least 10 or 5 determinations for basal or +TG, respectively.
a p < 0.001 (CMT1A versus the corresponding wild type).
Therefore, we first investigated whether a shortage of ATP could be responsible for dysfunction of active Ca2+ transport across cell membranes in CMT1A SC. The intracellular ATP concentration ([ATP]i) in CMT1A SC was in fact higher compared with wild type SC (3.13 ± 0.42 versus 1.95 ± 0.23 nmol/mg protein, respectively, n = 4, p < 0.01; Table 2). The driving force for mitochondrial ATP synthesis, i.e. the mitochondrial membrane potential (ΔΨm), was also compared in intact SC from control and CMT1A rats. The difference between the level of fluorescence (Δf) before and after addition of the uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone was again significantly higher (p < 0.01) in CMT1A SC than in control SC (41 ± 18, n = 75, and 32 ± 15, n = 61, respectively). These results rule out the possibility that the high basal [Ca2+]i in CMT1A SC could be due to low ATP levels impairing the ER Ca2+ re-uptake and/or extrusion of Ca2+ from the cytoplasm through ATP-dependent pumps on the plasma membrane.
TABLE 2.
Intracellular ATP, cADPR, and IP3 levels and ADP-ribosyl cyclase activity in SC from CMT1A and wild type rats
Intracellular ATP, IP3, and cADPR levels were determined in wild type and CMT1A SC as described under “Experimental Procedures.” Intact wild type and CMT1A SC were incubated in the presence of NAD+, and production of cADPR was evaluated for estimation of ADP-ribosyl cyclase activity, as described previously (15). Results are the mean ± S.D. of at least four determinations.
| [ATP]i | [IP3]i | [cADPR]i | ADP-ribosyl cyclase | |
|---|---|---|---|---|
| nmol/mg protein | nmol/mg protein | pmol/mg protein | pmol/min/mg protein | |
| Wild type SC | 1.95 ± 0.23 | 0.32 ± 0.05 | 4.72 ± 2.31 | 4.83 ± 1.01 |
| CMT1A SC | 3.13 ± 0.42a | 0.36 ± 0.07 | 2.23 ± 0.91b | 2.50 ± 0.35b |
a p < 0.01 (CMT1A versus wild type).
b p < 0.05.
Next, we investigated the state of replenishment of the intracellular calcium stores in SC from CMT1A rats, by testing the effect of thapsigargin (TG), which is known to empty the cADPR- and IP3-sensitive ER Ca2+ stores by inhibiting the ATP-dependent ER Ca2+ pumps (25). Upon addition of 10 μm TG, the [Ca2+]i increased less in CMT1A SC (315 ± 28 nm, n = 5) compared with wild type cells (417 ± 43 nm, n = 6) (Table 1), indicating that the Ca2+ content of the TG-sensitive stores was significantly reduced (p < 0.001) in SC from CMT1A rats compared with controls.
Ca2+ release from ER stores occurs mainly through the IP3 receptors and the ryanodine receptors (26), the latter being gated by cADPR (23, 27, 28). Therefore, to verify whether the reduced Ca2+ content of the TG-sensitive stores in CMT1A SC (Table 1) could be due to a sustained, either IP3- or cADPR-dependent, Ca2+ release from the ER, the intracellular concentration of these second messengers was determined in wild type and CMT1A SC. The intracellular IP3 concentration ([IP3]i) in CMT1A SC was not significantly different from the value found in wild type cells (0.36 ± 0.07 versus 0.32 ± 0.05 nmol/mg proteins, respectively, n = 4, p = not significant). The intracellular cADPR concentration ([cADPR]i) was significantly lower in CMT1A SC compared with wild type cells (2.23 ± 0.91 versus 4.72 ± 2.31 pmol/mg proteins, respectively, n = 5, p < 0.05) (Table 2). In line with the cADPR content, the ADP-ribosyl cyclase activity, responsible for cADPR synthesis from NAD+, was also lower in CMT1A SC than in the control ones (2.50 ± 0.35 versus 4.83 ± 1.01 pmol of cADPR/min/mg, respectively, n = 4, p < 0.05) (Table 2).
The fact that [IP3]i and [cADPR]i were not increased in CMT1A compared with wild type SC rules out a causal role of IP3 and cADPR in the enhanced basal [Ca2+]i of CMT1A. The lower concentration of cADPR in CMT1A SC might indicate the occurrence in these cells of compensatory mechanisms aimed at limiting intracellular Ca2+ release. Indeed, incubation of CMT1A SC with 50 μm ryanodine, a concentration inhibiting the calcium-induced calcium release (CICR) mechanism (29, 30), significantly reduced the [Ca2+]i in CMT1A SC from a basal value of 130 ± 10 nm in untreated CMT1A SC to 111 ± 3, 94 ± 4, and 93 ± 4 nm after 1, 3, or 6 h of treatment, respectively (data not shown). Altogether, these results suggest that the depletion of TG-sensitive Ca2+ stores in CMT1A SC might be due to a CICR mechanism from ryanodine-sensitive stores, possibly triggered by an increased influx of extracellular Ca2+ in CMT1A SC.
Causal Role of the P2X7 Purinoceptor in the Increased [Ca2+]i in CMT1A SC
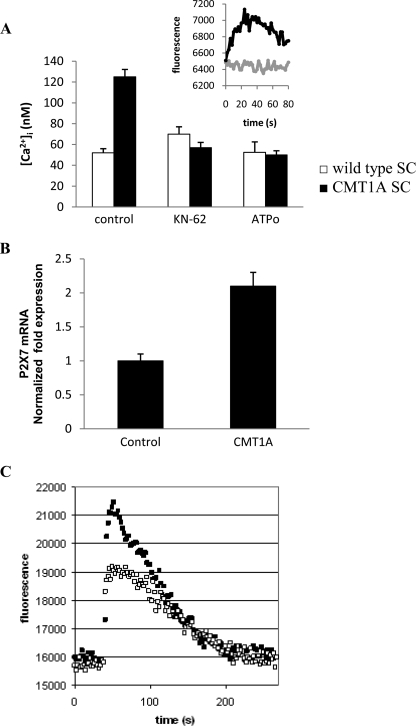
It has been reported that PMP22 interacts with the C-terminal domain of the P2X7 purinergic receptor, leading to receptor activation, channel opening, and influx of extracellular Ca2+ (31). Accordingly, to assess the possible involvement of P2X7 in the CICR mechanism leading to increased [Ca2+]i levels in CMT1A SC, we preincubated the cells with KN-62, a specific inhibitor of P2X7 receptor (32). Although KN-62 was described as a specific inhibitor of human P2X7 (32), it has been successfully used to inhibit P2X7 also in rat cells (33, 34). KN-62, at 20 μm (Fig. 2A, inset) but also at concentrations as low as 100 nm, indeed prevented the Ca2+ increase induced by ATP in SC. Preincubation of cells for 90 min with 20 μm KN-62 reduced the [Ca2+]i to levels in CMT1A SC comparable with those measured in SC from normal rats (Fig. 2A). Complete restoration of [Ca2+]i levels to normal values was also observed at much lower KN-62 concentrations by prolonging the incubation time to 24 h; specifically, the [Ca2+]i was reduced to 58 and to 52 nm with 100 nm and 1 μm KN-62, respectively (n = 3). The P2X7 receptor inhibitor oxidized ATP (oATP) (35) induced a similar decrease of the [Ca2+]i in CMT1A SC (Fig. 2A). Conversely, preincubation with KN-62 or with oATP did not significantly modify the [Ca2+]i of normal SC (Fig. 2A).
FIGURE 2.
Increased basal [Ca2+]i in CMT1A SC is because of P2X7 overexpression. A, wild type (white bars) and CMT1A (black bars) SC were preincubated in the absence (control) or in the presence of KN-62 (20 μm for 90 min) or of oATP (50 μm for 6 h) and loaded with Fura-2AM during the last 45 min of incubation; [Ca2+]i was measured as described (9). Results are the average ± S.D. of three determinations. p < 0.001 between KN-62- or oATP-treated CMT1A SC compared with control CMT1A SC. Inset, Fluo-3AM-loaded CMT1A were preincubated (gray trace), or not (black trace), with 20 μm KN-62 for 10 min and then challenged with 1 mm ATP, and [Ca2+]i changes were measured using a fluorescence plate reader (10). B, expression of P2X7 receptor in control and CMT1A SC was estimated by real time PCR analysis (average ± S.D. of six determinations). C, Fluo-3AM-loaded control (white squares) and CMT1A (black squares) SC were stimulated with 1 mm ATP, and [Ca2+]i changes were measured using the fluorescence plate reader. Traces are mean values recorded in different measurements (n = 3) (S.D. <15%).
Interestingly, real time PCR analysis revealed a significantly higher level of expression of the P2X7 receptor in CMT1A SC compared with the wild type SC (Fig. 2B). In line with this observation, stimulation of SC with millimolar ATP, a condition known to trigger P2X7-mediated Ca2+ influx (36), consistently induced a higher increase of the [Ca2+]i in CMT1A SC than in control SC (Fig. 2C).
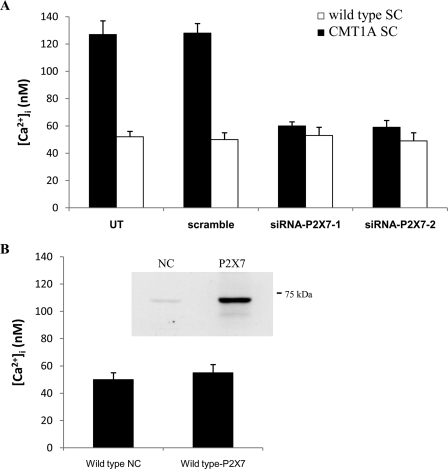
To demonstrate conclusively that Ca2+ influx through P2X7 is responsible for the higher basal [Ca2+]i recorded in CMT1A SC compared with control cells, P2X7 expression was down-regulated by means of specific siRNA transfection. Wild type and CMT1A SC were either electroporated in the absence of siRNA (UT) or transfected by nucleofection with two different siRNAs specific for P2X7 (siRNA-P2X7-1 and siRNA-P2X7-2) and in parallel with the StealthTM negative control (scramble), which does not match any sequence in the rat genome. P2X7 mRNA levels in wild type and CMT1A SC were then measured by real time PCR at 24 h after transfection. Cells (both CMT1A and wild type cells) transfected with the negative control (scramble) had P2X7 mRNA levels comparable with those of the corresponding SC electroporated in the absence of siRNA (UT) (Fig. 3A). Conversely, P2X7 mRNA levels were decreased to 20 and to 25% of control values (UT) in CMT1A cells transfected with siRNA1 and siRNA2, respectively (Fig. 3A), and to 25 and to 30% of UT values in wild type cells transfected with siRNA1 and siRNA2, respectively (data not shown). Nucleofection per se did not modify the relative expression level of P2X7 in wild type SC compared with CMT1A SC (Fig. 3A wild type UT and CMT1A UT), which was similar to the relative expression level observed in nonelectroporated cells (Fig. 2B). Down-regulation of P2X7 by siRNA transfection was accompanied by the restoration of [Ca2+]i levels similar to those recorded in control SC (Fig. 4A). Conversely, down-regulation of P2X7 in wild type SC did not significantly modify the [Ca2+]i levels (Fig. 4A).
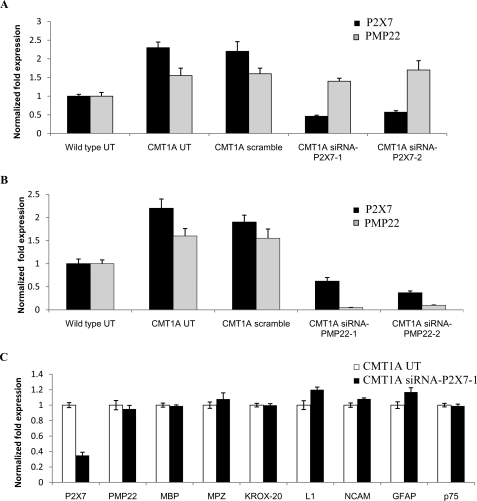
FIGURE 3.
Up-regulation of P2X7 in CMT1A SC is a consequence of PMP22 overexpression. Wild type and CMT1A SC were electroporated with P2X7-specific siRNAs (either siRNA-P2X7-1 or siRNA-P2X7-2) (A) or with PMP22-specific siRNAs (either siRNA-PMP22-1 or siRNA-PMP22-2) (B). Controls included the following : wild type and CMT1A SC electroporated without siRNA (UT) or with a negative control siRNA (scramble). C, CMT1A SC were electroporated with the siRNA-P2X7-1. After 24 h, cells were subjected to total RNA extraction, and gene expression was investigated by real time PCR. A and B, expression of P2X7 (black bars) and PMP22 (gray bars) is shown relative to the corresponding value in wild type UT SC; C, expression of the indicated genes in siRNA-P2X7-1-transfected CMT1A SC is shown relative to that in CMT1A UT SC. Results shown are the mean ± S.D. of three experiments.
FIGURE 4.
P2X7 expression down-regulation by siRNA restores normal basal [Ca2+]i in CMT1A SC. A, wild type and CMT1A SC were electroporated in the absence of siRNA (UT), in the presence of a negative control siRNA (scramble), or in the presence of the P2X7-specific siRNA (siRNA-P2X7-1 or siRNA-P2X7-2). After 24 h, cells were subjected to total RNA extraction and real time PCR for P2X7 expression (see Fig. 2A) or loaded with Fura-2AM. The basal [Ca2+]i was measured as described in Ref. 9. Results shown are the mean ± S.D. of three experiments. p < 0.001 between siRNAP2X7-1 or siRNAP2X7-1 CMT1A SC compared with untransfected CMT1A SC (UT). B, wild type cells were electroporated in the presence of a negative control (NC) plasmid or of the plasmid containing full-length rat P2X7 cDNA (P2X7). After 24 h, cells were loaded with Fura-2AM, and the basal [Ca2+]i was measured. Results shown are the mean ± S.D. of three experiments. Inset, overexpression of P2X7 was verified by Western blot analyses, using an anti-P2X7 polyclonal antibody (Calbiochem).
Next, we examined the effect of P2X7 knockdown on the expression of PMP22. The primers used for real time PCR detect total PMP22 levels, i.e. all transcripts arising from different promoters (37). Interestingly, P2X7 overexpression in CMT1A SC was even more pronounced than that of PMP22, which is known to be overexpressed in CMT1A SC and to be responsible for the disease (1.6-fold, Figs. 1 and 3). Real time PCR analysis confirmed the presence of higher levels of PMP22 in CMT1A SC compared with the wild type ones (Fig. 3). P2X7 knockdown did not affect PMP22 expression either in CMT1A (Fig. 3A) or in wild type SC (data not shown). Similarly, P2X7 knockdown by siRNA did not significantly modify the expression of a number of markers of the cell differentiation status, i.e. MBP, MPZ, and Krox-20 as markers of differentiated cells and L1, neural cellular adhesion molecules, GFAP, and p75 as markers of immature cells (Fig. 3C).
Interestingly, although P2X7 silencing did not affect PMP22 expression (Fig. 3), PMP22 down-regulation in CMT1A SC (Fig. 3B) and in wild type SC (data not shown), obtained by transfection with two different siRNAs specific for PMP22 (siRNA-PMP22-1 and siRNA-PMP22-2), also decreased P2X7 expression. Moreover, transfection of wild type and CMT1A SC with the StealthTM negative control (scramble) did not significantly modify either PMP22 or P2X7 expression (Fig. 3).
It is well established that expression of myelin proteins is down-regulated in SC cultured in the absence of neurons (18). Indeed, expression of PMP22, MBP, and MPZ was significantly lower in both wild type and CMT1A SC from 25-day-old cultures, compared with the respective SC from 5-day-old cultures. In agreement with the down-regulation of PMP22, expression of P2X7 was also significantly reduced in 25-day-old cultures, compared with 5-day-old cultures, in both wild type and CMT1A SC (data not shown). The [Ca2+]i recorded in SC from 25-day-old cultures was not significantly different in CMT1A and wild type SC (50 ± 5 versus 48 ± 6 nm, respectively, n = 4), suggesting that this basal [Ca2+]i value is maintained through P2X7-independent mechanisms.
Next, to verify whether P2X7 overexpression per se was sufficient to determine a higher basal [Ca2+]i, independently from PMP22 overexpression, we transfected wild type SC with P2X7. As shown in Fig. 4B, the [Ca2+]i was not significantly different in wild type SC overexpressing P2X7 compared with SC transfected with the empty plasmid.
Taken together, these results suggest that the abnormally high levels of basal [Ca2+]i observed in SC from CMT1A rats are due to PMP22-related overexpression and opening of functional P2X7 receptors, resulting in enhanced Ca2+ influx and in a reactive CICR mechanism.
Pharmacological Inhibition or Down-regulation of P2X7 in CMT1A SC Reverts Cell Functions to Normal
Next, we investigated whether down-regulation of the basal [Ca2+]i in CMT1A SC to values similar to those of wild type cells was able to restore the normal phenotype of CMT1A SC. Thus, selected functional parameters known to be affected by PMP22 overexpression in CMT1A SC, such as migration properties, release of CNTF, and myelination of neurons (8, 38–40), were studied in CMT1A SC upon treatment with the P2X7 antagonist KN-62 or following P2X7 down-regulation (by means of siRNA or shRNA transfection).
Migration
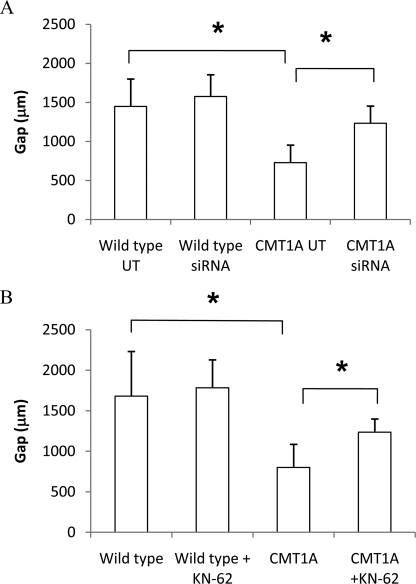
Migration of SC along the axon is a fundamental requirement for assembly of a correct myelin sheath. Both wild type and CMT1A SC were electroporated in the absence of siRNA (UT) or transfected with P2X7 receptor siRNA, and their migration properties were compared (8) (Fig. 5). Moreover, migration of CMT1A SC was also monitored after pharmacological inhibition of P2X7 with KN-62. As already shown (8), migration of CMT1A SC stimulated with FSK (which mimics the presence of the axon in primary cultures of SC, see Ref. 17) was significantly reduced compared with migration of FSK-treated wild type cells before treating the cells with siRNA experiments (728.4 ± 245 versus 1450.3 ± 350 μm; n = 3; p < 0.05) (Fig. 5A) or KN-62 (799.2 ± 286.1 versus 1681 ± 783 μm; n = 5 and n = 4, respectively; p < 0.05) (Fig. 5B). Transfection of CMT1A SC with a specific siRNA for P2X7 determined a significant (p < 0.05) increase of their migration ability (1234.1 ± 218.8 μm; n = 3) (Fig. 5A). In line with this observation, the pharmacological inhibition of P2X7 receptor with KN-62 was also able to restore migration of FSK-stimulated CMT1A SC to values similar to those measured in untreated CMT1A cells (1235.7 ± 461.8 μm; n = 5) (Fig. 5B). Conversely, silencing of P2X7 or treatment with KN-62 did not significantly affect migration of FSK-stimulated wild type SC (Fig. 5). Migration of FSK-stimulated wild type SC was not significantly modified by overexpression of P2X7 (896 ± 160 versus 1011 ± 87 μm in wild type transfected with the empty plasmid or with P2X7-containing plasmid, respectively; n = 5; p = not significant), in line with the observation that overexpression of P2X7 did not determine an increase of the basal [Ca2+]i in wild type cells (Fig. 4B).
FIGURE 5.
P2X7 siRNA silencing or pharmacological inhibition ameliorate migration of CMT1A SC. A, wild type and CMT1A SC were electroporated in the absence of siRNA (UT) or in the presence of a P2X7-specific siRNA. B, wild type and CMT1A SC were exposed, or not, to 100 nm KN-62 for 24 h. To test cell migration, SC movement within a 1-mm-wide gap on the cell monolayer was monitored following down-regulation of P2X7 expression and in the presence of FSK (see “Experimental Procedures”). A one-way analysis of variance was used to compare the different conditions, and results are expressed as means ± S.D. *, p < 0.05.
Production of CNTF
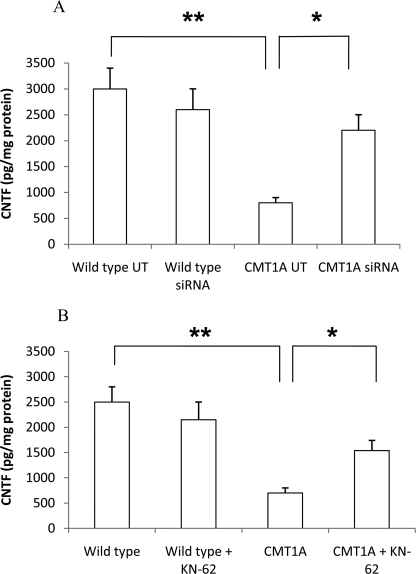
CNTF released by SC behaves as a trophic factor for sensory and motor neurons and stimulates glial cell proliferation and myelin formation. Therefore, impairment of CNTF expression, as demonstrated to occur in CMT1A SC, may be relevant to the pathogenesis of the disease (40–42). We analyzed CNTF release in CMT1A SC following both P2X7-specific silencing and pharmacological inhibition by KN-62. As shown in Fig. 6A, down-regulation of P2X7 determined an increased release of CNTF from CMT1A SC (CMT1A siRNA) compared with the release measured in CMT1A control cells (UT) (2193 ± 323 versus 767 ± 133 pg of CNTF/mg of protein; n = 3; p < 0.01), almost restoring the CNTF production to normal values, recorded in wild type SC (2992 ± 443 pg of CNTF/mg of protein). Likewise, release of CNTF from FSK-stimulated CMT1A SC increased upon treatment of cells with 100 nm KN-62 (from 727 ± 94 to 1540 ± 194 pg of CNTF/mg of protein; n = 3, p < 0.05) (Fig. 6B). Conversely, silencing of P2X7 or treatment with KN-62 did not significantly affect release of CNTF from FSK-stimulated wild type SC (Fig. 6).
FIGURE 6.
P2X7 siRNA silencing or pharmacological inhibition increase CNTF release of CMT1A SC after FSK stimulation. A, wild type and CMT1A SC were electroporated in the absence of siRNA (UT) or in the presence of a P2X7-specific siRNA. B, wild type and CMT1A SC were exposed, or not, to 100 nm KN-62 for 24 h. CNTF release in the culture medium was determined with a rat CNTF DuoSet kit and normalized for cell protein content. *, p < 0.05; **, p < 0.01.
Myelination
As there is no final proof that defective migration and CNTF release by CMT1A SC contribute to the pathogenesis of the disease, we also investigated the effect of P2X7 silencing on the myelination process, which is known to be markedly impaired in CMT1A SC co-cultured with DRG neurons (8). Wild type and CMT1A SC were transfected with shRNA-P2X7 to stably silence the expression of this purinergic receptor, and P2X7 mRNA levels in SC were measured by real time PCR 10 days after transfection. In cells transfected with shRNA-P2X7, P2X7 mRNA levels were decreased down to 30 ± 5% (n = 3) of control values, measured in cells transfected with the NC plasmid (data not shown). Wild type or CMT1A SC transfected with shRNA-P2X7 or with the NC plasmid were co-cultured with DRG neurons, and myelination was evaluated after 4 weeks. Morphologic examination on co-cultures immunostained for MBP clearly showed a lower number of segments devoid of myelin in CMT1A SC transfected with shRNA-P2X7 cells (Fig. 7A). Morphometric analysis of co-cultures stained with Sudan black confirmed a significant reduction of myelin segment density in the co-cultures containing NC-transfected CMT1A SC compared with control co-cultures, containing NC-transfected wild type SC (mean number of myelinated segments/mm2 ± S.E.: 0.000017 ± 0.000005 versus 0.000037 ± 0.000004; n = 4; p < 0.05; Fig. 7B). Notably, the density of myelinated segments in cultures containing shRNA-P2X7-transfected CMT1A SC was significantly increased, to values similar to those observed in cultures containing NC-transfected wild type SC (number of myelinated segments/mm2 ± S.E.: 0.000030 ± 0.000001; n = 4; Fig. 7B). Conversely, the density of myelinated segments in cultures containing shRNA-P2X7-transfected wild type SC was not significantly modified, compared with values observed in cultures containing NC-transfected wild type SC (Fig. 7B).
Finally, expression of MPZ was evaluated as a marker of myelin production. As determined by Western blot, MPZ was significantly reduced in co-cultures containing NC-transfected CMT1A SC compared with the control ones (7.4 ± 0.8 versus 22.3 ± 0.7; n = 3; p < 0.05; Fig. 7C). Expression of MPZ was increased in co-cultures containing shRNA-P2X7-transfected CMT1A SC to levels observed in control co-cultures (12.4 ± 0.942; n = 3; Fig. 7C). On the contrary, expression of MPZ was not modified in co-cultures containing wild type SC transfected with shRNA-P2X7, compared with levels observed in control co-cultures (Fig. 7C).
DISCUSSION
No effective pharmacological treatment is as yet available for CMT1A patients, and current research is aimed at reducing PMP22 levels. PMP22 expression was reported to be stimulated by progesterone and by cAMP (43, 44). Accordingly, recent studies have shown that progesterone antagonists and ascorbic acid, which inhibits the stimulatory effect of cAMP on PMP22 expression, reduce the levels of PMP22 and improve the phenotype in animal models of CMT1A (45, 46). Clinical trials are underway to investigate the efficacy of ascorbic acid on human CMT1A (47).
Correcting the biochemical alterations induced by PMP22 overexpression in SC may prove successful by itself or in combination with the above mentioned therapeutic strategies. In this respect, our finding that the [Ca2+]i in rat CMT1A SC is strikingly higher than in normal SC, because of overexpression of P2X7, opens a new and promising perspective on the control of the biochemical derangements characterizing CMT1A. Indeed, the [Ca2+]i is arguably the single most influential intracellular signal regulating several tissue-specific cell functions, including proliferation, differentiation, migration, and secretion (21, 22, 24). Therefore, elucidating both the molecular mechanisms that underlie an enhanced basal [Ca2+]i and the functional effects downstream of the [Ca2+]i perturbation may give insights into the development of specific therapies in several diseases, including CMT1A.
Several experimental conditions inducing an increased [Ca2+]i in SC are known to impair differentiation and myelination (5, 6). Incubation of nerves with Ca2+ and the divalent cation ionophores A23187 or ionomycin causes a rapid demyelination, which is dependent upon rise in SC [Ca2+]i (5). Addition of ATP to SC monocultures, which induces an immediate and large increase in the [Ca2+]i, prevents maturation and differentiation of SC, as well as MBP synthesis and production of compact myelin (6).
As regulation of the cell [Ca2+]i homeostasis depends on the interplay between Ca2+-extruding ATPases and permeable Ca2+ channels on the plasma membrane and on subcellular Ca2+ stores, we tried to elucidate the role of these Ca2+-transport mechanisms in the perturbation of the [Ca2+]i levels in CMT1A SC. Impairment of Ca2+ extrusion by plasma membrane or ER ATP-dependent pumps is unlikely because of the higher levels of intracellular ATP and of the mitochondrial membrane potential (ΔΨm, the driving force for respiratory chain-coupled ATP generation) in CMT1A compared with wild type SC (see under “Results” and Table 2).
Moreover, increased Ca2+ mobilization from IP3 or cADPR-sensitive Ca2+ stores is also apparently ruled out as a possible mechanism inducing high basal levels of [Ca2+]i in CMT1A SC by the normal or even lower than normal control levels of these Ca2+ mobilizers in CMT1A SC (Table 2). In fact, quite unexpectedly, the lower levels of the ryanodine receptor-gating second messenger cADPR and of the cADPR-generating enzyme ADP-ribosyl cyclase (see Table 2) in CMT1A compared with wild type SC suggest a negative feedback mechanism aimed at limiting further [Ca2+]i increases via CICR. A similar negative feedback between a ryanodine receptor-mediated Ca2+ release and ADP-ribosyl cyclase activity has been reported in Namalwa cells (48).
These results suggesting absence of abnormalities of both ATP-dependent Ca2+ transport and of intracellular Ca2+ release pointed to an exceedingly high influx of extracellular Ca2+ as the likely cause of the high basal [Ca2+]i in CMT1A SC. P2X7, a ligand-gated ion channel, was known to interact with members of the epithelial membrane protein family, which includes PMP22, in HEK293 cells overexpressing P2X7 and members of the epithelial membrane protein family (31). This interaction had been shown to induce opening of the purinergic receptor channel and influx of extracellular Ca2+, triggering several Ca2+-induced functional effects (31). In addition, Grafe et al. (49) reported the occurrence in the paranodal membrane of rat SC of an ionotropic ATP receptor with electrophysiological and pharmacological properties similar to those of the P2X7 receptor and showed its functional activation in myelinating SC. Based on these data, Grafe et al. (49) suggested a possible role of P2X7 in SC-dependent nerve injury or neuropathy. These findings prompted us to investigate whether P2X7 was responsible for the increased Ca2+ influx in CMT1A cells. Indeed, functional (i.e. Ca2+-permeable) P2X7 receptors are overexpressed in CMT1A SC compared with wild type cells (Fig. 2), and overexpression is causally related to PMP22 transcription, because knocking down PMP22 also down-regulates P2X7 (Fig. 3). Conversely, knocking down P2X7 does not affect PMP22 expression (Fig. 3). Thus, overexpression of P2X7 is downstream of the genetic abnormality responsible for CMT1A neuropathy, although the molecular mechanisms by which PMP22 overexpression leads to increased levels of P2X7 in CMT1A SC remain to be identified.
Interestingly, overexpression of P2X7 in HEK293 cells was recently found to increase both the resting mitochondrial potential (ΔΨm) and the intracellular ATP content (50). This effect is mediated by the increased extracellular Ca2+ influx across the receptor itself (50). Likewise, CMT1A SC, which overexpress P2X7, show an increase of both ΔΨm and the intracellular ATP content (see “Results” and Table 2).
Down-regulation of P2X7 expression and function through the use of P2X7 pharmacological antagonists and of siRNA silencing resulted in a decrease of the basal [Ca2+]i levels in CMT1A SC (Figs. 2 and 4), and in the recovery of several functional parameters known to be defective in these transgenic cells, i.e. cell migration, CNTF release, and most notably myelin synthesis (Figs. 5–7). Although it is not established how impaired migration and CNTF release contribute to CMT1A pathogenesis, both functions are defective in CMT1A SC (8, 38–40, 42). Restoration of myelination by down-regulation of P2X7 unequivocally establishes a pathogenetic link between high [Ca2+]i and impaired SC function in CMT1A.
P2X7 receptors are widely distributed in immunocompetent cells of the central and peripheral nervous system, including SC, and the pathogenetic role of P2X7 and the therapeutic potential of P2X7 antagonists are being increasingly recognized in several neurological disorders, including peripheral neuropathies (32, 36, 51). It is also noteworthy that overexpression of P2X7 and the consequent increase of Ca2+ influx have been implicated in the pathogenesis of Duchenne muscular dystrophy (52).
P2X7 antagonists, including oATP, which is effective in reducing the [Ca2+]i levels in SC from CMT1A rats (Fig. 2), have been utilized for chronic therapy of neuropathic and inflammatory pain in rats (53, 54). Therefore, in principle, these compounds can be administered for long term treatment of neurological disorders, thus representing promising pharmacological tools. Identification of mechanisms and related pharmacological tools for restoring normal SC Ca2+ levels in CMT1A SC could possibly lead to the development of new therapeutic approaches also for other neuropathies sharing a derangement of SC Ca2+ homeostasis, notably of those characterized by a functional hypertrophy of systems mediating primary Ca2+ influx. In this respect, demyelination occurring in CMT1X, a neuropathy clinically indistinguishable from CMT1A (55), could be causally related to an impairment of the [Ca2+]i in SC. In CMT1X, mutations of connexin 32 (a gap junctional protein forming hexameric hemichannels permeable to ions and low molecular weight molecules) leads to uncontrolled pore opening (56); the remarkably steep gradient of Ca2+ concentrations across the plasma membrane accounts for the rapid influx of extracellular Ca2+ upon opening of connexin hemichannels (57, 58). The correction of perturbed Ca2+ homeostasis seems to be a promising therapeutic strategy to treat different types of neuronal and glial derived disorders that are now being recognized as “calcium diseases.”
Acknowledgments
We are indebted to Dr. Klaus-Armin Nave for critical revision and helpful comments on this manuscript.
This work was supported in part by Telethon Grants GGP06178 and GUP05007 (to A. S.), by Regione Liguria, the Italian Ministry of Education, University and Scientific Research Grants MIUR-PRIN 2005, MIUR FIRB RBAUO19A3C, MIUR FIRB RBNE01ERXR, MIUR FIRB RBLA039LSF, and MIUR FIRB RBIP06LSS2, by the University of Genova, and by Fondazione Cassa di Risparmio di Genova e Imperia.
- CMT1
- Charcot-Marie-Tooth disease type 1
- SC
- Schwann cells
- [Ca2+]i
- intracellular concentration of calcium
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA expressing plasmid
- MBP
- myelin basic protein
- MPZ
- myelin protein zero
- GFAP
- glial fibrillary acidic protein
- DRG
- dorsal root ganglion
- NGF
- nerve growth factor
- cADPR
- cyclic ADP-ribose
- FSK
- forskolin
- CNTF
- ciliary neurotrophic factor
- CICR
- calcium-induced calcium release
- oATP
- oxidized ATP
- HBSS
- Hanks' balanced salt solution
- IP3
- inositol 1,4,5-trisphosphate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ER
- endoplasmic reticulum
- oligo
- oligonucleotide
- UT
- untransfected
- CNTF
- ciliary neurotrophic factor
- NC
- negative control
- TG
- thapsigargin.
REFERENCES
- 1.Dyck P. J., Karnes J. L., Lambert E. H. (1989) Neurology 39,1302–1308 [DOI] [PubMed] [Google Scholar]
- 2.Lupski J. R., de Oca-Luna R. M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B. J., Saucedo-Cardenas O., Barker D. F., Killian J. M., Garcia C. A., Chakravarti A., Patel P. I. (1991) Cell 66,219–232 [DOI] [PubMed] [Google Scholar]
- 3.Suter U., Scherer S. S. (2003) Nat. Rev. Neurosci. 4,714–726 [DOI] [PubMed] [Google Scholar]
- 4.Sereda M., Griffiths I., Pühlhofer A., Stewart H., Rossner M. J., Zimmerman F., Magyar J. P., Schneider A., Hund E., Meinck H. M., Suter U., Nave K. A. (1996) Neuron 16,1049–1060 [DOI] [PubMed] [Google Scholar]
- 5.Smith K. J., Hall S. M., Schauf C. L. (1985) J. Neurol. Sci. 71,19–37 [DOI] [PubMed] [Google Scholar]
- 6.Stevens B., Fields R. D. (2000) Science 287,2267–2271 [DOI] [PubMed] [Google Scholar]
- 7.Brockes J. P., Fields K. L., Raff M. C. (1979) Brain Res. 165,105–118 [DOI] [PubMed] [Google Scholar]
- 8.Nobbio L., Vigo T., Abbruzzese M., Levi G., Brancolini C., Mantero S., Grandis M., Benedetti L., Mancardi G., Schenone A. (2004) Neurobiol. Dis. 16,263–273 [DOI] [PubMed] [Google Scholar]
- 9.Zocchi E., Daga A., Usai C., Franco L., Guida L., Bruzzone S., Costa A., Marchetti C., De Flora A. (1998) J. Biol. Chem. 273,8017–8024 [DOI] [PubMed] [Google Scholar]
- 10.Moreschi I., Bruzzone S., Nicholas R. A., Fruscione F., Sturla L., Benvenuto F., Usai C., Meis S., Kassack M. U., Zocchi E., De Flora A. (2006) J. Biol. Chem. 281,31419–31429 [DOI] [PubMed] [Google Scholar]
- 11.Bradford M. M. (1976) Anal. Biochem. 72,248–254 [DOI] [PubMed] [Google Scholar]
- 12.Guida L., Franco L., Zocchi E., De Flora A. (1995) FEBS Lett. 368,481–484 [DOI] [PubMed] [Google Scholar]
- 13.Bruzzone S., Dodoni G., Kaludercic N., Basile G., Millo E., De Flora A., Di Lisa F., Zocchi E. (2007) J. Biol. Chem. 282,5045–5052 [DOI] [PubMed] [Google Scholar]
- 14.Graeff R., Lee H. C. (2002) Biochem. J. 361,379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruzzone S., De Flora A., Usai C., Graeff R., Lee H. C. (2003) Biochem. J. 375,395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meintanis S., Thomaidou D., Jessen K. R., Mirsky R., Matsas R. (2001) Glia 34,39–51 [PubMed] [Google Scholar]
- 17.Morgan L., Jessen K. R., Mirsky R. (1991) J. Cell Biol. 112,457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jessen K. R., Mirsky R. (2008) Glia 56,1552–1565 [DOI] [PubMed] [Google Scholar]
- 19.Woodhoo A., Sommer L. (2008) Glia 56,1481–1490 [DOI] [PubMed] [Google Scholar]
- 20.Niemann S., Sereda M. W., Suter U., Griffiths I. R., Nave K. A. (2000) J. Neurosci. 20,4120–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carafoli E. (2005) FEBS J. 272,1073–1089 [DOI] [PubMed] [Google Scholar]
- 22.Berridge M. J. (2002) Cell Calcium 32,235–249 [DOI] [PubMed] [Google Scholar]
- 23.Lee H. C. (ed) (2002) Cyclic ADP-ribose and NAADP: Structures, Metabolism and Functions, Kluwer Academic Publishers, Norwell, MA [Google Scholar]
- 24.Clapham D. E. (1995) Cell 80,259–268 [DOI] [PubMed] [Google Scholar]
- 25.Inesi G., Sagara Y. (1992) Arch. Biochem. Biophys. 298,313–317 [DOI] [PubMed] [Google Scholar]
- 26.Berridge M. J. (2005) Annu. Rev. Physiol. 67,1–21 [DOI] [PubMed] [Google Scholar]
- 27.Lee H. C., Walseth T. F., Bratt G. T., Hayes R. N., Clapper D. L. (1989) J. Biol. Chem. 264,1608–1615 [PubMed] [Google Scholar]
- 28.Guse A. H. (2005) FEBS J. 272,4590–4597 [DOI] [PubMed] [Google Scholar]
- 29.Galione A., Lee H. C., Busa W. B. (1991) Science 253,1143–1146 [DOI] [PubMed] [Google Scholar]
- 30.Sutko J. L., Airey J. A., Welch W., Ruest L. (1997) Pharmacol. Rev. 49,53–98 [PubMed] [Google Scholar]
- 31.Wilson H. L., Wilson S. A., Surprenant A., North R. A. (2002) J. Biol. Chem. 277,34017–34023 [DOI] [PubMed] [Google Scholar]
- 32.Sperlágh B., Vizi E. S., Wirkner K., Illes P. (2006) Prog. Neurobiol. 78,327–346 [DOI] [PubMed] [Google Scholar]
- 33.Miras-Portugal M. T., Díaz-Hernández M., Giráldez L., Hervás C., Gómez-Villafuertes R., Sen R. P., Gualix J., Pintor J. (2003) Neurochem. Res. 28,1597–1605 [DOI] [PubMed] [Google Scholar]
- 34.Liu C., Mather S., Huang Y., Garland C. J., Yao X. (2004) Am. J. Physiol. Heart Circ. Physiol. 286,H1688–H1695 [DOI] [PubMed] [Google Scholar]
- 35.Murgia M., Hanau S., Pizzo P., Rippa M., Di Virgilio F. (1993) J. Biol. Chem. 268,8199–8203 [PubMed] [Google Scholar]
- 36.Fields R. D., Burnstock G. (2006) Nat. Rev. Neurosci. 7,423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suter U., Snipes G. J., Schoener-Scott R., Welcher A. A., Pareek S., Lupski J. R., Murphy R. A., Shooter E. M., Patel P. I. (1994) J. Biol. Chem. 269,25795–25808 [PubMed] [Google Scholar]
- 38.Brancolini C., Marzinotto S., Edomi P., Agostoni E., Fiorentini C., Müller H. W., Schneider C. (1999) Mol. Biol. Cell 10,2441–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobbio L., Mancardi G., Grandis M., Levi G., Suter U., Nave K. A., Windebank A. J., Abbruzzese M., Schenone A. (2001) Ann. Neurol. 50,47–55 [DOI] [PubMed] [Google Scholar]
- 40.Vigo T., Nobbio L., Hummelen P. V., Abbruzzese M., Mancardi G., Verpoorten N., Verhoeven K., Sereda M. W., Nave K. A., Timmerman V., Schenone A. (2005) Mol. Cell. Neurosci. 28,703–714 [DOI] [PubMed] [Google Scholar]
- 41.ten Asbroek A. L., Verhamme C., van Groenigen M., Wolterman R., de Kok-Nazaruk M. M., Baas F. (2005) J. Neurosci. Res. 79,825–835 [DOI] [PubMed] [Google Scholar]
- 42.Nobbio L., Fiorese F., Vigo T., Cilli M., Gherardi G., Grandis M., Melcangi R. C., Mancardi G., Abbruzzese M., Schenone A. (2009) J. Neuropathol. Exp. Neurol. 68,441–455 [DOI] [PubMed] [Google Scholar]
- 43.Désarnaud F., Do Thi A. N., Brown A. M., Lemke G., Suter U., Baulieu E. E., Schumacher M. (1998) J. Neurochem. 71,1765–1768 [DOI] [PubMed] [Google Scholar]
- 44.Sabéran-Djoneidi D., Sanguedolce V., Assouline Z., Lévy N., Passage E., Fontés M. (2000) Gene 248,223–231 [DOI] [PubMed] [Google Scholar]
- 45.Sereda M. W., Meyer zu, Hörste G., Suter U., Uzma N., Nave K. A. (2003) Nat. Med. 9,1533–1537 [DOI] [PubMed] [Google Scholar]
- 46.Passage E., Norreel J. C., Noack-Fraissignes P., Sanguedolce V., Pizant J., Thirion X., Robaglia-Schlupp A., Pellissier J. F., Fontés M. (2004) Nat. Med. 10,396–401 [DOI] [PubMed] [Google Scholar]
- 47.Pareyson D., Schenone A., Fabrizi G. M., Santoro L., Padua L., Quattrone A., Vita G., Gemignani F., Visioli F., Solari A. (2006) Pharmacol. Res. 54,436–441 [DOI] [PubMed] [Google Scholar]
- 48.McCarthy T. V., Datar S., Mackrill J. J. (2003) FEBS Lett. 554,133–137 [DOI] [PubMed] [Google Scholar]
- 49.Grafe P., Mayer C., Takigawa T., Kamleiter M., Sanchez-Brandelik R. (1999) J. Physiol. 515,377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adinolfi E., Callegari M. G., Ferrari D., Bolognesi C., Minelli M., Wieckowski M. R., Pinton P., Rizzuto R., Di Virgilio F. (2005) Mol. Biol. Cell 16,3260–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Virgilio F. (2006) Novartis Found. Symp. 276,253–258; discussion 259–262, 275–281 [DOI] [PubMed] [Google Scholar]
- 52.Yeung D., Zablocki K., Lien C. F., Jiang T., Arkle S., Brutkowski W., Brown J., Lochmuller H., Simon J., Barnard E. A., Górecki D. C. (2006) FASEB J. 20,610–620 [DOI] [PubMed] [Google Scholar]
- 53.Dell'Antonio G., Quattrini A., Dal Cin E., Fulgenzi A., Ferrero M. E. (2002) Neurosci. Lett. 327,87–90 [DOI] [PubMed] [Google Scholar]
- 54.Honore P., Donnelly-Roberts D., Namovic M. T., Hsieh G., Zhu C. Z., Mikusa J. P., Hernandez G., Zhong C., Gauvin D. M., Chandran P., Harris R., Medrano A. P., Carroll W., Marsh K., Sullivan J. P., Faltynek C. R., Jarvis M. F. (2006) J. Pharmacol. Exp. Ther. 319,1376–1385 [DOI] [PubMed] [Google Scholar]
- 55.Suter U., Snipes G. J. (1995) J. Neurosci. Res. 40,145–151 [DOI] [PubMed] [Google Scholar]
- 56.Gómez-Hernández J. M., de Miguel M., Larrosa B., González D., Barrio L. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,16030–16035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abrams C. K., Bennett M. V., Verselis V. K., Bargiello T. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,3980–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodenough D. A., Paul D. L. (2003) Nat. Rev. Mol. Cell Biol. 4,285–294 [DOI] [PubMed] [Google Scholar]