Abstract
F1-ATPase is a rotary molecular motor in which the γ-subunit rotates against the α3β3 cylinder. The unitary γ-rotation is a 120° step comprising 80 and 40° substeps, each of these initiated by ATP binding and ADP release and by ATP hydrolysis and inorganic phosphate release, respectively. In our previous study on γ-rotation at low temperatures, a highly temperature-sensitive (TS) reaction step of F1-ATPase from thermophilic Bacillus PS3 was found below 9 °C as an intervening pause before the 80° substep at the same angle for ATP binding and ADP release. However, it remains unclear as to which reaction step the TS reaction corresponds. In this study, we found that the mutant F1(βE190D) from thermophilic Bacillus PS3 showed a clear pause of the TS reaction below 18 °C. In an attempt to identify the catalytic state of the TS reaction, the rotation of the hybrid F1, carrying a single copy of βE190D, was observed at 18 °C. The hybrid F1 showed a pause of the TS reaction at the same angle as for the ATP binding of the incorporated βE190D, although kinetic analysis revealed that the TS reaction is not the ATP binding step. These findings suggest that the TS reaction is a structural rearrangement of β before or after ATP binding.
F1-ATPase (F1)2 is an ATP-driven rotary motor protein. The subunit composition of the bacterial F1-ATPase is α3β3γδϵ, and the minimum complex of F1-ATPase as a rotary motor is α3β3γ subcomplex. This motor protein forms the FoF1-ATP synthase complex by binding to another rotary motor, namely, Fo, which is driven by the proton flux resulting from the proton motive force across the membranes (1–4). Under physiological conditions, where the proton motive force is sufficiently large, Fo forcibly rotates F1-ATPase in the reverse direction of F1-ATPase, leading the reverse reaction of ATP hydrolysis, i.e. ATP synthesis from ADP and inorganic phosphate (Pi). When the proton motive force diminishes or F1 is isolated from Fo, F1-ATPase hydrolyzes ATP to rotate the γ-subunit against the α3β3 stator ring in the counterclockwise direction as viewed from the Fo side (5). The catalytic sites are located at the interface of the α- and β-subunits, predominantly on the β-subunit (6). Each β-subunit carries out a single turnover of ATP hydrolysis during the γ-rotation of 360° following the common catalytic reaction pathway, whereas they are 120° different in the catalytic phase. In this manner, the three β-subunits undergo different reaction steps of ATP hydrolysis upon each rotational step. The rotary motion of the γ-subunit has been demonstrated by biochemical (7) and spectroscopic methods (8) and directly proved in single-molecule observation studies (5).
Since the establishment of the single-molecule rotation assay, the chemomechanical coupling scheme of F1 has been studied extensively by resolving the rotation into discrete steps. The stepping rotation was first observed under an ATP-limiting condition where F1 makes discrete 120° steps upon ATP binding (9). Then, high speed imaging of the rotation with a small probe of low friction was performed, which revealed that the 120° step comprises 80 and 40° substeps, each initiated by ATP binding, and two unknown consecutive reactions, respectively (10). This finding necessitated the identification of the two reactions that trigger the 40° substep. Hence, the rotation assay was performed using a mutant, namely F1(βE190D), and a slowly hydrolyzed ATP analog, namely ATPγS (11). Glutamate 190 of the β-subunit of F1, derived from thermophilic Bacillus PS3 and the corresponding glutamates from other F1-ATPases (Glu-181 of F1 from Escherichia coli and Glu-188 of F1 from bovine mitochondria), has been identified as one of the most critical catalytic residues for ATP hydrolysis (6, 12–15). When this glutamate was substituted with aspartic acid, which has a shorter side chain than that of glutamate, the ATP cleavage step of F1 was drastically slowed. In the rotation assay, this mutant showed a distinct long pause before the 40° substep. ATPγS also caused a long pause before the 40° substep. These observations established that the 40° substep is initiated by hydrolysis. Accordingly, the pause angles before the 80 and 40° substeps are referred to as to the binding angle and the catalytic angle, respectively. Then, the rotation assay was performed in the presence of a high amount of Pi in the solution. It was shown that Pi rebinding caused the long pause at the catalytic angle, suggesting that Pi is released before the 40° substep (16).
However, the reaction scheme of F1 cannot be established by simply assigning each reaction step to either the binding angle or the catalytic angle, because each reaction step must be assigned to one of the three binding or catalytic angles when considering the 360° cyclic reaction scheme of each β-subunit. Direct information about the timing of ADP release was obtained by simultaneous imaging of fluorescently labeled nucleotides and γ rotation, which showed that each β retains ADP until the γ rotates 240° after binding of the nucleotide as ATP and releases ADP between 240 and 320° (16, 17). Another powerful approach is the use of a hybrid F1 carrying a mutant β that causes a characteristic pause during the rotation. In a previous study, the hybrid F1 carrying a single copy of β(E190D), α3β2β(E190D)γ, showed a distinct pause caused by the slow hydrolysis of β(E190D) at +200° from the ATP binding angle of the mutant β (18). From this observation, it was confirmed that each β executes the chemical cleavage of the bound ATP at +200° from the angle where the ATP binds to β. The asymmetric feature of the pause of the hybrid F1 was also utilized in other experiments as a marker in the rotational trajectory to correlate the rotational angle and the conformational state of β (19) or to determine the state of F1 in the crystal structures as the pausing state at catalytic angle (20).
Recently, we have found a new reaction intermediate of F1 rotation as a clear intervening pause before the 80° substep in the rotation assay below 9 °C (21). Furuike et al. (22) also observed the TS reaction in a high speed imaging experiment. The rate constant of this reaction was remarkably sensitive to temperature, giving a Q10 factor around 19. When ADP was added to solution, the pause before the 80° substep was prolonged, whereas the solution Pi caused a longer pause before the 40° substep (21). Although this result can be explained by assuming that the temperature-sensitive (TS) reaction is ADP release, it was not decisive for the identification of the TS reaction.
In this study, we found that the mutant F1(βE190D) also exhibits the distinct pause of the TS reaction but at a higher temperature than for the wild-type F1, i.e. at 18 °C. This feature was advantageous in identifying the angle position of the TS reaction in the catalytic cycle for each β-subunit coupled with the 360° rotation. Taking advantage of the feature of the hybrid F1, we analyzed the rotational behavior of the hybrid F1 at 18 °C in order to assign the angle position of the TS reaction in the catalytic cycle of the 360° rotation, and we have shown that the TS reaction is not directly involved in the ADP release but in some conformational rearrangement before or after ATP binding step.
EXPERIMENTAL PROCEDURES
Preparation of F1-ATPase
The α(His6 at N terminus/C193S)3β(His10 at N terminus)3γ(S108C/I211C) subcomplex of F1-ATPase from thermophilic Bacillus PS3, which was modified for the rotation assay, and the α(His6 at N terminus/C193S)3β(His10 at N terminus/E190D)3γ(S108C/I211C) subcomplex were expressed in E. coli, purified, and biotinylated as described previously (20). For simplicity, the former was referred to as wild-type F1 because the effect of the mutations on the ATP hydrolysis activity was minor, and the latter was referred to as F1(βE190D). The ATP hydrolysis activity of F1(βE190D) was measured in bulk solution as described previously (21). As with the wild-type F1 (21), the mutant showed a lag phase followed by slow activation in the ATP hydrolysis assay. Therefore, the steady-state activity was determined as the rate of ATP hydrolysis. The hybrid F1, α3β2β(E190D)γ, was prepared by mixing solutions of F1(βE190D) and the wild-type F1 in a molar ratio of 1:2 and incubating the solution at 4 °C for longer than 1 week in the presence of 500 mm NaCl.
Rotation Assay
The rotary motion of F1 was visualized by attaching a magnetic bead (∼200 nm, Seradyn Inc., Indianapolis, IN) onto the γ-subunit of F1 and immobilizing the α3β3 ring on a nickel-nitrilotriacetic acid glass surface. Bright-field or phase-contrast images of the rotating beads were obtained using an inverted optical microscope (IX-70; Olympus, Tokyo) equipped with magnetic tweezers, which was used to reactivate pausing F1 molecules by forcibly rotating their γ subunits over 90° when the observed molecules lapsed into the ADP-inhibited form (23). For the data shown in Fig. 1, A and B, and Fig. 3, the images were captured at the video rate of 30 frames/s using a FC300M CCD camera (Takenaka System Co., Kyoto, Japan). The other image data were acquired at 500 or 1000 frames/s using a high speed HAS220 CCD camera (DETECT., Tokyo, Japan) or video rate at 30 frames/s with 1 ms exposure time/frame. All data for the dwell time analysis at 28 °C were acquired at 500 or 1000 frames/s. The recorded images were analyzed with image analysis software (Digimo, Tokyo) or a custom-made program (K. Adachi). The experimental procedures of the rotational assay were the same as those described previously (24) except for the buffer content. The assay solutions contained 20 mm HEPES, 50 mm KCl, 2 mm MgCl2, 5 mg/ml bovine serum albumin, and an indicated amount of ATP with an ATP-regenerating system (100 μg/ml pyruvate kinase, 1.0 mm phosphoenolpyruvate) at pH 8.0. For the rotation assay in the presence of ADP, the ATP-regenerating system was omitted, and the solution contained the indicated amount of MgCl2. For the assay above 18 °C, a microscope was set in a room at 18 °C, and a temperature control system (MATS-LH; Tokai Hit, Shizuoka, Japan) was attached to the objective lens. For the assay below 17 °C, a stable microscope stage with a temperature-controlled specimen holder (22) (Ikedarika, Japan) was used. The actual temperature of the specimen was monitored with a thermocouple probe attached to the sample chamber. The precision of temperature control was ±1 °C.
FIGURE 1.
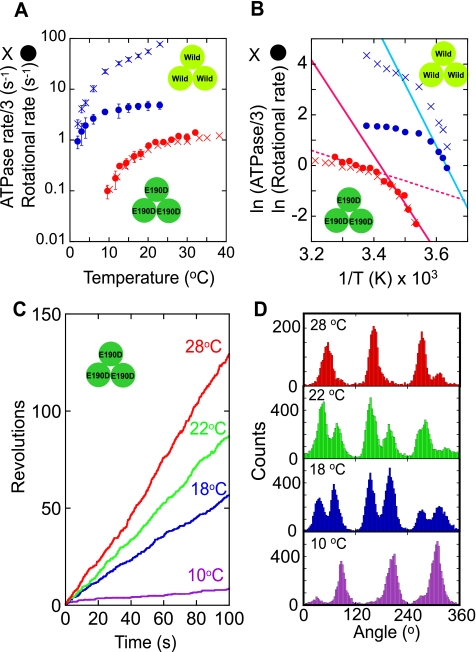
A, temperature dependence of the rotational rate (circles) and ATP hydrolysis rate in a bulk solution (crosses) of F1(βE190D) (red) and wild-type F1 (blue) (21) at saturating ATP concentration (2 mm for F1(βE190D) and 1 mm for wild-type F1). The ATP hydrolysis rate was divided by 3 for comparison with the rotational rate. B, Arrhenius plots of traces in A. The rate-limiting reactions of the rotational rates at low temperature (below 17 °C for the mutant F1 and below 4 °C for wild-type F1) were analyzed to determine the activation energy (Table 1) using linear regression (solid lines). The rotational rates of F1(βE190D) above 20 °C were also fitted by linear regression (dotted red line). C, time courses of rotation of F1(βE190D) at various temperatures. D, histograms of the angle from traces in C.
FIGURE 3.
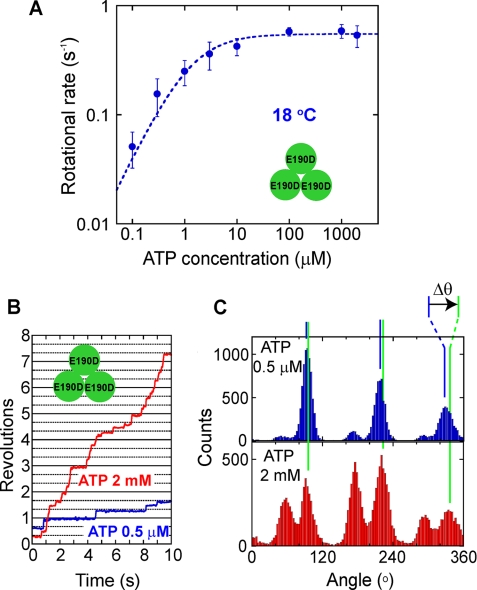
A, ATP concentration dependence of the rotational rate of F1(βE190D) at 18 °C. Curve shows the fitting line with the Michaelis-Menten equation, V = Vmax[ATP]/(Km+[ATP]); Vmax = 0.55 s−1, Km = 1.3 μm. B, time courses of the rotation at 18 °C of a molecule of F1(βE190D) at 0.5 μm ATP (blue) and 2 mm ATP (red). During observation, the buffer was exchanged to increase ATP from 0.5 μm to 2 mm. C, histograms of the angle from traces in B. Δθ represents angle distances of the TS angle at 2 mm ATP from the ATP binding angle at 0.5 μm ATP.
RESULTS
Temperature Dependence of the Rotational Rate of F1(βE190D)
The rotational rate of F1(βE190D) was measured at 2 mm ATP, which was later confirmed to be a saturation condition, at various temperatures ranging from 10 to 32 °C, by using a ∼200-nm magnetic bead as a rotation marker at the video rate (Fig. 1, A and C). Although the previously reported rotation rate of the wild-type F1 was limited by the viscous friction on the probe at room temperatures (>10 °C) (21), that of the mutant was not determined by the viscous friction but by the sluggish hydrolysis step of which the time constant is ∼300 ms at room temperature (11). Therefore, the rotational rate of the mutant was in good agreement with the estimated rotational rate, which was one-third of the ATP hydrolysis rate determined in the bulk solution based on the coupling ratio of 3 ATPs/turn; the wild-type F1 showed a large difference between two values around room temperature (Fig. 1A) (21). The rotational velocity of the mutant at 22–24 °C was 0.9 Hz, which was consistent with a previous report (11). At below 17 °C, F1(βE190D) decreased the rotational rate significantly, suggesting that a different reaction step dominates the pace of the catalytic turnover cycle. This point was clearly shown in the Arrhenius plots of the rotational rate and the bulk ATP hydrolysis rate, both of which showed a breakpoint around 18 °C (Fig. 1B). Thus, F1(βE190D) has a TS reaction step similar to that of the wild type; however, the temperature range at which the TS reaction of the mutant becomes prominent is different from that of the wild-type F1 (<9 °C). For a comparison of the temperature dependence between the mutant and the wild-type F1, the apparent activation energy of the TS reaction was determined instead of the Q10 factor from the slope of the Arrhenius plot, because the Q10 factor changes depending on the temperature range from which it is determined. The activation energy of the TS reaction of F1(βE190D) was 82–86 kJ/mol, which was remarkably high and essentially the same as the activation energy of the TS reaction of the wild-type F1, which was 97–98 kJ/mol (Table 1). However, the activation energy for the hydrolysis step of F1(βE190D) was 11–18 kJ/mol, which was much smaller than that of the TS reaction. The wild-type F1 also showed smaller activation energy for hydrolysis than that for the TS reaction. Thus, the TS reaction of F1(βE190D) showed the same temperature dependence as the wild-type F1.
TABLE 1.
Activation energy of the TS reaction and hydrolysis of F1(βE190D) and wild-type F1
Activation energy was calculated by the linear fitting of the Arrhenius plots of ATPase and the rotational rate as shown in Fig. 1B or by the comparison of rate constants at 18 and 28 °C determined from the dwell time histograms shown in Fig. 2, B and C, for βE190D and Fig. 4, C and D, for βE190D(hybrid).
| Reaction | F1 | Activation energy |
|---|---|---|
| kJ/mol | ||
| TS | βE190D | 86 (ATPase), 82 (rotation rate), 78 (dwell time) |
| βE190D(hybrid) | 62 (dwell time) | |
| Hydrolysis | βE190D | 11 (ATPase), 18 (rotation rate), 8.3 (dwell time) |
| βE190D(hybrid) | 16 (dwell time) | |
| TS | Wild type | 97a (ATPase), 98a (rotation rate) |
| Hydrolysis (ATPγS) | Wild type | 2.4b (dwell time) |
Substeps of F1(βE190D) at Saturating ATP Concentration
It has been shown that the wild-type F1 has a pause for the TS reaction at the ATP binding angle (21). This point was also confirmed for the TS reaction of F1(βE190D); the angle histogram of the rotation of F1(βE190D) at 28 and 22 °C showed three main peaks corresponding to the hydrolysis dwell time at the catalytic angles (Fig. 1D) (11). In addition, a small peak at the right side of each catalytic angle was observed, although some peaks were not clear. Below 18 °C, where the TS reaction limits the pace of the overall reaction cycle, these peaks appeared as clear main peaks at around +40° from the catalytic angle, suggesting that F1(βE190D) also creates a pause at the binding angle to wait for the TS reaction.
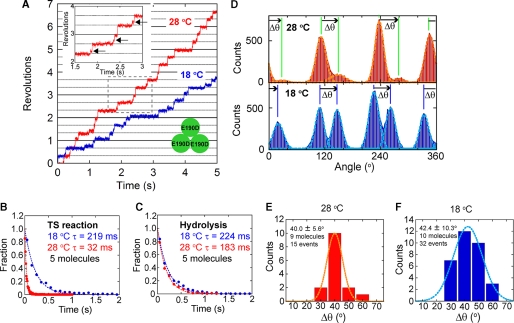
This point was confirmed by changing the specimen temperature from 28 to 18 °C during the observation of a rotating particle (Fig. 2). Even at 28 °C, F1(βE190D) showed short pauses concomitant with long hydrolysis (Fig. 2A, inset) when observed with a high speed camera at 500 or 1000 frame/s. In the angle histogram, these short pauses are shown as protuberant tails on the right side of the large peaks of the hydrolysis pauses (Fig. 2D). When the specimen was cooled to 18 °C, the short pauses were elicited as the major pauses, meaning that the short pause found at 28 °C corresponds to the waiting state for the TS reaction. The histograms of the dwell time of the short pause verified this point (Fig. 2B). By fitting the dwell time histogram of the short pause with an exponential function, the time constant of the short pause at the binding angle was determined to be 219 ms at 18 °C and 32 ms at 28 °C (Fig. 2B), giving the large activation energy of 78 kJ/mol, which coincides with the activation energy determined from the average rotational velocity and the bulk ATPase assay (Table 1). On the other hand, the time constant of the long hydrolysis pause was similar between 224 ms at 18 °C and 183 ms at 28 °C (Fig. 2C), giving the small activation energy of 8 kJ/mol (Table 1). Then, the angle distance between the catalytic pause and the TS pause was analyzed. As seen in Fig. 2D, the TS pause always appeared at the right side of the hydrolysis peaks in the angle histogram. The average angle distance of the TS pause from the catalytic pause (Δθ) was +40.0 ± 5.6° at 28 °C and +42.4 ± 10.3° at 18 °C (Fig. 2, E and F). Thus, it was verified that the TS reaction of F1(βE190D) occurs at the ATP binding angle, as does the TS reaction of the wild-type F1.
FIGURE 2.
A, time courses of rotations of a molecule of F1(βE190D) at different temperatures (18 and 28 °C). During observation, the specimen temperature was changed from 28 to 18 °C. The inset shows a magnified view of the trace at 28 °C to show the short pauses by the TS reaction (arrows). B and C, dwell time histograms of the pause for TS reaction (B) and hydrolysis (C) at 18 °C (blue) and 28 °C (red). The histogram was fitted with single exponential decay. D, histograms of the angle from traces in A at 28 °C (top) and 18 °C (bottom). Δθ represents angle distances from the pause angle for hydrolysis to that for the TS reaction. E and F, histograms of Δθat 28 °C (E) and 18 °C (F).
Rotation of F1(βE190D) at Various ATP Concentrations
For further characterization of the TS reaction of F1(βE190D), the rotational rate was measured at various ATP concentrations at 18 °C, and Michaelis-Menten curves were obtained (Fig. 3A). The Vmax and Km values at 18 °C were 0.55 s−1 and 1.3 μm, respectively. The binding rate of ATP determined from 3 × Vmax/Km was 1.3 × 106 m−1s−1. Therefore, the dwell time for ATP binding at 2 mm ATP should be 0.39 ms, which is too short to explain the dwell time of the TS reaction (219 ms). Thus, the TS reaction cannot be the ATP binding step from solution. To compare the angle positions for the TS reaction and the ATP binding, buffer exchange experiments were conducted in which the ATP level was increased from 0.5 μm to 2 mm during the observation of rotating molecules (Fig. 3, B and C). When ATP was increased, three major peaks of the ATP binding dwell diminished, but the pause at the binding angle was still clearly observed at 2 mm ATP due to the pause caused by the TS reaction (Fig. 3C). The angle distance (Δθ) from the ATP binding pause at 0.5 μm ATP to the TS pause determined at 2 mm ATP was −0.2 ± 14.1° (15 events for 5 molecules). This result also supports the above conclusion that the TS reaction occurs at the binding angle.
Temperature Dependence of the Rotation of Hybrid F1
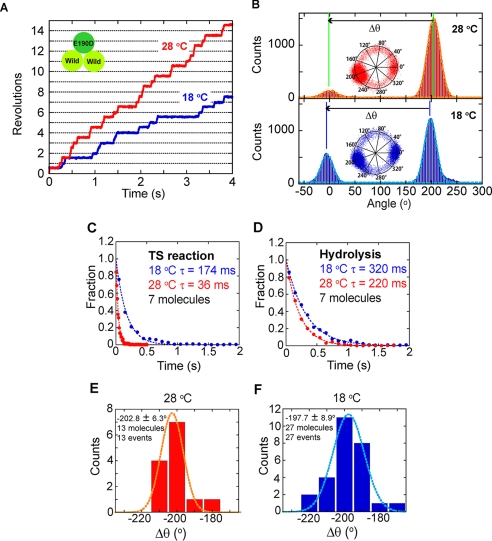
The above experiments established that the TS reaction of F1(βE190D) is essentially the same as that of the wild-type F1, and the dwell time of the TS reaction of the mutant is long enough to be detected in the conventional rotation assay, even at 18 °C where the wild type does not show the distinct pause of the TS reaction. Taking advantage of this feature of the mutant, the hybrid F1 carrying a single copy of β(E190D), α3β2β(E190D)γ, was analyzed in order to identify the angle position of the TS pause in the 360° cyclic reaction scheme. The hybrid F1 was prepared by mixing the F1(βE190D) and the wild-type F1 in a 1:2 ratio. The rotation of the hybrid F1 was found at 28 °C and 2 mm ATP under optical microscope. Although other complexes, such as the wild-type F1, α3ββ(E190D)2γ, or F1(βE190D), were also contained in the sample, α3β2β(E190D)γ was able to be distinguished from the others as a molecule rotating with long hydrolysis pauses at a single angle, as reported in previous works (18–20). We defined the hydrolysis pause as the pause of which dwell time was longer than 150 ms. Although most of molecules showed the concomitant TS pause as mentioned below, the dwell time of TS pause (36 ms) was much shorter and was able to be distinguished from the hydrolysis pause. More than half (59%) of the rotating molecules showed the hydrolysis pause at a single angle (α3β2β(E190D)γ), whereas 27% showed no pause (wild), 12% had two pause positions (α3ββ(E190D)2γ), and 2% had three pause positions (F1(βE190D)). When observed with a high speed camera, a single short pause was also found in addition to the single hydrolysis pause in the rotation of α3β2β(E190D)γ. The angle distance of the short pause from the hydrolysis pause (Δθ) was determined to be −200.5 ± 11.0° at 28 °C (supplemental Fig. S1). For hybrid F1, α3β2β(E190D)γ, the temperature change experiment was conducted to determine the angle position of the TS reaction of βE190D in relation to the long hydrolysis pause of β(E190D). Selected molecules responded well to the temperature change by extending the dwell time of the short pause unless the experiment was obstructed by nonspecific surface immobilization or the detachment of molecules. The time constant of the short pause of the hybrid F1 was determined to be 36 ms at 28 °C and 174 ms at 18 °C from the dwell time analysis, both of which were consistent with the time constant of the TS reaction determined for F1(βE190D), suggesting that the short pause corresponded to the TS pause of the incorporated βE190D in the hybrid F1 (Fig. 4C). The time constant of the hydrolysis dwell was less sensitive (220 ms at 28 °C and 320 ms at 18 °C), which was also consistent with the values determined for F1(βE190D) (Fig. 4D).
FIGURE 4.
A, time courses of the rotation of a molecule of the hybrid F1, α3β2β(E190D)γ, at 18 °C (blue) and 28 °C (red). The specimen temperature was first set at 28 °C and changed to 18 °C during the observation. B, histograms of the angle from traces in A. The inset shows the centroid of the rotation. Δθ represents the angle distances of TS pause from the hydrolysis pause. C and D, dwell time histograms of the pause of the TS reaction (C) and hydrolysis (D). Blue and red circles show the histogram at 18 and 28 °C, respectively. The histogram was fitted with single exponential decay. E and F, histogram of Δθat 28 °C (E) and 18 °C (F).
The angular distance of the TS pause from the hydrolysis pause (Δθ in Fig. 4B) was analyzed. The rotational data at 28 °C that was recorded at the video rate was not analyzed because the TS pause was not clearly resolved at this time resolution. The centroid traces of rotating molecules before and after the temperature change are shown in supplemental Fig. S2 for all cases that demonstrated a clear short pause at 28 °C. The angle distance was −202.8 ± 6.3° at 28 °C, and −197.7 ± 8.9°at 18 °C (Fig. 4, E and F). The angular deviation of the hydrolysis pause before and after the temperature change was also determined to be 0.6 ± 8.9° (data not shown), supporting the validity of the temperature change experiment. Thus, it was confirmed that the TS reaction occurs at −200° from the pause angle for hydrolysis. This angle corresponds to the ATP binding angle of βE190D.
DISCUSSION
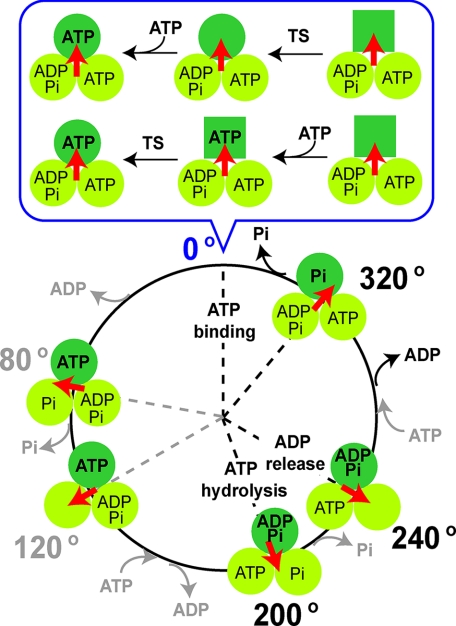
The TS reaction of F1(βE190D) was shown to have the same characteristics as the TS reaction of wild-type F1. (i) The reaction occurs at the ATP binding angle (Figs. 2 and Fig. 3). (ii) The reaction strongly depends on temperature; the apparent activation energy of the reaction (78–86 kJ/mol) is as high as that of wild-type F1 (97–98 kJ/mol; B, Table 1). (iii) Finally, the reaction does not depend on the ATP concentration (Fig. 3). Thus, the temperature-sensitive reaction of F1(βE190D) is not the ATP binding step from solution. However, the rate constant of the TS reaction of F1(βE190D) was 150 times less than that of the wild type; therefore, it could be discriminated from the TS reaction of the wild type. Using this advantageous feature, the rotation of the hybrid F1, α3β2β(E190D)γ, which has a single copy of βE190D, was analyzed. It was clearly shown that the TS reaction occurs at the ATP binding angle, that is −200° (+160°) from the hydrolysis angle of the mutant β. This position corresponds to 0° in the 360° cyclic reaction scheme of the β-subunit. Based on this new finding, the reaction scheme including the TS reaction step was proposed (Fig. 5). Here, the TS reaction is assumed to be a conformational change of the β-subunit not coupled with a chemical reaction at the catalytic site because the β-subunit only carries out the ATP binding step at 0°, and there is strong evidence that the TS reaction is not the ATP binding step. There are at least two possible reaction schemes; the TS reaction could occur before or after ATP binding. The former model assumes that the TS reaction is the conformational change of the β-subunit to develop into the ATP binding site. The latter model assumes that the TS reaction is the conformational change of the β-subunit for the affinity change to bound ATP. The present study does not provide crucial data to discriminate between these models. One possible experimental approach for this task is the simultaneous imaging of the binding and release of a fluorescently labeled nucleotide and the γ-rotation at 18 °C.
FIGURE 5.
Proposed reaction scheme of F1 and the TS reaction. The β-subunit that binds ATP at 0° is shown as a dark green circle. The scheme at 0° where the TS reaction and ATP binding occur is shown in the upper panel (expanded). Two possible models are shown in parallel, the case of ATP binding following the TS reaction (upper model) and that in which the TS reaction follows ATP binding (lower model).
Considering the exceptionally high temperature dependence of the TS reaction, it is likely that this reaction is a conformational rearrangement of the β-subunit, which accompanies a break of strong interaction. Therefore, it would seem that the TS reaction involves a global conformational rearrangement of the β-subunit. However, such a large conformational change during the 320–360° rotation has not been reported in a recent single-molecule work by Masaike et al. (19), who measured the angular displacement of the C-terminal domain of the β-subunit with advanced total internal reflection fluorescence microscopy. A possible explanation is that the TS reaction occurs after ATP binding and it was too fast for their time resolution (19) or that the TS reaction is accompanied by a conformational change in the horizontal plane of the rotational axis, which was not detectable with their method. Another idea of the TS reaction is a localized conformational rearrangement around the catalytic site. It seems to be reasonable considering that a single mutation introduced at glutamate 190 of the β-subunit causes the remarkable suppression in the rate constant of the TS reaction.
In a previous paper, we suggested that the TS reaction is relevant to ADP release based on the observation that solution ADP prolongs the dwell time at the binding angle (21). However, the results in the present study indicate that the TS reaction is not directly related to the ADP release, which is thought to occur at +40° from the hydrolysis angle (+240° from the binding angle). The previous kinetic analysis was based on the assumption that solution ADP binds to the site for ADP release; however, it is also possible that solution ADP competes with ATP for the ATP binding site. This contention also explains the inhibitory effect of solution ADP on the TS reaction. In fact, although the pause of F1(βE190D) at the binding angle was prolonged by the addition of a large amount of ADP (20 mm) in solution, the same as the wild-type F1, the effect of ADP was largely suppressed by the addition of an excess amount of ATP (supplemental Fig. S3). These results indicate that for F1, solution ADP acts not as a product inhibitor but as a competitive inhibitor with ATP.
It should be noted that there is a critical inconsistency between the present results and the previous report by Ariga et al. (18), who reported that the hybrid F1, α3β2β(E190D)γ rotates with one long pause at 200° caused by ATP hydrolysis and an additional short pause, as found in the present study. However, the angle position was assigned to be 320°, not 360°, although the time constant of their short pause was 20 ms (18), which is very close to the TS pause at 28 °C determined in the present experiment. Supplemental Fig. 2 shows the centroid plots for all hybrid F1 molecules for which the temperature change experiment was successfully carried out. In the data set, all molecules showed the short pause not at 320 but at 360°. Statistical analysis confirms this conclusion. The reason for this inconsistency is not clear. Although there are some small differences in the experimental procedures and materials, these do not seem to explain this critical inconsistency. Even if there is another short pause in addition to that of the TS reaction, the pause of the TS reaction should have been included in their data set.
Supplementary Material
Acknowledgments
We thank Dr. S. Nishikawa, Mr. K. Ikezaki, Dr. M. Sugawa, and Dr. T. Yanagida for technical help, Dr. K. Adachi for the custom image analysis program, Dr. T. Okamoto for critical discussion, and members of the Noji laboratory for help and advice.
This work was supported by Grants-in-aid 18074005 and 18201025 for Scientific Research (to H. N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant from the Post-Silicon Materials and Devices Research Alliance, Institute of Scientific and Industrial Research at Osaka University (to H. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- F1
- F1-ATPase
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- TS
- temperature-sensitive.
REFERENCES
- 1.Boyer P. D. ( 1997) Annu. Rev. Biochem. 66,717–749 [DOI] [PubMed] [Google Scholar]
- 2.Cross R. L. ( 2000) Biochim. Biophys. Acta 1458,270–275 [DOI] [PubMed] [Google Scholar]
- 3.Senior A. E., Nadanaciva S., Weber J. ( 2002) Biochim. Biophys. Acta 1553,188–211 [DOI] [PubMed] [Google Scholar]
- 4.Yoshida M., Muneyuki E., Hisabori T. ( 2001) Nat. Rev. Mol. Cell Biol. 2,669–677 [DOI] [PubMed] [Google Scholar]
- 5.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. ( 1997) Nature 386,299–302 [DOI] [PubMed] [Google Scholar]
- 6.Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. ( 1994) Nature 370,621–628 [DOI] [PubMed] [Google Scholar]
- 7.Duncan T. M., Bulygin V. V., Zhou Y., Hutcheon M. L., Cross R. L. ( 1995) Proc. Natl. Acad. Sci. U.S.A. 92,10964–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabbert D., Engelbrecht S., Junge W. ( 1996) Nature 381,623–625 [DOI] [PubMed] [Google Scholar]
- 9.Yasuda R., Noji H., Kinosita K., Jr., Yoshida M. ( 1998) Cell 93,1117–1124 [DOI] [PubMed] [Google Scholar]
- 10.Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. ( 2001) Nature 410,898–904 [DOI] [PubMed] [Google Scholar]
- 11.Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100,14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amano T., Tozawa K., Yoshida M., Murakami H. ( 1994) FEBS Lett. 348,93–98 [DOI] [PubMed] [Google Scholar]
- 13.Ohtsubo M., Yoshida M., Ohta S., Kagawa Y., Yohda M., Date T. ( 1987) Biochem. Biophys. Res. Commun. 146,705–710 [DOI] [PubMed] [Google Scholar]
- 14.Park M. Y., Omote H., Maeda M., Futai M. ( 1994) J. Biochem. 116,1139–1145 [DOI] [PubMed] [Google Scholar]
- 15.Senior A. E., al-Shawi M. K. ( 1992) J. Biol. Chem. 267,21471–21478 [PubMed] [Google Scholar]
- 16.Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. ( 2007) Cell 130,309–321 [DOI] [PubMed] [Google Scholar]
- 17.Nishizaka T., Oiwa K., Noji H., Kimura S., Muneyuki E., Yoshida M., Kinosita K., Jr. ( 2004) Nat. Struct. Mol. Biol. 11,142–148 [DOI] [PubMed] [Google Scholar]
- 18.Ariga T., Muneyuki E., Yoshida M. ( 2007) Nat. Struct. Mol. Biol. 14,841–846 [DOI] [PubMed] [Google Scholar]
- 19.Masaike T., Koyama-Horibe F., Oiwa K., Yoshida M., Nishizaka T. ( 2008) Nat. Struct. Mol. Biol. 15,1326–1333 [DOI] [PubMed] [Google Scholar]
- 20.Okuno D., Fujisawa R., Iino R., Hirono-Hara Y., Imamura H., Noji H. ( 2008) Proc. Natl. Acad. Sci. U.S.A. 105,20722–20727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe R., Iino R., Shimabukuro K., Yoshida M., Noji H. ( 2008) EMBO Rep. 9,84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuike S., Adachi K., Sakaki N., Shimo-Kon R., Itoh H., Muneyuki E., Yoshida M., Kinosita K., Jr. ( 2008) Biophys. J. 95,761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirono-Hara Y., Ishizuka K., Kinosita K., Jr., Yoshida M., Noji H. ( 2005) Proc. Natl. Acad. Sci. U.S.A. 102,4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rondelez Y., Tresset G., Nakashima T., Kato-Yamada Y., Fujita H., Takeuchi S., Noji H. ( 2005) Nature 433,773–777 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.