Abstract
SYBR Green I (SG) is widely used in real-time PCR applications as an intercalating dye and is included in many commercially available kits at undisclosed concentrations. Binding of SG to double-stranded DNA is non-specific and additional testing, such as DNA melting curve analysis, is required to confirm the generation of a specific amplicon. The use of melt curve analysis eliminates the necessity for agarose gel electrophoresis because the melting temperature (Tm) of the specific amplicon is analogous to the detection of an electrophoretic band. When using SG for real-time PCR multiplex reactions, discrimination of amplicons should be possible, provided the Tm values are sufficiently different. Real-time multiplex assays for Vibrio cholerae and Legionella pneumophila using commercially available kits and in-house SG mastermixes have highlighted variability in performance characteristics, in particular the detection of only a single product as assessed by Tm analysis but multiple products as assessed by agarose gel electrophoresis. The detected Tm corresponds to the amplicon with the higher G+C% and larger size, suggesting preferential binding of SG during PCR and resulting in the failure to detect multiple amplicons in multiplex reactions when the amount of SG present is limiting. This has implications for the design and routine application of diagnostic real-time PCR assays employing SG.
INTRODUCTION
Real-time PCR technologies and chemistries have undergone intensive development since the original description of the principles of monitoring PCR kinetics using ethidium bromide (1,2). Since then, several manufacturers have developed operating systems, and detection chemistries such as Taqman (3), FRET (4), iFRET (5), scorpions (6) and molecular beacons (7) that are rapidly advancing the use of real-time PCR in both diagnostic and research fields. Various double-stranded DNA (dsDNA)-specific intercalating dyes have been evaluated (8,9), but the most commonly used is SYBR Green I (SG) (10) in both commercial and in-house mastermixes.
A reason that SG is often used is that it is relatively inexpensive compared with other detection chemistries. Additionally, SG allows confirmation of amplicons by melting curve analysis, producing a characteristic melting temperature (Tm) for an amplicon that is analogous to the detection of a specific sized fragment by electrophoresis. The Tm, therefore, can be considered diagnostic so long as the reproducibility of the assay is demonstrated under defined conditions, because Tm values may vary as a function of the concentration of DMSO (11), magnesium chloride, template DNA, SG, and the rate of temperature transition during the melting analysis (12).
Optimisation of PCRs using SG is relatively simple with commercially available SG kits. The exact formulation of these kits is proprietary information and the amount of SG present is undisclosed. In-house SG recipes are widely used and compare favourably with some commercial kits (13). In situations where amplification of multiple loci is required for a diagnostic result, multiplexing such assays would streamline workflow and improve turnaround times. Provided the Tm values of the amplicons are sufficiently different, it should be possible to analyse multiplex PCR in a real-time format. We describe the development of two such real-time multiplex applications—simultaneous detection of ctxAB and hylA genes for Vibrio cholerae and mip and 16S rRNA genes for Legionella pneumophila. Our results highlight the variability in performance characteristics of the various commercial and in-house SG mastermix formulations and the difficulties encountered when taking established multiplex reactions and transferring them to real-time PCR detection using SG, largely due to the apparent preferential binding of SG to selected amplicons.
MATERIALS AND METHODS
Bacterial strains
Vibrio cholerae serotype O1, El-tor biotype (kindly supplied by the Department of Microbiology and Immunology, University of Adelaide) and L.pneumophila serotype 1 ATCC 43111 strains were used for this study. Vibrio cholerae was subcultured onto Columbia blood agar (Oxoid) and incubated at 35°C aerobically for 24 h. Legionella pneumophila serotype 1 was subcultured onto BCYEα and incubated at 35°C aerobically for 3 days.
DNA template preparations
Vibrio cholerae template DNA was prepared by resuspending two to three colonies in 200 µl of Instagene (Bio-Rad, USA) and following manufacturer’s instructions. A 0.5 McFarland suspension of L.pneumophila was used as template for L.pneumophila reactions.
In-house SG PCR conditions
Vibrio cholerae multiplex PCR was performed as described previously (14) except that AmpliTaq Gold was used as the DNA polymerase, the reaction volume was decreased to 20 µl and the cycling conditions were changed to an initial hold at 95°C for 10 min, followed by 40 cycles consisting of 94°C for 20 s, 60°C for 45 s and 72°C for 45 s. Legionella pneumophila multiplex PCR included 16S rRNA (genus specific) primers as previously described (15) and newly designed mip primers (L.pneumophila specific): mip 99F, 5′-TGTCTTATAGCATTGGTGCC-3′; mip 213R, 5′-CAATTGAGCGCCACTCATAG-3′. Four microlitres of template DNA was used in a 20 µl reaction mixture that included 1× PCR buffer II (Applied Biosystems, NJ), 4 mM MgCl2, 200 µM dNTP mix (Promega Corporation, Madison, WI), 0.5 µM each of forward and reverse 16S rRNA primer, 0.1 µM of forward and reverse mip primers, and 1 U AmpliTaq Gold (Applied Biosystems). Cycling conditions began with an initial hold at 95°C for 10 min, followed by 30 cycles consisting of 94°C for 10 s, 50°C for 20 s and 72°C for 25 s. For both multiplex assays SG (supplied as 10 000×; Molecular Probes, OR) was used at final concentrations of either 0.2× or 0.4×. Multiplex PCR conditions were used for amplification of individual amplicons using the appropriate primer pair, except that mip primer concentrations were raised to 0.5 µM. All reactions were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia) with data acquisition at 72°C on the SYBR channel (excitation at 470 nm, detection at 585 nm using a high pass filter) at gains of 2 and 5.
Commercial SYBR Green I kits
LightCycler (LC)–FastStart DNA Master SYBR Green I (catalogue no. 2239264; Roche Diagnostics, Germany), Applied Biosystems SYBR Green PCR mastermix (catalogue no. 4309155; Applied Biosystems) and Quantitect SYBR Green PCR mastermix (catalogue no. 204143; Qiagen, CA) were evaluated under the conditions defined (i.e. the same MgCl2 and primer concentrations, and cycling conditions as for the in-house mastermixes), except that Quantitect assays had an initial hold at 95°C for 15 min.
DNA melting curve analysis
Following amplification, melting curves were acquired on the SYBR channel (at gains of 2 and 5) using a ramping rate of 1°C/60 s for 75–99°C. The differentiated data were analysed using RotorGene software V4.6.94. with the digital filter set as ‘none’.
Gel electrophoresis
When required, samples were analysed by 1% agarose gel electrophoresis with the addition of a Gelstar nucleic acid stain (Cambrex Bio Science; Rockland Inc.) using standard methods (16).
RESULTS
Legionella multiplex PCR
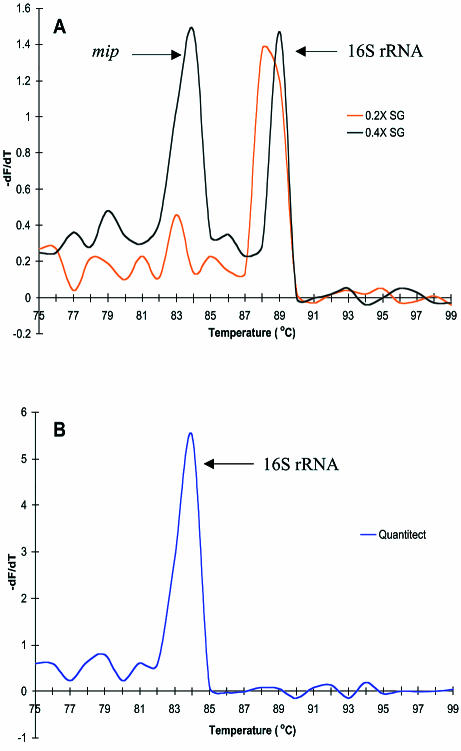
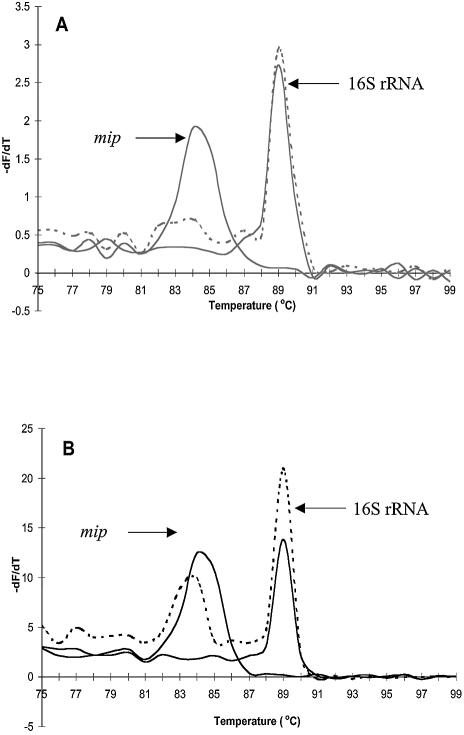
The results of L.pneumophila multiplex PCR differed when using in-house and commercially available kits under the defined conditions. For the commercially available kits, detection of only a single product, as analysed by Tm, was evident. The Tm values for Quantitect, LC FastStart and the Applied Biosystems kits were 84.0, 86.9 and 84.1°C, respectively (Fig. 1B and C). These Tm values correspond to the 16S rRNA gene product (386 bp) as determined by melt curve analysis of simplex reactions (data not shown). In-house SG mastermix with a final concentration of 0.2× SG produced a single Tm of 88.7°C, also corresponding to the 16S rRNA gene product, while the 0.4× SG mastermix produced multiple Tm values of 89.1 and 83.9°C, corresponding to 16S rRNA and mip gene (114 bp) products, respectively (Fig. 1A).
Figure 1.
multiplex melting curves. (A) In-house SG mastermixes. (B) Quantitect. (C) LC FastStart and Applied Biosystems.
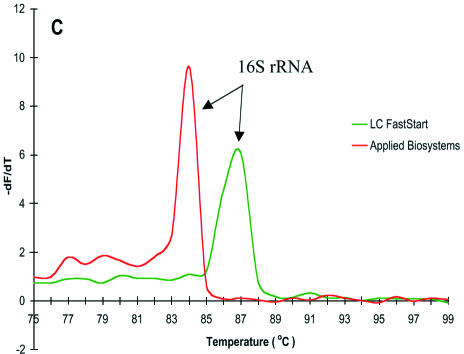
Analysis of the products of multiplex PCR by agarose gel electrophoresis demonstrated that both gene targets were being amplified (Fig. 2). This conflicts with the melt curve analysis, which suggested that only the 16S rRNA gene was being amplified for most of the reaction conditions tested. This is especially clear for the LC FastStart and in-house 0.2× SG mastermixes, which have two clearly defined electrophoretic bands with only a single Tm identified by melt curve analysis. The Quantitect and Applied Biosystems SG mastermixes appeared to perform sub-optimally under the given conditions, with variable generation of mip gene product that could not be detected by Tm when detected by gel electrophoresis.
Figure 2.
Agarose gel electrophoresis of L.pneumophila multiplex reactions. Lane M, 100 bp molecular marker; lanes 1 and 2, 0.2× in-house SG; lanes 3 and 4, 0.4× in-house SG; lanes 5 and 6, LC FastStart; lanes 7 and 8, Quantitect; lanes 9 and 10, Applied Biosystems.
Vibrio multiplex PCR
Similar discrepancies between melt curve analysis and gel electrophoresis were observed for the ctxAB and hylA multiplex PCR. Again, only a single Tm (corresponding to hylA) in Quantitect, LC FastStart and 0.2× in-house SG mastermixes was detected, contrasting with agarose gel electrophoresis, which revealed two distinct bands for both amplified fragments in these reactions (data not shown).
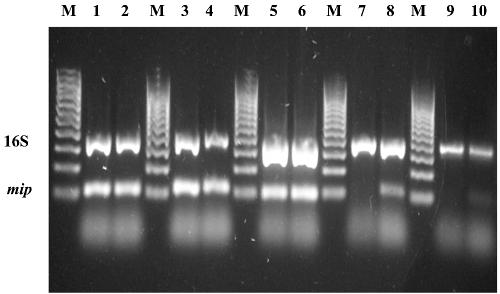
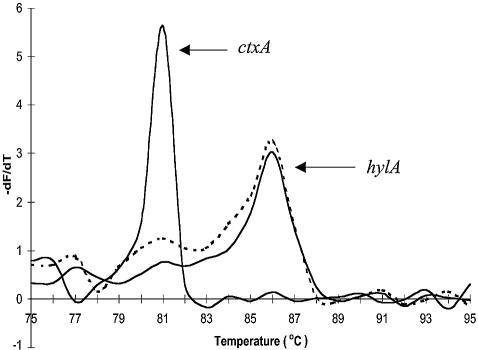
To investigate the possible mechanism for this observation, multiplex PCR using the LC FastStart kit was monitored at 10 cycle intervals by melt curve analysis (at increments of 1°C with a 15 s hold). After 20 cycles, both the ctxAB (385 bp, Tm of 80.8°C) and the hylA (497 bp, Tm of 86.1°C) amplicons could be clearly detected by melt curve analysis (Fig. 3A). However, Tm analysis after 30 and 40 cycles detected only a single Tm for the hylA amplicon (Fig. 3B and C). The production of both amplicons was confirmed by gel electrophoresis (data not shown), indicating end-point detection failure of ctxAB by melt curve analysis.
Figure 3.
Sequential melting curve analysis for V.cholerae multiplex reaction using LC FastStart mastermix. (A) After 20 cycles. (B) After 40 cycles.
DNA melting curve analysis of mixed amplicons
Amplicons were amplified separately, combined (1:1 mixture, 10 µl of each) and the subsequent Tm values determined for the separate and mixed amplicons. When the Legionella products amplified using either the Quantitect, LC FastStart or 0.2× in-house SG mastermixes were combined, only the 16S rRNA Tm was detectable. However, both amplicons were detected for mixes that had been amplified using the Applied Biosystems and in-house 0.4× SG mastermixes. Figure 4 clearly illustrates the difference in the detection when altering the final concentration of SG. In contrast with the Legionella results, mixtures of the ctxAB and hylA amplicons from V.cholerae resulted in the sole detection of the hylA Tm for all mastermixes tested (Fig. 5).
Figure 4.
DNA melting curve analysis of separate and mixed amplicons for L.pneumophila. Solid lines represent simplex melting profiles and dashed lines represent mixed reactions. (A) 0.2× in-house SG. (B) 0.4× in-house SG.
Figure 5.
DNA melting curve analysis of separate and mixed amplicons for V.cholerae using Quantitect. Solid lines represent simplex melting profiles and dashed lines represent mixed reactions.
DISCUSSION
Little is known regarding the precise mechanisms of SG binding to dsDNA. Studies comparing other dsDNA-specific dyes initially suggested that SG exhibits fluorescence via surface or groove binding (17), but further work suggests that fluorescence from SG interaction with dsDNA is dependent on the ratio of base pair/SG molecule complexes, and SG initially intercalates and then probably groove binds when the ratio exceeds 0.15 (http://www.wissenschaft-online.de/gbm/homepage/abstract_detail.php?artikel_id=).
Here we demonstrate discrepancies between agarose gel electrophoresis and detection of amplicons by DNA melting curve analysis using SG. The exact nature of this phenomenon remains to be fully elucidated and the lack of information regarding the structure and initial concentration of SG in commercial kits makes comparison difficult. Furthermore, commercial SG kits often include additional components that increase shelf life or enhance PCR (18), potentially resulting in altered binding characterises of SG.
Data for multiplex reactions and mixtures of amplicons highlight the variability in the performance of commercial and in-house SG mastermixes. For the V.cholerae multiplex assay using a final concentration of 0.2× SG, melt curve analysis failed to detect the ctxAB amplicon at the end of 40 cycles of PCR even though it could be detected after 20 cycles (Fig. 3). This result suggests that the loss of the ctxAB signal may be due to a combination of dye relocation (19–21), preferential binding of SG to the hlyA amplicon and the limited amount of SG available. This is supported by the data from the melt curve experiments combining the individually amplified Legionella products, which found that both mip and 16S rRNA amplicons were detected when amplified with the 0.4× SG mastermix but only the 16S rRNA amplicon was detected with the 0.2× SG mastermix.
Sequence analysis (Primer premier v5.0; Premier Biosoft International, USA) of 16S rRNA, hylA, mip and ctxAB gene products show that G+C% are 50.78, 46.28, 40.40 and 30.65, respectively. In multiplex reactions, amplicons not detected by melt curve analysis correspond to those amplicons with a lower G+C% (ctxAB and mip). Coincidentally, the fragment with higher G+C% is also larger, and previous studies have shown that SG fluorescence strongly correlates with amplicon size (17,22).
Recent publications on heteroduplex discrimination utilising labelled primers (20) and LCGreen (21) have also shown discrepancies in the detection of multiple targets when using SG (in these cases heteroduplexes and homoduplexes). The authors attributed this to the redistribution of SG from products with a lower Tm to a higher Tm during melt curve analysis. The structure and binding characteristic of LCGreen, like SG, is not publicly available and direct comparison cannot be made. However, SG ‘strongly favoured species that melted at higher temperatures’ when compared with LCGreen (21), indicating a preference to G+C-rich areas. In this study, an increase in final SG concentration allowed detection of the two Legionella amplicons (Fig. 4A and B). This may be due to a saturation effect of SG intercalation and groove binding to G+C-rich areas when SG is in excess, which as a result allows detection of the amplicon with the lower Tm. Whilst both the redistribution and the preferential binding of SG can explain our observations, the experimental differentiation of these competing hypotheses may be difficult and needs to be examined systematically in further detail.
In conclusion, our data support previous findings that SG appears to bind preferentially to amplicons with higher G+C% and larger size (17,22). However, those studies have been limited to examining DNA quantitation in vitro and not examining the kinetic effects during real-time PCR as described here. Whilst sequence-specific interaction cannot be eliminated, the data suggest SG will preferentially bind to selected amplicons during multiplex real-time PCR, resulting in the failure to detect an amplicon. With an increasing demand for high-throughput and multiplexing assays, the characteristics of SG described here may reduce optimisation times by avoiding the use of limiting concentrations of SG in assays that target G+C-rich templates.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge the financial support from the Australian Water Quality Centre and the South Australian Water Corporation.
REFERENCES
- 1.Higuchi R., Dollinger,G., Walsh,P.S. and Griffith,R. (1992) Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY), 10, 413–417. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY), 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 3.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 4.Chen X. and Kwok,P.Y. (1999) Homogeneous genotyping assays for single nucleotide polymorphisms with fluorescence resonance energy transfer detection. Genet. Anal., 14, 157–163. [DOI] [PubMed] [Google Scholar]
- 5.Howell W.M., Jobs,M. and Brookes,A.J. (2002) iFRET: an improved fluorescence system for DNA-melting analysis. Genome Res., 12, 1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solinas A., Brown,L.J., McKeen,C., Mellor,J.M., Nicol,J., Thelwell,N. and Brown,T. (2001) Duplex Scorpion primers in SNP analysis and FRET applications. Nucleic Acids Res., 29, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis.Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 8.Skeidsvoll J. and Ueland,P.M. (1995) Analysis of double-stranded DNA by capillary electrophoresis with laser-induced fluorescence detection using the monomeric dye SYBR green I. Anal. Biochem., 231, 359–365. [DOI] [PubMed] [Google Scholar]
- 9.Zipper H., Buta,C., Lammle,K., Brunner,H., Bernhagen,J. and Vitzthum,F. (2003) Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res., 31, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–131, 134–138. [DOI] [PubMed] [Google Scholar]
- 11.Jung M., Muche,J.M., Lukowsky,A., Jung,K. and Loening,S.A. (2001) Dimethyl sulfoxide as additive in ready-to-use reaction mixtures for real-time polymerase chain reaction analysis with SYBR Green I dye. Anal. Biochem., 289, 292–295. [DOI] [PubMed] [Google Scholar]
- 12.Ririe K.M., Rasmussen,R.P. and Wittwer,C.T. (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem., 245, 154–160. [DOI] [PubMed] [Google Scholar]
- 13.Karsai A., Muller,S., Platz,S. and Hauser,M.T. (2002) Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques, 32, 790–792, 794–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shangkuan Y.H., Show,Y.S. and Wang,T.M. (1995) Multiplex polymerase chain reaction to detect toxigenic Vibrio cholerae and to biotype Vibrio cholerae O1. J. Appl. Bacteriol., 79, 264–273. [DOI] [PubMed] [Google Scholar]
- 15.Cloud J.L., Carroll,K.C., Pixton,P., Erali,M. and Hillyard,D.R. (2000) Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol., 38, 1709–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 17.Vitzthum F., Geiger,G., Bisswanger,H., Brunner,H. and Bernhagen,J. (1999) A quantitative fluorescence-based microplate assay for the determination of double-stranded DNA using SYBR Green I and a standard ultraviolet transilluminator gel imaging system. Anal. Biochem., 276, 59–64. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti R. and Schutt,C.E. (2001) The enhancement of PCR amplification by low molecular-weight sulfones. Gene, 274, 293–298. [DOI] [PubMed] [Google Scholar]
- 19.Aktipis S., Martz,W.W. and Kindelis,A. (1975) Thermal denaturation of the DNA-ethidium complex. Redistribution of the intercalated dye during melting. Biochemistry, 14, 326–331. [DOI] [PubMed] [Google Scholar]
- 20.Gundry C.N., Vandersteen,J.G., Reed,G.H., Pryor,R.J., Chen,J. and Wittwer,C.T. (2003) Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin. Chem., 49, 396–406. [DOI] [PubMed] [Google Scholar]
- 21.Wittwer C.T., Reed,G.H., Gundry,C.N., Vandersteen,J.G. and Pryor,R.J. (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem., 49, 853–860. [DOI] [PubMed] [Google Scholar]
- 22.Schneeberger C., Speiser,P., Kury,F. and Zeillinger,R. (1995) Quantitative detection of reverse transcriptase–PCR products by means of a novel and sensitive DNA stain. PCR Methods Appl., 4, 234–238. [DOI] [PubMed] [Google Scholar]