Abstract
A common difficulty in mapping quantitative trait loci (QTLs) is that QTL effects may show environment specificity and thus differ across environments. Furthermore, quantitative traits are likely to be influenced by multiple QTLs or genes having different effect sizes. There is currently a need for efficient mapping strategies to account for both multiple QTLs and marker-by-environment interactions. Thus, the objective of our study was to develop a Bayesian multi-locus multi-environmental method of QTL analysis. This strategy is compared to (1) Bayesian multi-locus mapping, where each environment is analysed separately, (2) Restricted Maximum Likelihood (REML) single-locus method using a mixed hierarchical model, and (3) REML forward selection applying a mixed hierarchical model. For this study, we used data on multi-environmental field trials of 301 BC2DH lines derived from a cross between the spring barley elite cultivar Scarlett and the wild donor ISR42-8 from Israel. The lines were genotyped by 98 SSR markers and measured for the agronomic traits “ears per m²,” “days until heading,” “plant height,” “thousand grain weight,” and “grain yield”. Additionally, a simulation study was performed to verify the QTL results obtained in the spring barley population. In general, the results of Bayesian QTL mapping are in accordance with REML methods. In this study, Bayesian multi-locus multi-environmental analysis is a valuable method that is particularly suitable if lines are cultivated in multi-environmental field trials.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-009-1021-6) contains supplementary material, which is available to authorized users.
Introduction
Detecting favourable exotic quantitative trait loci (QTLs) and introducing them into elite lines could greatly enhance breeding success. Tanksley and Nelson (1996) proposed an advanced backcross QTL analysis combining QTL discovery and variety development in a single step. Using advanced backcross populations derived from a cross of an elite cultivar with an exotic donor, it is possible to identify superior exotic QTLs, whereas the number of negative alleles from the unadapted material is reduced.
In order to map QTLs, the plant material is genotyped by DNA markers and measured on agronomic traits in multi-environmental field trials. In the following statistical analysis, significant associations between DNA markers and phenotypic traits are determined. As quantitative traits are influenced by multiple genes having effects of different magnitudes, it is of primary interest in QTL mapping to select the appropriate model and to estimate the effects and locations of the QTLs (Broman and Speed 2002; Sillanpää and Corander 2002). A common difficulty in QTL mapping is that QTLs may show environment specificity, i.e., QTL effects may significantly differ across environments (Kang and Gauch 1996).
Several authors have examined multi-environmental data in composite interval mapping (Jansen et al. 1995), where selection of background markers is performed in several steps. Usually, uncorrelated residuals, i.e., no genetic (background) correlation among environments, are assumed in these models. Tinker and Mather (1995) implemented composite interval mapping to multi-environmental data using the least-squares estimation (Haley and Knott 1992). They included a test for QTL-by-environment interaction and used partial regression coefficients from background markers to control genetic variance due to non-target QTLs. Recently, Yandell et al. (2007) presented a software package called “R/qtlbim” providing Bayesian interval mapping by accounting for gene-by-environment interaction. Verbyla et al. (2003) computed a multiplicative mixed model for QTL-by-environment interaction of the factorial analysis type. The mixed-model method and the least-squares estimation were used by Piepho (2000). In this study, the genetic correlation among environments was also taken into account. In order to consider genetic correlations, Jiang et al. (1999) used a multi-trait approach of Jiang and Zeng (1995) and regarded expressions of the same trait in different environments as different traits. Fixed effects were pre-corrected by SAS software prior to the QTL analysis. Also, Boer et al. (2007) proposed a modeling approach of genotype-by-environment interactions accounting for genetic correlations between environments and error structure within environments of F5 maize testcross progenies. A multi-locus analysis was applied by Crossa et al. (1999). In this study, partial least-squares regression and factorial regression models were used utilizing genetic markers and environmental covariables for studying QTL-by-environment interaction. Korol et al. (1998) presented an approach where the dependence of a putative QTL effect on environmental conditions is expressed as a function of environmental mean value of the regarded trait. This strategy allows for considering QTL-by-environment interactions across a large number of environments.
Concerning the known literature, a multi-locus QTL mapping approach that simultaneously considers model selection in multi-environmental data has not been fully developed. Since the magnitude of QTL effects can depend on the specific environmental conditions, it is important to account for these effects in the model.
The objective of our research was to compare different approaches of multi-environmental QTL detection considering Bayesian and Restricted Maximum Likelihood (REML) methods: (i) REML single-locus analysis using a mixed hierarchical model, (ii) REML multi-locus analysis by a forward selection approach applying a mixed hierarchical model, (iii) Bayesian multi-locus mapping analyzing one environment at a time, and (iv) Bayesian multi-locus mapping in all environments jointly.
For this purpose, we used field data for an advanced backcross BC2DH population derived from a cross of the malting barley cultivar Scarlett with the wild barley accession ISR42-8 from Israel (von Korff et al. 2006). In order to verify the results obtained with the real dataset, additionally a simulation study was performed. First, in a REML single-locus analysis, a mixed hierarchical model was computed in the Mixed procedure of the software package SAS 9.1 (SAS Institute 2004). Then, the same statistical model was applied by using a forward selection approach where the most significant marker of the current one-dimensional search round was always taken as a fixed cofactor in the model of the next estimation round. Furthermore, we applied a Bayesian multi-locus approach that was extended to handle multi-environmental data. In this approach, only marker points were considered as putative QTLs. In all cases, it was possible to account for QTL effects in multiple environments. This was compared to a Bayesian model where separate single-environmental analyses were executed for one environment at a time. In all analyses, we assumed the absence of genetic (background) correlation among environments.
Materials and methods
Real dataset of a spring barley population
A population with 301 BC2DH lines originating from the cross of the German spring barley variety Scarlett with the Israeli wild barley accession ISR42-8 was developed. The BC2DH population was genotyped by 98 SSR markers. Phenotypic evaluation of the traits “ears per m2” (Ear), “days until heading” (Hea), “plant height” (Hei), “thousand grain weight” (Tgw), and “grain yield” (Yld) was carried out under field conditions in unreplicated experiments at four different locations during the seasons 2003 and 2004. Data on the parental lines were collected but, as we considered the BC2DH lines for QTL mapping, not included in the analysis. A detailed description is given in von Korff et al. (2004, 2006).
Simulation study
In the computer simulation, the real marker data of 301 BC2DH lines were used by imposing known simulated genetic effects influencing the quantitative phenotype. For the genetic effects, marker main, marker interaction (crossover and non-crossover), and markers having both, a main and an interaction effect, were simulated. The positions and effect sizes of the simulated markers are presented in Table 5. As in the field dataset, a population of 301 DH lines was assumed being cultivated in six different environments. Normally distributed phenotypic values of a trait with a heritability of 0.59 were simulated. In the simulation, residuals were assumed to be independent (no correlation structure) with a standard deviation of 1.2 [N(0, 1.2)] and variance was considered to be the same for all environments. Also, no additional environmental effects were generated.
Table 5.
Results from Bayesian single- and multi-environmental QTL mapping of the simulated dataset
| Marker | Chr.a | Pos.b | Effect typec | Env.d | True effect | Bayes singlee | Bayes multif | |
|---|---|---|---|---|---|---|---|---|
| Estimated effect | Estimated effect (unstandardized) | Occ. prob.g | ||||||
| 16 | 1H | 144 | M | 1 | 3.5 | 3.27 | 3.39 | 1 |
| 2 | 3.5 | 3.30 | 3.39 | 1 | ||||
| 3 | 3.5 | 3.38 | 3.39 | 1 | ||||
| 4 | 3.5 | 3.63 | 3.39 | 1 | ||||
| 5 | 3.5 | 3.53 | 3.39 | 1 | ||||
| 6 | 3.5 | 3.64 | 3.39 | 1 | ||||
| 22 | 2H | 80 | M + Ic | 1 | −2.1 | −2.41 | −1.98 | 1 |
| 2 | −3.5 | −3.45 | −3.33 | 1 | ||||
| 3 | −2.1 | −1.90 | −1.98 | 1 | ||||
| 4 | −2.1 | −2.11 | −1.98 | 1 | ||||
| 5 | 0.4 | 0 | 0.21 | 0.98 | ||||
| 6 | −2.1 | −2.14 | −1.98 | 1 | ||||
| 31 | 3H | 25 | Ic | 1 | 0 | 0 | 0.24 | 0 |
| 2 | −3.3 | −3.25 | −2.85 | 1 | ||||
| 3 | 2.8 | 2.94 | 0.24 | 0 | ||||
| 4 | 1.7 | 2.16 | 2.12 | 0.90 | ||||
| 5 | −2.3 | 0 | 0.24 | 0 | ||||
| 6 | 0 | 0 | 0.24 | 0 | ||||
| 32 | 3H | 30 | – | 1 | 0 | 0 | −0.14 | 0 |
| 2 | 0 | 0 | −0.14 | 0 | ||||
| 3 | 0 | 0 | 2.04 | 0.99 | ||||
| 4 | 0 | 0 | −0.14 | 0 | ||||
| 5 | 0 | −1.71 | −1.94 | 0.99 | ||||
| 6 | 0 | 0 | −0.14 | 0 | ||||
| 40 | 3H | 155 | Inc | 1 | 1.2 | 1.01 | 1.22 | 0.99 |
| 2 | 0 | 0 | −0.06 | 0 | ||||
| 3 | 0 | 0 | −0.06 | 0 | ||||
| 4 | 2.1 | 2.29 | 2.56 | 1 | ||||
| 5 | 0 | 0 | −0.06 | 0 | ||||
| 6 | 0.8 | 0 | 0.77 | 0.59 | ||||
| 45 | 4H | 21 | M | 1 | −1.3 | −1.37 | −1.08 | 0.50 |
| 2 | −1.3 | −0.96 | −1.08 | 0.50 | ||||
| 3 | −1.3 | −1.53 | −1.09 | 0.50 | ||||
| 4 | −1.3 | −1.77 | −1.09 | 0.51 | ||||
| 5 | −1.3 | −1.09 | −1.08 | 0.49 | ||||
| 6 | −1.3 | −1.38 | −1.08 | 0.50 | ||||
| 53 | 4H | 125 | Inc | 1 | 0 | 0 | 0.16 | 0 |
| 2 | −3.1 | −3.21 | −3.13 | 1 | ||||
| 3 | 0 | 0 | 0.16 | 0 | ||||
| 4 | −2.2 | −2.18 | −2.13 | 1 | ||||
| 5 | 0 | 0 | 0.16 | 0 | ||||
| 6 | −2.8 | −2.77 | −2.68 | 1 | ||||
| 65 | 5H | 43 | Ic | 1 | 0.3 | 0 | −0.21 | 0.06 |
| 2 | 0.9 | 0.92 | 0.98 | 0.86 | ||||
| 3 | −0.8 | −0.06 | −0.43 | 0.01 | ||||
| 4 | 0 | 0 | −0.42 | 0 | ||||
| 5 | −0.2 | 0 | −0.43 | 0 | ||||
| 6 | −0.6 | 0 | −0.42 | 0 | ||||
| 75 | 6H | 96 | M | 1 | 1.8 | 1.99 | 1.58 | 1 |
| 2 | 1.8 | 1.62 | 1.58 | 1 | ||||
| 3 | 1.8 | 1.83 | 1.58 | 1 | ||||
| 4 | 1.8 | 1.86 | 1.58 | 1 | ||||
| 5 | 1.8 | 1.86 | 1.58 | 1 | ||||
| 6 | 1.8 | 1.28 | 1.58 | 1 | ||||
| 79 | 6H | 135 | M | 1 | 0.2 | 0 | 0.08 | 0 |
| 2 | 0.2 | 0 | 0.08 | 0 | ||||
| 3 | 0.2 | 0 | 0.08 | 0 | ||||
| 4 | 0.2 | 0 | 0.08 | 0 | ||||
| 5 | 0.2 | 0 | 0.08 | 0 | ||||
| 6 | 0.2 | 0 | 0.07 | 0 | ||||
| 92 | 7H | 146 | M + Inc | 1 | 2.5 | 2.45 | 2.06 | 1 |
| 2 | 2.5 | 2.50 | 2.06 | 1 | ||||
| 3 | 0.6 | 0 | 0.84 | 0.02 | ||||
| 4 | 1.2 | 1.24 | 2.06 | 1 | ||||
| 5 | 2.5 | 2.32 | 2.06 | 1 | ||||
| 6 | 2.5 | 2.71 | 2.06 | 1 | ||||
Significant markers with chromosomal positions, effect type, environment, true and estimated effects and occupancy probabilities of marker effects being higher than the significance threshold are shown
aChromosomal location of the markers
bPosition of the markers on the chromosome in centiMorgan (cM)
cEffect type of the marker: M = marker main effect; Ic = marker interaction effect (crossover); Inc = marker interaction effect (non-cross-over); M + Ic = marker main and interaction (crossover) effect; M + Inc = marker main and interaction (non-crossover) effect
dIn the computer simulation, the environment where the lines were cultivated
eBayesian single-environmental QTL mapping
fBayesian multi-environmental QTL mapping
gOccupancy probability of marker effects being higher than the significance threshold in Bayesian multi-environmental mapping
QTL mapping strategies
In our study, we compared different approaches of multi-locus multi-environmental QTL detection in the real and in the simulated dataset:
REML single-locus analysis
The single-locus analysis was performed with SAS 9.1 software (SAS Institute 2004) using REML method of the Mixed procedure. Then, the applied mixed hierarchical model was as follows:
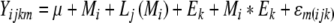 |
With phenotypic observations Yijkm, general mean μ, fixed effect Mi of the ith marker, random effect Lj(Mi) of the jth BC2DH line nested in the ith marker, random effect Ek of the kth environment, random interaction effect Mi * Ek of the ith marker with the kth environment, and residue εm(ijk) of Yijkm.
In this analysis, the random factor Lj(Mi) can be interpreted as a genetic background effect. The residuals were assumed to be identically and independently normally distributed. For each marker, a value of F-statistic, used to test the marker effect, is computed considering the residual mean of squares as an error term. The marker-by-environment interactions are tested by the value of t-statistic.
Missing marker data are handled by omitting each observation with a missing marker value from the dataset. Thus, the amount of phenotypic information is reduced due to missing marker data.
The relative performance of the homozygous exotic genotype (RP[Hsp]) was calculated by 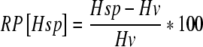 , where Hsp represents the least square mean of the homozygous exotic genotype and Hv the least square mean of the elite genotype.
, where Hsp represents the least square mean of the homozygous exotic genotype and Hv the least square mean of the elite genotype.
The computing time was about 1 min for one trait of both, the spring barley population and the simulated dataset on a Pentium IV 2.0 GHz processor.
REML multi-locus analysis using a forward selection approach
The same mixed hierarchical model as described above was applied here for stepwise variable selection in SAS Proc Mixed. The stepwise variable selection strategy is described in Sillanpää and Corander (2002) and has been applied for example in Kilpikari and Sillanpää (2003). The first round of forward selection procedure corresponds to the single-locus analysis. Next, the marker with the most significant effect (based on the P value of hypothesis test Type III F-statistic) is chosen as a fixed cofactor in the model of the following estimation rounds. Using this extended model, the marker effects are estimated again. This procedure is repeated until no further significant markers can be found. The computing time for this method was about 20 min for one trait of the real and of the simulated dataset.
Bayesian multi-locus analysis using multi-environmental data
Additionally, we performed Bayesian multi-locus QTL mapping using multi-environmental data.
The statistical model for phenotypic trait values Yjk was as follows:
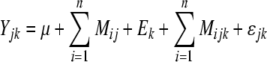 |
where μ is the overall sample mean of the phenotypes, Mij is the effect of the ith marker genotype of the jth line, Ek is the effect of the kth environment, Mijk is the effect of the ith marker genotype of the jth line in the kth environment (i.e. genotype-by-environment interaction), and, n is the number of markers.
Residuals are assumed to be independently and identically normally distributed as ɛjk∼N(0, σ20), where σ20 = residual variance common to all environments.
In the Bayesian setting, we parametrized the statistical model so that for each marker one genotype effect was assigned a value of zero; thus, for each marker we only needed to estimate one main effect Mij. Similarly, in each environment for each marker, one environment-specific genotype effect was assigned a value of zero, resulting in one estimable coefficient Mijk at each environment. By denoting the genotype (A or B) of line j at marker i with Xij, the effects can be written as 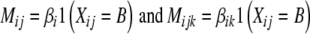 . The parameters βi and βik are interpreted as the difference of the main genotype effects and the differences of environment-specific genotype effects. Note, however, that unlike REML, this model is still oversaturated. The prior densities of the unknown marker effect differences in a K-environment model, θ = (β1,…, βn, β11,…, βnk), were specified following Xu (2003); Hoti and Sillanpää (2006), where each effect θr, r = 1,…, (K + 1)n, in the statistical model is assigned a zero mean normal distribution with its own variance parameter σ2r combined with Jeffreys’ scale invariant prior
. The parameters βi and βik are interpreted as the difference of the main genotype effects and the differences of environment-specific genotype effects. Note, however, that unlike REML, this model is still oversaturated. The prior densities of the unknown marker effect differences in a K-environment model, θ = (β1,…, βn, β11,…, βnk), were specified following Xu (2003); Hoti and Sillanpää (2006), where each effect θr, r = 1,…, (K + 1)n, in the statistical model is assigned a zero mean normal distribution with its own variance parameter σ2r combined with Jeffreys’ scale invariant prior 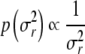 . The prior of the overall mean was p(μ) ∝ 1, and the priors of the environmental effects were
. The prior of the overall mean was p(μ) ∝ 1, and the priors of the environmental effects were 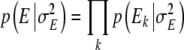 , where
, where 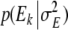 is a normal distribution with zero mean and a common variance
is a normal distribution with zero mean and a common variance 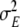 . The variance of the environmental effects and the variance of the residual term were assigned improper uniform priors,
. The variance of the environmental effects and the variance of the residual term were assigned improper uniform priors, 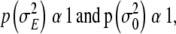 respectively.
respectively.
In order to obtain Markov Chain Monte Carlo (MCMC) samples of the joint posterior distribution of marker effects, Gibbs sampling (Geman and Geman 1984) and Metropolis-Hastings algorithms (Hastings 1970) were used. Here, we give the fully conditional posterior distributions of the environmental effects Ek and the effect variance σ2E. The sampling distributions/updating steps of the remaining parameters and handling of missing data are described in Hoti and Sillanpää (2006). The fully conditional posterior distribution of the environmental effect Ek is a normal distribution with mean 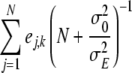 and variance
and variance 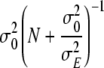 , where N is the total number of lines. For the effect variance
, where N is the total number of lines. For the effect variance 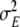 , the fully conditional posterior distribution is the scaled inverted chi-squared distribution with the degree of freedom parameter K and the scale parameter
, the fully conditional posterior distribution is the scaled inverted chi-squared distribution with the degree of freedom parameter K and the scale parameter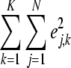 .
.
The Bayesian analysis was implemented using Matlab 7 (2007). The missing values were randomly assigned initial values from their empirical distributions. The MCMC algorithm was run for 400,000 rounds in the field dataset and 50,000 rounds in the simulated dataset. In order to reduce autocorrelation, only every 10th round was stored. In all cases, in the field data the first 380,000 MCMC-rounds (simulated data: 20,000 rounds) were considered to be “burn-in” rounds and were thus not considered in the final results. Computing time was about 33 h for one trait of the real dataset and about 20 min for the simulated dataset.
In the QTL analysis, we obtained estimates of marker main (Mij) and interaction (Mijk) effects. For each MCMC-sample, the sum of main and corresponding environment interaction effect was calculated. Then, to interpret the results at each marker locus the median of the posterior distribution of marker effect over all MCMC-rounds was computed. If this median was non-zero in all environments, this marker had a main effect on the specific trait value. Otherwise, if the median was non-zero in some environments only, it was interpreted as a specific kind of a marker-by-environment interaction effect.
Bayesian multi-locus analysis using single-environmental data
In order to determine whether multi-environmental QTL testing improves the results, we also conducted the same Bayesian multi-locus mapping as described above, but used data of each environment separately. Thus, the statistical model was reduced to:
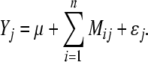 |
In this analysis, computing time of the main analysis was about 9 h for one trait of the real dataset and about 15 min for the simulated dataset.
Analogous to the multi-environmental Bayesian QTL analysis, the posterior median of the marker effects over all MCMC-rounds was computed. In both single- and multi-environmental Bayesian analysis, model selection and parameter estimation were based on adaptive shrinkage (Xu 2003; Hoti and Sillanpää 2006). Note that this approach is closely related to the so-called genome-wide selection (Meuwissen et al. 2001). In the genome-wide selection approach, breeding values are predicted based on molecular markers covering the whole genome. This strategy is in contrast to the use of genetic similarities, which are calculated based on the molecular marker data, in the prediction of breeding values (Bauer et al. 2006, 2008).
Convergence of the MCMC—chain
The convergence assessment of the Bayesian mapping strategies was performed by plotting the MCMC paths for the markers with estimated non-zero effects as suggested by Kass et al. (1998).
Significance threshold of estimated marker effects
In order to determine whether the detected QTL effects were due to spurious effects, we estimated an experimentwise critical value following Churchill and Doerge (1994). In this estimation, the data are shuffled by computing random permutations of the phenotypic observation vector. The ith observation is assigned to the ith line whose index is given by the ith element of the permutation. Thus, the association between marker data and observations is destroyed. The shuffled data were analysed for Bayesian single-environmental and REML single-locus analysis. Overall, 50 permutations were calculated. In order to obtain the experimentwise critical value for a trait analysed by Bayesian single-environmental mapping, first the maximum median of the marker effects of every QTL analysis of permuted data is selected. In REML single-locus analysis, the maximum F value (for marker main effects) and the maximum t value (for marker interaction effects) of all permuted QTL analyses are chosen. In each mapping strategy, these values are ordered. The experimentwise critical value then corresponds to the 100(1 − α) percentile, where α equals 0.05. In order to detect QTL effects in the original data and to determine statistical significance, the results of the QTL analysis can be compared to this critical value.
The forward selection approach utilizes the significance threshold obtained from REML single-locus analysis. As computing time was demanding for a Bayesian multi-environmental analysis, the calculation of a permutation analysis was not possible. Therefore, following Hoti and Sillanpää (2006) the MCMC-samples of all traits and markers were standardized to a common scale by multiplying each MCMC-sample with 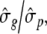 where
where 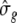 is the empirical standard deviation of each marker and
is the empirical standard deviation of each marker and 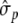 corresponds to the empirical standard deviation of phenotypic data. In the field dataset, a marker was defined to be significant if its standardized effect was greater than +0.17 or smaller than −0.17. In the simulated dataset, a significance threshold of ±0.10 was chosen, thus, all markers having an effect greater than −0.10 or smaller than +0.10 are not considered to be significant.
corresponds to the empirical standard deviation of phenotypic data. In the field dataset, a marker was defined to be significant if its standardized effect was greater than +0.17 or smaller than −0.17. In the simulated dataset, a significance threshold of ±0.10 was chosen, thus, all markers having an effect greater than −0.10 or smaller than +0.10 are not considered to be significant.
Putative QTLs
Following Pillen et al. (2003), for each QTL mapping strategy linked significant markers that had a distance of ≤20 cM were interpreted as a single putative QTL.
Bin marker map
A Bin marker class was assigned to all used SSR markers following Kleinhofs and Graner (2001); Costa et al. (2001). Additionally, for the markers HVM62, GBM1015, HVM67, HVLTPPB, HVM36, and GBM1052, Bin classes were also available from the high-density consensus map recently published by Marcel et al. (2007). In the following, Bin classes obtained from Marcel et al. (2007) are given in italics.
Genetic variance explained by a marker
The genetic variance explained by a marker (R²) was computed by:
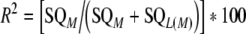 |
with SQM = sum of squares of markers obtained from hypothesis test Type I; SQL(M) = Type I sum of squares of lines nested in markers.
In order to obtain SQM and SQL(M) we calculated the following mixed model in SAS Proc Mixed:
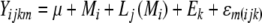 |
where all parameters have been fixed factors.
Heritability
The heritability of the traits was obtained by REML variance component estimation using the Varcomp procedure in the SAS software package:
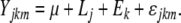 |
Then the heritability follows from 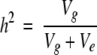 , where Vg = genetic variance of the BC2DH lines and Ve = residual variance.
, where Vg = genetic variance of the BC2DH lines and Ve = residual variance.
Results
Field data of a spring barley population
In general, similar QTLs were detected using REML single-locus analysis, the REML forward selection approach, Bayesian multi-locus multi-environmental method considering all environments jointly in the analysis, and Bayesian multi-locus single-environmental mapping where each environment is analysed separately (Table 1). Depending on the heritability of the trait, some QTLs could be found to have a significant effect in all four mapping strategies. For example, considering “plant height,” a trait with a high heritability h² of 0.76, three of nine QTLs were detected with all analyses. In contrast, regarding “ears per m²,” a trait with a low heritability of 0.21, only one of 11 QTLs could be found to be significant with all approaches. In addition, only marker main effects were detected using a REML mapping method, whereas both marker main and interaction effects could be found by using a Bayesian approach.
Table 1.
Detected QTLs of REML single-locus analysis (I), REML forward selection (II), Bayesian single-environmental (III), and Bayesian multi-environmental (IV) mapping for several traits in the spring barley population
| QTLa | Coded marker numberb | SSR-marker group | Chr.c | Pos.d | Bine | R² (%)f | RP(%)g | QTL mapping strategies | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ih | IIi | IIIj | IVk | ||||||||
| Days until heading (h² ≈ 0.77) | |||||||||||
| QHea.S42-1H.1 | 5 | GBM1042 | 1H | 39 | 6 | 4.4 | 2.7 | M*l | MxEm | ||
| QHea.S42-1H.2 | 14 | GBM1061 | 1H | 115 | 13 | 0.7 | 0.9 | Mm | |||
| QHea.S42-1H.3 | 16 | HVABAIP | 1H | 144 | 13 | 4.0 | 1.8 | MxE | MxE | ||
| QHea.S42-2H.1 | 20 | GBM1052 | 2H | 42 | 4 (4) | 21.6 | −9.0 | M* | M* | M | MxE |
| QHea.S42-2H.2 | 22–23 | EBmac684, GMS3 | 2H | 80-86 | 7–8 | 4.9, 5.0 | −1.9, −2.0 | M* | MxE | MxE | |
| QHea.S42-2H.3 | 29 | EBmac415 | 2H | 146 | 13 | 4.2 | −3.3 | MxE | |||
| QHea.S42-3H.1 | 31–32 | HVLTPPB, EBmac705 | 3H | 25–30 | 3 (3) | 4.9, 4.5 | −4.4, −4.3 | M* | MxE | ||
| QHea.S42-3H.2 | 39 | GBM1043 | 3H | 130 | 10 | 4.3 | −1.9 | M* | M* | MxE | |
| QHea.S42-3H.3 | 40–41 | HV13GEIII, HVM62 | 3H | 155–165 | 13–14 (15) | 7.3, 4.6 | −2.9, −2.2 | M* | M* | MxE | M |
| QHea.S42-4H.1 | 54 | EBmac701 | 4H | 130 | 10 | 0.5 | 0.7 | M | |||
| QHea.S42-4H.2 | 59–61 | HVJASIP, HVM67, HDAMYB | 4H | 180–190 | 12–13 (12) | 4.3, 4.9, 6.1 | 1.8, 1.9, 2.2 | M* | M* | MxE | |
| QHea.S42-5H.1 | 65 | Bmag337 | 5H | 43 | 5 | 0.3 | −0.6 | MxE | |||
| QHea.S42-5H.2 | 68 | MGB338 | 5H | 85 | 8 | 0.4 | 1.0 | MxE | |||
| QHea.S42-6H.1 | 77 | EBmac624 | 6H | 107 | 7 | 4.5 | −2.1 | M* | |||
| QHea.S42-7H.1 | 92 | BMS64 | 7H | 146 | 8 | 4.1 | −2.6 | M* | M* | ||
| Plant height (h² ≈ 0.76) | |||||||||||
| QHei.S42-2H.1 | 18–20 | HVM36, GBM1035, GBM1052 | 2H | 17–42 | 2–4 (3–4) | 6.0, 5.3, 3.0 | −8.9, −8.6, −11.5 | M* | M* | MxE | |
| QHei.S42-2H.2 | 22–24 | EBmac684, GMS3, HVTUB | 2H | 80–92 | 7–8 | 13.5, 16.3, 9.9 | −10.8, −12.0, −8.9 | M* | M* | M | |
| QHei.S42-3H.1 | 31–35 | HVLTPPB, EBmac705, HVITR1, MGB410, Bmag603 | 3H | 25–70 | 3–6 (3) | 5.4, 5.4, 8.2, 6.1, 6.8 | 16.7, 16.7, 24.0, 12.9 | M* | M* | MxE | MxE |
| QHei.S42-3H.2 | 39 | GBM1043 | 3H | 130 | 10 | 7.6 | 8.9 | M* | M* | M | MxE |
| QHei.S42-3H.3 | 40–43 | HV13GEIII, HVM62, MGB358, Bmac29 | 3H | 155–190 | 13–16 (15) | 22.0, 16.9, 10.5, 3.9 | 17.9, 15.1, 11.7, 7.3 | M* | M* | MxE | MxE |
| QHei.S42-4H.1 | 53–56 | TACMD, EBmac701, EBmac635, EBmac679 | 4H | 125–132 | 9-10 | 4.8, 6.3, 4.7, 5.1 | −7.3, −8.4, −7.0, −7.3 | M* | |||
| QHei.S42-4H.2 | 58–61 | GBM1015, HVJASIP, HVM67, HDAMYB | 4H | 170–190 | 12–13 (11–12) | 9.2, 14.6, 15.2, 5.8 | −8.8, −11.0, −11.0,−7.1 | M* | M* | MxE | |
| QHei.S42-5H.1 | 65 | Bmag337 | 5H | 43 | 5 | 6.4 | 9.6 | M* | |||
| QHei.S42-7H.1 | 83 | Bmag7 | 7H | 27 | 2 | 4.6 | 12.7 | M* | |||
| Grain yield (h² ≈ 0.70) | |||||||||||
| QYld.S42-1H.1 | 6 | MGB325 | 1H | 52 | 6 | 5.0 | −13.0 | M* | |||
| QYld.S42-1H.2 | 8–11 | HVM20, Bmag211, Bmag149, Bmag105 | 1H | 65–75 | 7–8 | 4.9, 4.1, 4.8, 7.2 | −12.9, −11.8, −12.8, −18.3 | M* | M* | MxE | M |
| QYld.S42-1H.3 | 16 | HVABAIP | 1H | 144 | 13 | 5.8 | −9.3 | MxE | MxE | ||
| QYld.S42-2H.1 | 25–26 | Bmag381, Bmag125 | 2H | 107–122 | 9–10 | 3.0, 4.2 | −6.5, −8.4 | M* | MxE | ||
| QYld.S42-3H.1 | 31–35 | HVLTPPB, EBmac705, HVITR1, MGB410, Bmag603 | 3H | 25-70 | 3–6 (3) | 13.8, 13.9, 13.7, 10.7, 12.4 | −32.6, −32.5, −39.8, −22.8, −24.7 | M* | M* | MxE | MxE |
| QYld.S42-3H.2 | 39 | GBM1043 | 3H | 130 | 10 | 2.0 | −5.3 | M* | MxE | MxE | |
| QYld.S42-3H.3 | 40–41 | HV13GEIII, HVM62 | 3H | 155-165 | 13–14 (15) | 8.2, 6.1 | −12.8, −10.7 | M* | MxE | MxE | |
| QYld.S42-3H.4 | 43 | Bmac29 | 3H | 190 | 16 | 0.1 | −1.1 | MxE | |||
| QYld.S42-5H.1 | 65 | Bmag337 | 5H | 43 | 5 | 3.6 | −9.6 | MxE | |||
| QYld.S42-5H.2 | 69 | GMS61 | 5H | 126 | 10 | 2.6 | −12.9 | MxE | |||
| QYld.S42-7H.1 | 92 | BMS64 | 7H | 146 | 8 | 4.2 | −11.1 | M* | |||
| Thousand grain weight (h² ≈ 0.54) | |||||||||||
| QTgw.S42-1H.1 | 8 | HVM20 | 1H | 65 | 7 | 1.5 | −2.1 | MxE | |||
| QTgw.S42-1H.2 | 16 | HVABAIP | 1H | 144 | 13 | 0.5 | 1.0 | MxE | |||
| QTgw.S42-2H.1 | 23 | GMS3 | 2H | 86 | 8 | 1.9 | 1.9 | M* | MxE | MxE | |
| QTgw.S42-2H.2 | 25–26 | Bmag381, Bmag125 | 2H | 107–122 | 9–10 | 0.3, 0.5 | 0.9, 0.9 | MxE | MxE | ||
| QTgw.S42-3H.1 | 38 | HVM60 | 3H | 110 | 9 (8) | 4.5 | 3.1 | M* | |||
| QTgw.S42-3H.2 | 40–42 | HV13GEIII, HVM62, MGB358 | 3H | 155–175 | 13–15 (15) | 3.3, 4.3, 6.5 | −2.8, −3.1, −3.8 | M* | M* | MxE | |
| QTgw.S42-4H.1 | 53–56 | TACMD, EBmac701, EBmac635, EBmac679 | 4H | 125–132 | 9–10 | 16.6, 16.3, 17.1, 16.8 | −5.6, −5.7, −5.5, −5.5 | M* | M* | MxE | MxE |
| QTgw.S42-4H.2 | 59–61 | HVJASIP, HVM67, HDAMYB | 4H | 180–190 | 12–13 (12) | 8.8, 8.4, 8.7 | −3.7, −3.6, −3.7 | M* | M* | MxE | |
| QTgw.S42-6H.1 | 75 | GMS6 | 6H | 96 | 5 | 1.4 | −1.8 | MxE | |||
| QTgw.S42-6H.2 | 79 | GBM1008 | 6H | 135 | 10 | 1.6 | 1.8 | MxE | |||
| QTgw.S42-7H.1 | 92–95 | BMS64, Bmag120, MGB317, EBmac755 | 7H | 146–166 | 8–11 | 4.6, 4.7, 4.4, 4.2 | −4.2, −4.3, −4.6, −3.6 | M* | M* | MxE | |
| Ears per m² (h² ≈ 0.21) | |||||||||||
| QEar.S42-1H.1 | 16 | HVABAIP | 1H | 144 | 13 | 6.0 | 8.4 | M* | |||
| QEar.S42-2H.1 | 18 | HVM36 | 2H | 17 | 2 (3) | 5.8 | 9.5 | M* | |||
| QEar.S42-2H.2 | 21–24 | MGB391, EBmac684, GMS3, HVTUB | 2H | 67–92 | 6–8 | 6.4, 21.5, 24.2, 21.2 | 11.1, 15.3, 16.5, 14.7 | M* | MxE | MxE | |
| QEar.S42-3H.1 | 34–35 | MGB410, Bmag603 | 3H | 65–70 | 5–6 | 6.7, 6.5 | −13.7, −13.8 | M* | M* | ||
| QEar.S42-3H.2 | 40–41 | HV13GEIII, HVM62 | 3H | 155–165 | 13–14 (15) | 5.2, 4.6 | −8.8, −7.9 | M* | |||
| QEar.S42-4H.1 | 45–46 | HVOLE, HVB23D | 4H | 21–25 | 3 | 5.2, 4.5 | −9.7, −9.3 | M* | |||
| QEar.S42-4H.2 | 53–56 | TACMD, EBmac701, EBmac635, EBmac679 | 4H | 125–132 | 9–10 | 10.0, 9.6, 9.5, 9.1 | 11.4, 11.3, 10.6, 10.6 | M* | |||
| QEar.S42-4H.3 | 58–61 | GBM1015, HVJASIP, HVM67, HDAMYB | 4H | 170–190 | 12–13 (11–12) | 13.6, 23.9, 22.7, 9.0 | 12.0, 16.0, 15.4, 9.8 | M* | M* | MxE | MxE |
| QEar.S42-5H.1 | 62 | MGB384 | 5H | 0 | 2 | 6.8 | −10.8 | M* | M* | ||
| QEar.S42-5H.2 | 65 | Bmag337 | 5H | 43 | 5 | 4.6 | −8.3 | M* | |||
| QEar.S42-6H.1 | 79 | GBM1008 | 6H | 135 | 10 | 5.1 | −7.6 | M* | |||
aNames of the QTLs consisting of the qualifier “Q”, the trait abbreviation, the determined population, the mapped chromosome and a QTL number to distinguish between QTLs on the same chromosome. Markers with a distance ≤20 cM were considered to be a single QTL
bThe markers were coded according to their chromosomal location
cChromosomal location of the SSR markers
dPosition of the SSR markers on the chromosome in centiMorgan (cM) (von Korff et al. 2004). If several markers were significant, the cM range is given
eBin marker class following Kleinhofs and Graner (2001); Costa et al. (2001). In the case of several significant markers, the Bin class range is displayed. An additional number in brackets gives the Bin class following Marcel et al. (2007). As the Bin class of Marcel et al. (2007) was available only for some markers, in the case of several significant markers belonging to the same QTL, these markers are given in italics
fGenetic variance explained by a marker calculated using REML method in SAS Proc Mixed
gRelative performance of the homozygous exotic genotype
hREML single-locus QTL mapping
iREML forward selection approach
jBayesian multi-locus single-environmental analysis
kBayesian multi-locus multi-environmental analysis
lMarker main effect obtained from REML analysis
mM Marker main effect, MxE Marker interaction effect. Both effects are derived from Bayesian analysis and have to be interpreted differently as in REML method. In Bayesian mapping the model was oversaturated so that marker main and interaction effects are not independently identifiable
In the following, detailed results of the QTL analyses will be described for every trait separately, where traits are grouped according to their heritability (Table 1):
“Days until heading” (h² ≈ 0.77)
Overall, 15 QTLs distributed over all chromosomes were found to be significant for the trait “days until heading.” Two QTLs were significant for four analyses, three QTLs for three analyses, four QTLs for two analyses, and six QTLs were found in one analysis.
“Plant height” (h² ≈ 0.76)
For “plant height,” nine QTLs on the chromosomes 2H, 3H, 4H, 5H, and 7H were detected. Three QTLs were found in all four approaches, in three analyses, and with only one strategy, respectively.
“Grain yield” (h² ≈ 0.70)
Eleven QTLs for the trait “grain yield” were located on the chromosomes 1H, 2H, 3H, 5H, and 7H. Two QTLs were found with all QTL mapping strategies, two QTLs were detected with three analyses, two QTLs with two analyses, and five QTLs were found in only one mapping strategy.
“Thousand grain weight” (h² ≈ 0.54)
For the trait “thousand grain weight” the analyses revealed 11 QTLs on all chromosomes with the exception of 5H. One QTL was detected with all mapping approaches, four QTLs with three analyses, one QTL with two analyses, and five QTLs were found to be significant in only one analysis.
“Ears per m2” (h² ≈ 0.21)
For the trait “ears per m²,” overall, 11 QTLs could be detected on all chromosomes except 7H. One QTL was found to be significant in all four mapping strategies, one QTL in three analyses, two QTLs in two analyses, and seven QTLs were detected in only one approach.
In order to illustrate the QTL mapping strategies, the results of all statistical analyses will be presented in more detail for the trait “grain yield.” Considering REML single-locus analysis, overall, 14 markers on chromosomes 1H, 2H, 3H, and 7H (Table 2) showed a F value greater than the significance threshold (obtained from permutation test). The P value of F test ranged between 0.001 and 0.017, and the estimated marker effects of the exotic allele ranged between −11.46 and −2.46. If a REML forward selection approach was performed, only four markers had a value of F-statistic greater than the significance threshold. These markers showed a P value of F test ranging from 0.001 to 0.009 and estimated effects from −7.35 to −3.35.
Table 2.
Significant SSR markers with their chromosomal positions, estimated effects of the exotic allele, F and P values from REML single-locus analysis and the REML forward selection approach for the trait “grain yield” of the spring barley population
| SSR marker | Chromosome | Position (cM) | Effect | F value | P value |
|---|---|---|---|---|---|
| REML single-locus analysis | |||||
| MGB325 | 1H | 52 | −3.80 | 15.8 | 0.011 |
| HVM20 | 1H | 65 | −3.78 | 15.5 | 0.011 |
| Bmag211 | 1H | 68 | −3.45 | 12.2 | 0.017 |
| Bmag149 | 1H | 70 | −3.74 | 14.5 | 0.013 |
| Bmag105 | 1H | 75 | −5.36 | 19.7 | 0.007 |
| Bmag125 | 2H | 122 | −2.46 | 12.8 | 0.016 |
| HVLTPPB | 3H | 25 | −9.54 | 39.3 | 0.002 |
| EBmac705 | 3H | 30 | −9.48 | 40.4 | 0.001 |
| HVITR1 | 3H | 49 | −11.46 | 28.7 | 0.003 |
| MGB410 | 3H | 65 | −6.69 | 42.5 | 0.001 |
| Bmag603 | 3H | 70 | −7.26 | 48.7 | 0.001 |
| HV13GEIII | 3H | 155 | −3.76 | 18.3 | 0.008 |
| HVM62 | 3H | 165 | −3.15 | 14.4 | 0.013 |
| BMS64 | 7H | 146 | −3.25 | 12.2 | 0.017 |
| REML forward selection | |||||
| Bmag603 | 3H | 70 | −7.26 | 48.75 | 0.001 |
| HVLTPPB | 3H | 25 | −7.35 | 23.46 | 0.005 |
| Bmag105 | 1H | 75 | −5.34 | 24.84 | 0.004 |
| GBM1043 | 3H | 130 | −3.35 | 17.39 | 0.009 |
Significance threshold obtained from permutation test: 12.14 (F value)
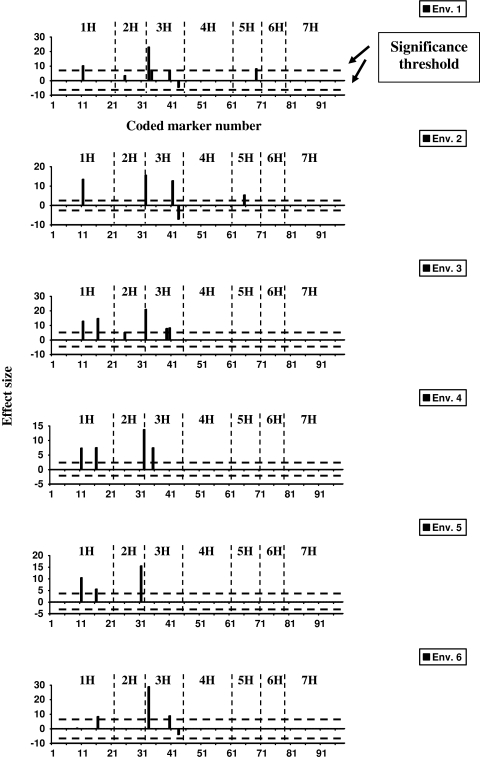
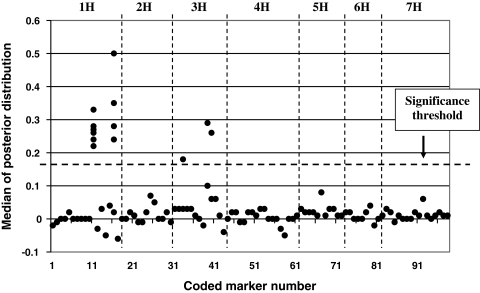
In Bayesian single-environmental mapping, overall, 14 markers showed a significant effect resulting in nine QTLs (Fig. 1; Table 1). None of the markers showed a significant effect in all six environments (Fig. 1). In contrast, considering Bayesian multi-environmental analysis, only five markers having a significant effect were mapped, yielding five QTLs (Fig. 2; Table 1). In Bayesian multi-environmental mapping markers flanking a significant QTL on the same chromosome often showed negligible effects (Fig. 2). For example, the marker 11 has estimated (standardized) effects between 0.22 and 0.33, and is hence defined to be significant. The flanking markers with the numbers 12–15 have small (standardized) effects ranging from −0.05 to +0.04.
Fig. 1.
Locus-specific point-estimates (posterior medians) of effect sizes of Bayesian multi-locus single-environmental QTL mapping for the trait “grain yield” in the spring barley population. The posterior medians are displayed for all environments separately. The significance threshold was obtained from permutation tests. The markers were coded according to their chromosomal location
Fig. 2.
Locus-specific point-estimates (posterior medians) of standardized effects of Bayesian multi-locus multi-environmental QTL mapping for the trait “grain yield” in the spring barley population. The posterior medians of all environments are displayed jointly in the graph. The markers were coded according to their chromosomal location
Simulation study
In the computer simulation, based on the real molecular marker data known genetic effects were assumed. Marker main and interaction (non-crossover and crossover) effects and combinations of both of them were simulated having different effect sizes (Table 5).
In Bayesian single- and multi-environmental QTL mapping, except for one marker with a main effect, all other markers with true (simulated) effects were detected regardless of the marker having an effect in all or in only some environments (Table 5). The marker that was not found in Bayesian analyses (marker 79), has a true effect of 0.2 but estimated effect of 0 which could be due to the small effect size. The false-positive marker 32 was supported by both Bayesian methods (in one or two environments) although the marker has a true effect of zero.
Considering REML single-locus analysis and the REML forward selection approach, all markers with an interaction effect (non-crossover or crossover) were not detected (Table 6). Also, only two of the four markers having a marker main effect were found. However, using REML single-locus mapping in several cases on the same chromosome, a marker near to a marker with a true effect was found to be significant although this marker has an effect of zero in the simulation. In the forward selection approach, a less number of false-positive markers were detected than in REML single-locus analysis.
Table 6.
Significant markers with their chromosomal positions, effect type, true (simulated) and estimated effects, F and P values from REML single-locus analysis and the REML forward selection approach of the simulated dataset
| Marker | Chromosome | Position (cM) | Effect typea | True effect | Estimated effect | F value | P value |
|---|---|---|---|---|---|---|---|
| REML single-locus analysis | |||||||
| 15 | 1H | 130 | – | 0 | −0.89 | 47.41 | 0.001 |
| 16 | 1H | 144 | M | 3.5 | −1.44 | 130.22 | 0 |
| 21 | 2H | 67 | – | 0 | 1.00 | 13.36 | 0.015 |
| 22 | 2H | 80 | M + Ic | −3.5 to 0.4 | – | – | – |
| 27 | 2H | 139 | – | 0 | −0.90 | 10.64 | 0.022 |
| 31 | 3H | 25 | Ic | −3.3 to 2.8 | – | – | – |
| 40 | 3H | 155 | Inc | 0 to 2.1 | – | – | – |
| 45 | 4H | 21 | M | −1.3 | – | – | – |
| 53 | 4H | 125 | Inc | −3.1 to 0 | – | – | – |
| 57 | 4H | 150 | – | 0 | 0.83 | 11.13 | 0.020 |
| 65 | 5H | 43 | Ic | −0.8 to 0.9 | – | – | – |
| 75 | 6H | 96 | M | 1.8 | −0.84 | 29.80 | 0.003 |
| 76 | 6H | 103 | – | 0 | −0.90 | 34.66 | 0.002 |
| 77 | 6H | 107 | – | 0 | −0.84 | 26.44 | 0.004 |
| 79 | 6H | 135 | M | 0.2 | – | – | – |
| 91 | 7H | 133 | – | 0 | −0.57 | 12.00 | 0.018 |
| 92 | 7H | 146 | M + Inc | 0.6 to 2.5 | −0.87 | 16.87 | 0.009 |
| REML forward selection | |||||||
| 16 | 1H | 144 | M | 3.5 | −1.44 | 130.22 | 0 |
| 22 | 2H | 80 | M + Ic | −3.5 to 0.4 | 1.09 | 13.25 | 0.015 |
| 31 | 3H | 25 | Ic | −3.3 to 2.8 | – | – | – |
| 40 | 3H | 155 | Inc | 0 to 2.1 | – | – | – |
| 45 | 4H | 21 | M | −1.3 | – | – | – |
| 47 | 4H | 31 | – | 0 | 0.43 | 35.09 | 0.002 |
| 53 | 4H | 125 | Inc | −3.1 to 0 | – | – | – |
| 65 | 5H | 43 | Ic | −0.8 to 0.9 | – | – | – |
| 75 | 6H | 96 | M | 1.8 | −1.07 | 74.70 | 0 |
| 79 | 6H | 135 | M | 0.2 | – | – | – |
| 92 | 7H | 146 | M + Inc | 0.6 to 2.5 | −0.96 | 34.38 | 0.002 |
For the true marker effect, the range over all environments is given if the marker has an interaction effect
Significance threshold obtained from permutation test: 9.69 (F value)
aEffect type of the marker: M = marker main effect; Ic = marker interaction effect (crossover); Inc = marker interaction effect (non-cross-over); M + Ic = marker main and interaction (crossover) effect; M + Inc = marker main and interaction (non-crossover) effect
Discussion
In this study, a QTL mapping approach was developed that accounts for both multiple marker loci and marker-by-environment interactions simultaneously in the statistical model. For comparison, a Bayesian multi-locus single-environmental analysis, a REML single-locus approach, and REML forward selection analysis were computed. In order to determine which markers showed significant effects, a permutation test was calculated for the REML single-locus and Bayesian single-environmental analysis. In general, this permutation test is used in frequentist approaches and only rarely in Bayesian data analysis. In a permutation test, multiple hypothetical datasets obtained from shuffling the phenotypic observations that could have given rise to the observations are considered. The objective here is to determine how extreme our observed dataset is (in the sense of producing the mapping signal). This means we examine the strength of the mapping signal in the observed dataset compared to the random mapping signals in the shuffled datasets.
Using REML single-locus QTL mapping, each marker locus was considered separately in the analysis. That means that, as the lines were genotyped by 98 SSR markers, 98 analyses had to be performed. All markers with an F value greater than the significance threshold were considered to be significant. In the single-locus analysis, many significant QTLs were detected in the real and in the simulated dataset (Tables 2, 6). This could be due to the consideration of only a single marker point at a time, which complicates the detection of a marker with a significant effect at the exact position on the chromosome. Hence, it can be observed that in several cases not the marker with a non-zero simulated effect itself (i.e., marker 22 in Table 6), but a marker with an effect of zero located near to the true marker on the same chromosome was found instead (i.e., marker 21). As both markers are within an interval of 20 cM, in the field dataset both markers were interpreted to belong to the same QTL. Compared to the REML single-locus mapping, using a REML forward selection approach, fewer markers were found to be significant (Tables 2, 6). Thus, as expected, the forward selection analysis seems to be more powerful for QTL mapping. Since the markers with the most significant effect in previous estimation rounds are included as fixed cofactors in the statistical model of the next estimation cycle, similar to composite interval mapping, the forward selection approach accounts for multiple marker loci in the analysis.
In Bayesian multi-locus analysis using multi-environmental data of the spring barley population, several significant QTLs were found (Table 1). However, some of these QTLs showed only negligible effects (Fig. 2). Remarkably, the observed small peaks were found around larger ones in the same region on the chromosome. One reason for detecting several candidates can be the increased power, in particular for identifying markers with low effects. Also, having multiple coefficients at single marker in the model is likely to improve mixing properties of the MCMC sampler, especially in the case of closely linked loci. Aside from this, we assumed an independent residual structure. According to Piepho (2000), omitting genetic (background) correlations among observations of the same genotype measured in different environments can cause spurious QTL signals. Accounting for this genetic correlation, however, was not possible in our study, as phenotypic observations of the traits were not replicated within each combination of line and environment in the dataset leading to confounded polygenic and residual variation and covariation. Nevertheless, compared to Bayesian single-environmental mapping, a Bayesian approach using multi-environmental data seems to be more stringent (based on our subjective significance criterion) (Table 1). This is in contrast to the simulation study where both, Bayesian single- and multi-environmental mapping yielded comparable results (Table 5). Based on the experimenting with different significance thresholds (around the true threshold value), it seems that the results of Bayesian multi-environmental analysis are not very sensitive to our choice of the threshold value (results not shown). Considering the estimated marker effects, the true marker effect was estimated more accurately using a Bayesian mapping strategy than a REML approach.
In a multi-locus analysis a potential complication is that in the case of a strong correlation between markers it could be difficult to determine which marker is significant. It can be assumed that the higher the number of markers, the stronger this correlation among markers could be. In the present study, 98 SSR markers were used in total. Still, on chromosome 6H there was a gap where no polymorphic markers could be found. Thus, in this research a strong correlation between markers is not probable, but this situation could arise in future studies where the lines are genotyped by hundreds of markers. This problem could be alleviated by collecting a large number of individuals.
Marker interaction effects were detected to a greater extent in Bayesian QTL mapping; whereas, using the REML method, all found marker effects were interpreted as a main effect in the analysis (Tables 5, 6). In the REML forward selection approach of the simulated dataset, all markers having a combined main and interaction effect were found to be a significant marker main effect (Table 6). Markers with an effect in some environments only were not found by REML methods. However, marker main and interaction effects have to be interpreted in different ways using the REML and Bayesian methods. In REML analysis, it is possible to interpret marker main and interaction effects separately. In contrast, in Bayesian implementation an oversaturated model was used, which means that more parameters were actually estimated than would be necessary. This oversaturated model was used because all environments were treated equally in the prior distribution during the model selection process. For each MCMC iteration, the sum of main effect and corresponding environmental effect was calculated. Thus, marker main and interaction effects were not independently identifiable. If the estimated effects in all environments were greater than the significance threshold, this effect was interpreted as a marker main effect; otherwise, there would be an interaction effect. This fact should be considered when comparing the QTL results from REML and Bayesian methods (Table 1).
As Bayesian multi-environmental mapping was computationally demanding, the calculation of a permutation test was not possible, and therefore a significance threshold was derived subjectively for this QTL analysis. This raises the question of whether the threshold is comparable/realistic, or if it is still too low or too high. Additionally, for each QTL locus, the probability of marker effects being higher than the chosen significance threshold was computed over all MCMC-rounds. If our significance threshold was too low, leading to “significant” QTLs that in reality are false-positives, then there would be a high number of markers that showed an effect greater than the threshold in each MCMC round. In this case, the probability of marker effects being higher than the threshold would be equal to 1 for most of the QTLs. In contrast, if the significance threshold was too high, then the markers would have effects greater than the threshold only in some MCMC rounds, so the probability would be much lower than 1. In our study, some QTLs show probabilities of 1, but a number of QTLs also show reduced probabilities in both, real and simulated datasets (Tables 3, 5). Thus, it seems that by applying the chosen significance threshold, the occurrence of false-positive QTLs is minimized. Tests with slightly different significance thresholds showed the chosen threshold to be most appropriate in reducing the number of false-positive QTLs. In the simulation study, with the chosen significance threshold only one false-positive marker with a true effect of zero was detected to be significant, whereas another marker with a true effect of 0.2 was not found due to the small effect size.
Table 3.
Occupancy probabilities P of marker effects being higher than the significance threshold in Bayesian multi-environmental mapping for all traits in the spring barley population
| Ears per m² | Days until heading | Plant height | Thousand grain weight | Grain yield | |||||
|---|---|---|---|---|---|---|---|---|---|
| QTL | P | QTL | P | QTL | P | QTL | P | QTL | P |
| QEar.S42-2H.2 | 1 | QHea.S42-1H.2 | 0.55 | QHei.S42-3H.1 | 0.63−0.8 | QTgw.S42-2H.1 | 0.91 | QYld.S42-1H.2 | 0.83−1 |
| QEar.S42-4H.3 | 0.53−1 | QHea.S42-1H.3 | 1 | QHei.S42-3H.2 | 0.89 | QTgw.S42-2H.2 | 0.63 | QYld.S42-1H.3 | 0.94−1 |
| QHea.S42-2H.1 | 0.98−1 | QHei.S42-3H.3 | 0.49−1 | QTgw.S42-4H.1 | 0.99 | QYld.S42-3H.1 | 0.66 | ||
| QHea.S42-2H.2 | 0.91 | QYld.S42-3H.2 | 1 | ||||||
| QHea.S42-2H.3 | 0.52 | QYld.S42-3H.3 | 0.98 | ||||||
| QHea.S42-3H.3 | 1 | ||||||||
| QHea.S42-4H.1 | 0.63 | ||||||||
| QHea.S42-5H.1 | 1 | ||||||||
For each QTL, the range of probabilities over all environments and significant markers is given
Accounting for missing marker data in the analysis is handled differently in REML and Bayesian mapping. In REML analysis, each observation with a missing marker value is omitted from the dataset. Thus, the amount of phenotypic information is reduced due to missing marker data. Considering the REML forward selection approach where several markers are accounted for simultaneously as cofactors in the statistical model, the number of phenotypic observations omitted from the dataset is increased due to the larger probability that missing marker data occur in one or more of the markers. In contrast, in a Bayesian analysis, missing markers are imputed according to their posterior distribution (Hoti and Sillanpää 2006; Yu and Schaid 2007). There, the distance between the missing locus and the flanking markers is calculated based on the recombination frequency and the missing marker is more frequently assigned the genotypic value of the flanking marker with the smallest distance.
Compared to QTL mapping studies using other barley populations and different molecular markers, several QTLs could be verified in our spring barley population (Table 4). Except for the trait “ears per m²,” for all other traits several QTLs detected in the spring barley population could be mapped in the same region on the chromosome by other authors. A detailed comparison of QTL positions detected here and in other barley QTL mapping studies is given in the Supplementary Material.
Table 4.
Detected QTLs by REML and Bayesian analyses in the spring barley population compared to QTL mapping studies using other barley populations and different molecular markers
| QTLa | Chr.b | Pos.c | Bind | Candidate gene | Literature |
|---|---|---|---|---|---|
| Days until heading (h² = 0.77) | |||||
| QHea.S42-1H.1 | 1H | 39 | 6 | Thomas et al. (1995) (DH-lines) | |
| QHea.S42-1H.2 | 1H | 115 | 13 | ||
| QHea.S42-1H.3 | 1H | 144 | 13 | Vrn-H3 | Laurie et al. (1995) (DH-lines), Sameri and Komatsuda (2004) (RILs), Sameri et al. (2006) (RILs; mapped in Bin class 14) |
| QHea.S42-2H.1 | 2H | 42 | 4 (4) | Ppd-H1 | Laurie et al. (1995) (DH-lines), Sameri and Komatsuda (2004) (RILs), Li et al. (2005) (BC3DH), Emebiri and Moody (2006) (DH-lines), Sameri et al. (2006) (RILs) |
| QHea.S42-2H.2 | 2H | 80–86 | 7–8 | Pillen et al. (2003) (BC2F2) | |
| QHea.S42-2H.3 | 2H | 146 | 13 | Pillen et al. (2003) (BC2F2) | |
| QHea.S42-3H.1 | 3H | 25–30 | 3 (3) | ||
| QHea.S42-3H.2 | 3H | 130 | 10 | ||
| QHea.S42-3H.3 | 3H | 155–165 | 13–14 (15) | Denso | Barua et al. (1993) (NILs and DH-lines), Laurie et al. (1995) (DH-lines), Thomas et al. (1995) (DH-lines), Tinker et al. (1996) (DH-lines) |
| QHea.S42-4H.1 | 4H | 130 | 10 | ||
| QHea.S42-4H.2 | 4H | 180–190 | 12–13 (12) | Vrn-H2 | Laurie et al. (1995) (DH-lines), Pillen et al. (2003) (BC2F2) |
| QHea.S42-5H.1 | 5H | 43 | 5 | Thomas et al. (1995) (DH-lines), Pillen et al. (2003) (BC2F2) | |
| QHea.S42-5H.2 | 5H | 85 | 8 | Pillen et al. (2003) (BC2F2) | |
| QHea.S42-6H.1 | 6H | 107 | 7 | ||
| QHea.S42-7H.1 | 7H | 146 | 8 | eps7HS | Pillen et al. (2003) (BC2F2), Emebiri and Moody (2006) (DH-lines) |
| Plant height (h² = 0.76) | |||||
| QHei.S42-2H.1 | 2H | 17–42 | 2–4 (3–4) | Ppd-H1 | Laurie et al. (1994) (DH-lines) |
| QHei.S42-2H.2 | 2H | 80–92 | 7–8 | sdw3 | Gottwald et al. (2004) (F4-progenies), Kraakman et al. (2006) (cultivars) |
| QHei.S42-3H.1 | 3H | 25–70 | 3–6 (3) | Sameri et al. (2006) (RILs) | |
| QHei.S42-3H.2 | 3H | 130 | 10 | Yin et al. (1999) (RILs; mapped in Bin class 11) | |
| QHei.S42-3H.3 | 3H | 155–190 | 13–16 (15) | Denso | Thomas et al. (1995) (DH-lines), Bezant et al. (1996) (DH-lines) |
| QHei.S42-4H.1 | 4H | 125-132 | 9–10 | ||
| QHei.S42-4H.2 | 4H | 170–190 | 12–13 (11–12) | ||
| QHei.S42-5H.1 | 5H | 43 | 5 | ||
| QHei.S42-7H.1 | 7H | 27 | 2 | ||
| Grain yield (h² = 0.70) | |||||
| QYld.S42-1H.1 | 1H | 52 | 6 | Li et al. (2005) (BC3DH; mapped in Bin class 8) | |
| QYld.S42-1H.2 | 1H | 65–75 | 7–8 | Thomas et al. (1995) (DH-lines), Tinker et al. (1996) (DH-lines) | |
| QYld.S42-1H.3 | 1H | 144 | 13 | ||
| QYld.S42-2H.1 | 2H | 107–122 | 9–10 | Yin et al. (1999) (RILs) | |
| QYld.S42-3H.1 | 3H | 25–70 | 3–6 (3) | Thomas et al. (1995) (DH-lines), Pillen et al. (2003) (BC2F2), Kraakman et al. (2004) (cultivars) | |
| QYld.S42-3H.2 | 3H | 130 | 10 | Yin et al. (1999) (RILs) | |
| QYld.S42-3H.3 | 3H | 155–165 | 13–14 (15) | Denso | Thomas et al. (1995) (DH-lines), Tinker et al. (1996) (DH-lines), Li et al. (2005) (BC3DH) |
| QYld.S42-3H.4 | 3H | 190 | 16 | ||
| QYld.S42-5H.1 | 5H | 43 | 5 | Pillen et al. (2003) (BC2F2) | |
| QYld.S42-5H.2 | 5H | 126 | 10 | Kraakman et al. (2004) (cultivars; mapped in Bin class 12-13) | |
| QYld.S42-7H.1 | 7H | 146 | 8 | Pillen et al. (2003) (BC2F2) | |
| Thousand grain weight (h² = 0.54) | |||||
| QTgw.S42-1H.1 | 1H | 65 | 7 | ||
| QTgw.S42-1H.2 | 1H | 144 | 13 | ||
| QTgw.S42-2H.1 | 2H | 86 | 8 | Pillen et al. (2003) (BC2F2), Sameri and Komatsuda (2007) (RILs; mapped in Bin class 10) | |
| QTgw.S42-2H.2 | 2H | 107–122 | 9–10 | Li et al. (2005) (BC3DH; mapped in Bin class 15) | |
| QTgw.S42-3H.1 | 3H | 110 | 9 (8) | ||
| QTgw.S42-3H.2 | 3H | 155–175 | 13–15 (15) | ||
| QTgw.S42-4H.1 | 4H | 125–132 | 9–10 | ||
| QTgw.S42-4H.2 | 4H | 180–190 | 12–13 (12) | ||
| QTgw.S42-6H.1 | 6H | 96 | 5 | ||
| QTgw.S42-6H.2 | 6H | 135 | 10 | ||
| QTgw.S42-7H.1 | 7H | 146–166 | 8–11 | Pillen et al. (2003) (BC2F2) | |
| Ears per m² (h² = 0.21) | |||||
| QEar.S42-1H.1 | 1H | 144 | 13 | ||
| QEar.S42-2H.1 | 2H | 17 | 2 (3) | ||
| QEar.S42-2H.2 | 2H | 67–92 | 6–8 | ||
| QEar.S42-3H.1 | 3H | 65–70 | 5–6 | ||
| QEar.S42-3H.2 | 3H | 155–165 | 13–14 (15) | ||
| QEar.S42-4H.1 | 4H | 21–25 | 3 | ||
| QEar.S42-4H.2 | 4H | 125–132 | 9–10 | ||
| QEar.S42-4H.3 | 4H | 170–190 | 12–13 (11–12) | ||
| QEar.S42-5H.1 | 5H | 0 | 2 | ||
| QEar.S42-5H.2 | 5H | 43 | 5 | ||
| QEar.S42-6H.1 | 6H | 135 | 10 | ||
In brackets, the population used for QTL mapping is displayed (BC backcross, DH doubled haploid, NILs near-isogenic lines, RILs recombinant inbred lines). A detailed description of the QTLs is given in the Supplementary Material
aNames of the QTLs consisting of the qualifier “Q”, the trait abbreviation, the determined population, the mapped chromosome and a QTL number to distinguish between QTLs on the same chromosome. Markers with a distance ≤20 cM were considered to be a single QTL
bChromosomal location of the SSR markers
cPosition of the SSR markers on the chromosome in centiMorgan (cM) (von Korff et al. 2004). If several markers were significant, the cM range is given
dBin marker class following Kleinhofs and Graner (2001) and Costa et al. (2001). In the case of several significant markers, the Bin class range is displayed. An additional number in brackets gives the Bin class following Marcel et al. (2007). As the Bin class of Marcel et al. (2007) was available only for some markers, in the case of several significant markers belonging to the same QTL, these markers are given in italics
QTLs that were detected in three or four of our QTL mapping strategies and are not yet described in the literature might be “new” QTLs. These are the QTLs QEar.S42-4H.3 (“ears per m²”), QHea.S42-3H.2 (“days until heading”), QHei.S42-4H.2 (“plant height”), and the QTLs QTgw.S42-3H.2, QTgw.S42-4H.1, and QTgw.S42-4H.2 for the trait “thousand grain weight.” In general, the power to detect a significant QTL with several statistical analyses is higher with increasing heritability of the regarded trait.
In conclusion, Bayesian multi-locus multi-environmental QTL mapping seems to be a valuable strategy accounting for both multiple loci and marker-by-environment interactions. This QTL analysis is suitable, especially if the lines are cultivated in multi-environmental field trials.
In comparing REML and Bayesian QTL mapping strategies, a Bayesian analysis can be computationally demanding. In this study, REML analysis took about 20 min, whereas Bayesian analysis needed about 33 h for the same trait in the spring barley population on a Pentium IV 2.0 GHz processor. So, when performing Bayesian QTL mapping, it is important to use efficiently programmed MCMC estimation methods. On the other hand, in a Bayesian framework all marker loci were considered jointly in a single analysis which resulted in a valuable method. Thus, the user has to decide if the gain of Bayesian QTL mapping over other methods justifies the computional burden.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-009-1021-6) contains supplementary material, which is available to authorized users.
References
- Barua UM, Chalmers KJ, Hackett CA, Thomas WTB, Powell W, Waugh R (1993) Identification of RAPD markers linked to a Rhynchosporium secalis resistance locus in barley using near-isogenic lines and bulked segregant analysis. Heredity 71:177–184 [DOI] [PubMed]
- Bauer AM, Reetz TC, Léon J (2006) Estimation of breeding values of inbred lines using best linear unbiased prediction (BLUP) and genetic similarities. Crop Sci 46:2685–2691 [DOI]
- Bauer AM, Reetz TC, Léon J (2008) Predicting breeding values of spring barley accessions by using the singular value decomposition of genetic similarities. Plant Breed 127:274–278 [DOI]
- Bezant J, Laurie D, Pratchett N, Chojecki J, Kearsey M (1996) Marker regression mapping of QTL controlling flowering time and plant height in a spring barley (Hordeum vulgare L.) cross. Heredity 77:64–73 [DOI]
- Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, van Eeuwijk FA (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177:1801–1813 [DOI] [PMC free article] [PubMed]
- Broman KW, Speed TP (2002) A model selection approach for identification of quantitative trait loci in experimental crosses. J R Stat Soc B 64(641–656):737–775 (with discussion)
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed]
- Costa JM, Corey A, Hayes PM, Jobet C, Kleinhofs A, Kopisch-Obusch A, Kramer SF, Kudrna D, Li M, Riera-Lizarazu O, Sato K, Szucs P, Toojinda T, Vales MJ, Wolfe RI (2001) Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor Appl Genet 103:415–424 [DOI]
- Crossa J, Vargas M, van Eeuwijk FA, Jiang C, Edmeades GO, Hoisington D (1999) Interpreting genotype × environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet 99:611–625 [DOI] [PubMed]
- Emebiri LC, Moody DB (2006) Heritable basis for some genotype-environment stability statistics: inferences from QTL analysis of heading date in two-rowed barley. Field Crops Res 96:243–251 [DOI]
- Geman S, Geman D (1984) Stochastic relaxation, Gibbs distributions and Bayesian restoration of images. IEEE Trans Pattern Anal Mach Intell 6:721–741 [DOI] [PubMed]
- Gottwald S, Börner A, Stein N, Sasaki T, Graner A (2004) The gibberellic-acid insensitive dwarfing gene sdw3 of barley is located on chromosome 2HS in a region that shows high colinearity with rice chromosome 7L. Mol Gen Genomics 271:426–436 [DOI] [PubMed]
- Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324 [DOI] [PubMed]
- Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57:97–109 [DOI]
- Hoti F, Sillanpää MJ (2006) Bayesian mapping of genotype × expression interactions in quantitative and qualitative traits. Heredity 97:4–18 [DOI] [PubMed]
- Jansen RC, van Ooijen JW, Stam P, Lister C, Dean C (1995) Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor Appl Genet 91:33–37 [DOI] [PubMed]
- Jiang C, Zeng Z-B (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127 [DOI] [PMC free article] [PubMed]
- Jiang C, Edmeades GO, Armstead I, Lafitte HR, Hayward MD, Hoisington D (1999) Genetic analysis of adaptation differences between highland and lowland tropical maize using molecular markers. Theor Appl Genet 99:1106–1119 [DOI] [PubMed]
- Kang MS, Gauch HG Jr (1996) Genotype-by-environment interaction. CRC Press, Boca Raton, FL
- Kass RE, Carlin BP, Gelman A, Neal RM (1998) Markov Chain Monte Carlo in practice: a roundtable discussion. Am Stat 52:93–100 [DOI]
- Kilpikari R, Sillanpää MJ (2003) Bayesian analysis of multilocus association in quantitative and qualitative traits. Genet Epidemiol 25:122–135 [DOI] [PubMed]
- Kleinhofs A, Graner A (2001) An integrated map of the barley genome. Kluwer, Dordrecht, The Netherlands, pp 187–199
- Korol AB, Ronin YI, Nevo E (1998) Approximate analysis of QTL-environment interaction with no limits on the number of environments. Genetics 148:2015–2028 [DOI] [PMC free article] [PubMed]
- Kraakman ATW, Niks RE, van den Berg PMMM, Stam P, van Eeuwijk FA (2004) Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168:435–446 [DOI] [PMC free article] [PubMed]
- Kraakman ATW, Martínez F, Mussiraliev B, van Eeuwijk FA, Niks RE (2006) Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Mol Breed 17:41–58 [DOI]
- Laurie DA, Pratchett N, Bezant JH, Snape JW (1994) Genetic analysis of a photoperiod-response gene on the short arm of chromosome 2 (2H) of Hordeum vulgare (barley). Heredity 72:619–627 [DOI]
- Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome 38:575–585 [DOI] [PubMed]
- Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS (2005) Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley. Theor Appl Genet 110:356–363 [DOI] [PubMed]
- Marcel TC, Varshney RK, Barbieri M, Jafary H, de Kock MJD, Graner A, Niks RE (2007) A high-density consensus map of barley to compare the distribution of QTLs of partial resistance to Puccinia hordei and of defense gene homologues. Theor Appl Genet 114:487–500 [DOI] [PubMed]
- Matlab (2007) High-performance numeric computation and visualization software, vol 7. The Math Works Inc, Natick, Mass, USA
- Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genomic value using genome-wide dense marker map. Genetics 157:1819–1829 [DOI] [PMC free article] [PubMed]
- Piepho H-P (2000) A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156:2043–2050 [DOI] [PMC free article] [PubMed]
- Pillen K, Zacharias A, Léon J (2003) Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theor Appl Genet 107:340–352 [DOI] [PubMed]
- Sameri M, Komatsuda T (2004) Identification of quantitative trait loci (QTLs) controlling heading time in the population generated from a cross between oriental and occidental barley cultivars (Hordeum vulgare L.). Breed Sci 54:327–332 [DOI]
- Sameri M, Komatsuda T (2007) Localization of quantitative trait loci for yield components in a cross oriental × occidental barley cultivar (Hordeum vulgare L.). JARQ 41:195–199
- Sameri M, Takeda K, Komatsuda T (2006) Quantitative trait loci controlling agronomic traits in recombinant inbred lines from a cross of oriental- and occidental-type barley cultivars. Breed Sci 56:243–252 [DOI]
- SAS Institute (2004) The SAS system for Windows, Release 9.1. SAS Inst, Cary, NC, USA
- Sillanpää MJ, Corander J (2002) Model choice in gene mapping: what and why. Trends Genet 18:301–307 [DOI] [PubMed]
- Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203 [DOI] [PubMed]
- Thomas WTB, Powell W, Waugh R, Chalmers KJ, Barua UM, Jack P, Lea V, Forster BP, Swanston JS, Ellis RP, Hanson PR, Lance RCM (1995) Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). Theor Appl Genet 91:1037–1047 [DOI] [PubMed]
- Tinker NA, Mather DE (1995) Methods for QTL analysis with progeny replicated in multiple environments. J Quant Trait Loci 1
- Tinker NA, Mather DE, Rossnagel BG, Kasha KJ, Kleinhofs A, Hayes PM, Falk DE, Ferguson T, Shugar LP, Legge WG, Irvine RB, Choo TM, Briggs KG, Ullrich SE, Franckowiak JD, Blake TK, Graf RJ, Dofing SM, Saghai Maroof MA, Scoles GJ, Hoffmann D, Dahleen LS, Kilian A, Chen F, Biyashev RM, Kudrna DA, Steffenson BJ (1996) Regions of the genome that affect agronomic performance in two-row barley. Crop Sci 36:1053–1062
- Verbyla AP, Eckermann PJ, Thompson R, Cullis BR (2003) The analysis of quantitative trait loci in multi-environmental trias using a multiplicative mixed model. Aust J Agric Res 54:1395–1408 [DOI]
- von Korff M, Wang H, Léon J, Pillen K (2004) Development of candidate introgression lines using an exotic barley accession (H. vulgare ssp. spontaneum) as donor. Theor Appl Genet 109:1736–1745 [DOI] [PubMed]
- von Korff M, Wang H, Léon J, Pillen K (2006) AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 112:1221–1231 [DOI] [PubMed]
- Xu S (2003) Estimating polygenic effects using markers of the entire genome. Genetics 163:789–801 [DOI] [PMC free article] [PubMed]
- Yandell BS, Mehta T, Banerjee S, Shriner D, Venkataraman R, Moon JY, Neely WW, Wu H, von Smith R, Yi N (2007) R/qtlbim: QTL with Bayesian interval mapping in experimental crosses. Bioinformatics 23:641–643 [DOI] [PMC free article] [PubMed]
- Yin X, Stam P, Johan Dourleijn C, Kropff MJ (1999) AFLP mapping of quantitative trait loci for yield-determining physiological characters in spring barley. Theor Appl Genet 99:244–253 [DOI]
- Yu Z, Schaid DJ (2007) Methods to impute missing genotypes for population data. Hum Genet 122:495–504 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.