Abstract
Micro-RNAs (miRNAs) are short, single-stranded, noncoding RNAs that are involved in the regulation of protein-coding genes at the level of messenger RNA (mRNA). They are involved in the regulation of numerous traits, including developmental timing, apoptosis, immune function, and neuronal development. To better understand how the expression of the miRNAs themselves is regulated, we looked for miRNA expression differences among four mouse inbred strains, A/J, BALB/cJ, C57BL/6J, and DBA/2J, in one tissue, the hippocampus. A total of 166 miRNA RT-PCR assays were used to screen RNA pools for each strain. Twenty miRNA species that were markedly different between strains were further investigated using eight individual samples per strain, and 11 miRNAs showed significant differences across strains (p < 0.05). This is the first observation of miRNA expression differences across inbred mice strains. We conducted an in silico correlation analysis of the expression of these differentially expressed miRNAs with phenotype data and mRNA expression to better characterise the effects of these miRNAs on both phenotype and the regulation of mRNA expression. This approach has allowed us to nominate miRNAs that have potential roles in anxiety, exploration, and learning and memory.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-008-9116-y) contains supplementary material, which is available to authorized users.
Keywords: False Discovery Rate, miRNA Expression, miRNA Binding Site, DiGeorge Syndrome, Reverse Transcription Primer
Introduction
Much research has focused on a class of functional untranslated RNA transcripts referred to as microRNAs (miRNAs). MiRNAs play a pivotal role in the expression of potentially one third of all messenger RNAs (mRNAs) (Bartel 2004; Griffiths-Jones et al. 2006). Mature miRNAs cause post-transcriptional gene silencing by base pairing, as part of a larger molecular complex, with target mRNAs, which in turn can lead to mRNA cleavage or translational repression (Kloosterman et al. 2006; Yekta et al. 2004). A single miRNA can have target sites on hundreds of different mRNAs (Bartel 2004); therefore, changing the expression of a single miRNA can have wide-reaching effects on gene expression.
While the full potential of the influence of miRNAs is not yet known, they have already been shown to be involved in the regulation of numerous processes, including developmental timing, apoptosis, immune function, and neuronal development (Chen et al. 2004; Rodriguez et al. 2007; Thai et al. 2007; Zhang et al. 2007b). In addition, miRNA function and biogenesis have been implicated in disease, including Tourette’s syndrome, DiGeorge syndrome, cancer, and stress-related heart growth (Ableson et al. 2005; Gregory and Shiekhattar 2005; van Rooij et al. 2007). The involvement of miRNAs in these phenotypes and disease states most likely occurs through the transcriptional regulation of associated mRNAs.
Little is known about the regulation of miRNA expression, although it is commonly thought that transcriptional regulation is the main control mechanism (Johnson et al. 2003). Regulation can occur at other stages of miRNA biogenesis (Ambros et al. 2003), in particular, maturation (Ambros et al. 2003). It is thus important to distinguish between mature miRNA and pre-miRNA expression levels.
We investigated mature miRNA abundance in hippocampus of four common inbred strains, A/J, BALB/cJ, C57BL/6J, and DBA/2J, using 166 miRNA TaqMan® real-time PCR assays to see if miRNA expression is polymorphic, at least in hippocampus. We then correlated the expression of the differentially expressed miRNAs with phenotypes from an extensive phenotype database, as well as mRNA expression for those genes with predicted miRNA pairing sites. This analysis allowed us to characterise the effects of miRNA expression on phenotypes and gene expression to gain a better understanding of the effects of these miRNAs.
Methods
Animals
Hippocampal tissue was collected from four strains of inbred mice, A/J, BALB/cJ, C57BL/6J, and DBA/2J (4 males and 4 females for each strain). All animals were between 100 and 110 days old. After cervical dislocation bilateral hippocampi were dissected in their entirety within 2 min from the time of death, and any connecting tissue was trimmed. The hippocampi were immediately snap-frozen on dry ice and stored at −80°C. All housing and experimental procedures were performed in accordance with the UK Home Office’s Animals (Scientific Procedures) Act 1986 under License PPL No. 70/5113.
miRNA isolation
To obtain reliable levels of miRNA, total RNA was isolated from hippocampus using an miRNA isolation kit (Ambion, Austin, TX, USA). The total RNA concentration was determined using a nanodrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA).
Screening strategy
For each of the four strains investigated we made one male pool and one female pool, each consisting of four animals. These pools were used to screen the 166 assays investigated in this study. These 166 assays comprised the entire list of mouse TaqMan® microRNA assays available from the complete list of multiplexed reverse transcription reactions. Those miRNAs that had at least a 1.5-fold difference in gene expression across strains were followed up in the individual samples.
Reverse transcription reactions
TaqMan microRNA Assays (Applied Biosystems, Foster City, CA, USA) were used in this study. The reverse transcription (RT) reactions were conducted using eight different sets of miRNA RT primers (Supplementary Table 1). The RT primer pools were compiled by selecting all mouse TaqMan microRNA assays available from the complete list of Multiplex TaqMan microRNA assays available for various species. For the pooled RNAs we used pooled RT primers for each of these eight miRNA RT primer sets. For the assays investigated using RNA from individual animals, we conducted the RT reactions using unpooled RT primers.
RT primer pools were concentrated using a Speed Vac 110 (Savant and Refrigerated Vapor Trap RVT) to a final concentration of 62.5 nM for each primer. The RT reactions using pooled RT primers were conducted using the following modified protocol: 4 μl of 62.5 nM pooled RT primers, 0.4 μl of 25 mM dNTPs, 4 μl of Multiscribe Reverse Transcriptase (50 U/μl), 2 μl of 10 × RT buffer, 0.25 μl of AB RNase Inhibitor (20 U/μl), 5.35 μl of nuclease-free water, and 4 μl of miRNA sample (20 ng per reaction). The reaction was then incubated at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min in a PTC-100 thermal cycler (MJ Research, Bio-Rad, Hercules, CA, USA). The RT reactions using individual RT primers were conducted using the standard TaqMan microRNA RT protocol (http://www.appliedbiosystems.com, document No. 4364031).
TaqMan real-time polymerase chain reactions (RT-PCR)
Mature miRNA expression was quantified using TaqMan RT-PCR. Reactions were conducted on the RT products from the pooled RNA samples (a total of 166 assays) and using the RT products from individual samples (a total of 20 assays). The RT-PCR reactions were conducted in triplicate using 0.5 μl of 20 × PCR Probe/Primer Mix, 1.5 μl of product from the RT reaction (diluted 1:10), 5 μl of 2 × TaqMan Master Mix (without uracil-N-glycosylase), and 3 μl of nuclease-free water. A sample minus RT enzyme was used as a negative control for the RT-PCR reactions. All of the negative controls used failed to reach threshold by 45 cycles.
Reactions were run on a 7900HT Fast Real-Time PCR System (Applied Biosystems) in 384-well format. RNU19 was used as the only control for the assays run on the pooled samples, while RNU19, RNU6B, and miR-16 were used as controls for each of the test assays investigated in the individual samples. These assays were chosen as controls because their expression did not differ across strains and they had low variability within strain (unpublished data).
TaqMan RT-PCR statistical analysis
Relative expression was calculated using the standard 2−Δ(Ct) relative expression method in Microsoft Excel. Relative expression levels were normalized to RNU19 for the pooled samples. The relative expression of the individual samples was normalized to the geometric mean of RNU6B, RNU19, and miR-16. Statistica v6.1 (StatSoft, Inc., Tulsa, OK, USA) was used to conduct a two-way analysis of variance (ANOVA) to detect differences in miRNA expression across strains and sex. Fisher’s least significant difference post-hoc analysis test was conducted on the statistically significant main effects and interactions. Bonferonni corrections and false discovery rate (FDR) control (Benjamini and Hochberg 1995) were used to account for multiple testing.
Correlations of behaviour and miRNA expression
Correlations were conducted between the 252 phenotypic measures from the Mouse Phenome Database (http://www.jax.org/phenome) that had data available for the four investigated strains, and the relative expression of those miRNAs that were differentially expressed across strains. Correlations were conducted using Statistica v6.1. FDR control (Benjamini and Hochberg 1995) was used to account for multiple testing.
Correlations of mRNA and miRNA expression
Lists of genes with MIRANDA-predicted miRNA sites (Betel et al. 2008) were downloaded from an online miRNA database, miRBase (http://microrna.sanger.ac.uk/). Hippocampal mRNA expression for the inbred mice was obtained from the data series GSE5055 (Nadler et al. 2006) of the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo/). Data were available for the A/J, C57BL/6J, and DBA/2J mice, but we had to use BALB/cByJ to approximate BALB/cJ expression. Correlations between miRNA and mRNA expression for those mRNAs with MIRANDA-predicted miRNA target sites were conducted using the open source statistical program R (Ihaka and Gentleman 1996). FDR control (Benjamini and Hochberg 1995) was used to account for multiple testing. Gene lists, consisting of all genes whose expressions were significantly correlated with miRNA expression (p < 0.05, uncorrected), were generated for each miRNA and were used to generate summaries of gene function using the KEGG pathway module in WebGestalt (Zhang et al. 2005).
Results
Results of screening
Of the 166 miRNAs we investigated, 130 miRNAs had detectable levels of mature miRNA expression in the hippocampus (Ct < 40). Expression levels varied across miRNAs, with 18 highly expressed (Ct ≤ 28) and 24 with relatively low expression (Ct ≥ 35).
Twenty of the assays investigated in the pooled RNAs had at least 1.5-fold differences in expression across strains. These 20 assays were further investigated using the RNA from individual animals. A two-way ANOVA for strain, sex, and interactions of strain and sex was conducted for each of these 20 assays (Table 1).
Table 1.
Summary of two-way ANOVA for strain and sex
| miRNA | Strain | Sex | Strain × sex | |||||
|---|---|---|---|---|---|---|---|---|
| F(3, 24) | p | Bonf. p | FDR q | F(1, 24) | p | F(3, 24) | p | |
| miR-203 | 121 | 1e-14 | 2.5E-13 | 2.5E-13 | 3.8 | 0.06 | 10.7 | 1e-4 |
| miR-451 | 34 | 8e-9 | 1.7E-07 | 8.4E-08 | 0.0 | 0.98 | 0.3 | 0.80 |
| miR-378 | 12 | 6e-5 | 0.001 | 4.0E-04 | 0.8 | 0.39 | 0.2 | 0.90 |
| miR-195 | 5.4 | 0.005 | 0.113 | 0.028 | 0.0 | 0.99 | 0.7 | 0.56 |
| miR-34a | 5.0 | 0.008 | 0.163 | 0.033 | 0.1 | 0.76 | 0.3 | 0.85 |
| miR-34c | 4.8 | 0.009 | 0.185 | 0.031 | 0.1 | 0.81 | 1.4 | 0.27 |
| miR-15b | 4.6 | 0.01 | 0.230 | 0.033 | 0.4 | 0.51 | 0.3 | 0.86 |
| miR-323 | 4.5 | 0.01 | 0.242 | 0.030 | 0.9 | 0.36 | 0.9 | 0.47 |
| miR-301a | 3.4 | 0.03 | 0.661 | 0.073 | 1.0 | 0.32 | 0.8 | 0.53 |
| miR-212 | 3.2 | 0.04 | 0.793 | 0.079 | 5.7 | 0.03 | 0.7 | 0.58 |
| miR-31 | 3.1 | 0.05 | 0.924 | 0.083 | 0.0 | 0.84 | 0.9 | 0.47 |
| miR-7a | 2.2 | 0.12 | 2.378 | 0.208 | 0.0 | 0.97 | 0.5 | 0.71 |
| miR-139 | 2.1 | 0.12 | 2.473 | 0.190 | 0.4 | 0.55 | 0.5 | 0.67 |
| miR-30e | 1.6 | 0.22 | 4.412 | 0.315 | 0.0 | 0.92 | 1.1 | 0.37 |
| miR-187 | 1.5 | 0.23 | 4.628 | 0.309 | 1.7 | 0.21 | 0.2 | 0.90 |
| miR-150 | 1.2 | 0.34 | 6.817 | 0.426 | 1.9 | 0.18 | 0.6 | 0.64 |
| miR-425 | 0.8 | 0.49 | 9.848 | 0.579 | 0.3 | 0.59 | 0.6 | 0.62 |
| miR-26a | 0.7 | 0.60 | 11.79 | 0.655 | 1.0 | 0.34 | 0.8 | 0.53 |
| let-7g | 0.5 | 0.71 | 14.12 | 0.743 | 0.5 | 0.47 | 0.4 | 0.77 |
| miR-9 | 0.1 | 0.96 | 19.20 | 0.960 | 0.2 | 0.68 | 0.3 | 0.84 |
Results of the two-way ANOVA for strain and sex conducted on the 20 miRNA RT-PCR TaqMan assays that were investigated in individual animals. The Bonferonni corrected p values and q values are shown for the main effect of strain. The underlines in the p-value, Bonferonni corrected p-value, and FDR q-value columns represent the thresholds for these measurements; thus, any assay above this threshold is deemed significant
F = 2-way ANOVA; p = p value; Bonf. p = Bonferonni corrected p value; FDR q = q value of false discovery rate
Strain differences
The strain means and chromosomal locations of the 20 miRNA species investigated in individual samples are given in Table 2. Eleven of these miRNAs had nominally significant strain differences (p < 0.05) (Fig. 1). All 11 of these nominally significant differences survived multiple-testing corrections by controlling the FDR (Benjamini and Hochberg 1995) to q < 0.1, while eight survived at an FDR q < 0.05. Three of these assays survived the more stringent Bonferonni correction (summarized in Table 1).
Table 2.
Means of relative expression across strain
| Relative expression by strain (mean) | ||||||
|---|---|---|---|---|---|---|
| Assay | Chr (X Mb) | Set | A/J | BALB/cJ | C57/6J | DBA/2J |
| miR-15b | 3 (69 Mb) | 1 | 1.2 | 0.9 | 0.9 | 0.7 |
| miR-34a | 4 (150 Mb) | 2 | 0.7 | 0.9 | 0.5 | 0.6 |
| miR-7a | 13 (58 Mb) | 3 | 9.1 | 7.3 | 6.2 | 6.3 |
| let-7g | 9 (106 Mb) | 3 | 29.7 | 24.2 | 26.2 | 27.6 |
| miR-195 | 11 (70 Mb) | 3 | 4.7 | 6.2 | 5.4 | 5.3 |
| miR-26a | 9 (119 Mb)a | 4B | 170 | 190 | 170 | 160 |
| miR-34c | 9 (51 Mb) | 4B | 1.5 | 2.1 | 1.2 | 0.7 |
| miR-150 | 7 (52 Mb) | 4B | 10.5 | 11.8 | 10.2 | 8.9 |
| miR-139 | 7 (109Mb) | 5 | 4.4 | 4.7 | 3.4 | 3.3 |
| miR-301a | 11 (87 Mb) | 5 | 3.0 | 3.1 | 3.1 | 2.1 |
| miR-378 | 18 (62 Mb) | 5 | 0.6 | 0.9 | 0.4 | 0.4 |
| miR-425 | 9 (108 Mb) | 5 | 0.2 | 0.2 | 0.2 | 0.3 |
| miR-9 | 3 (88 Mb)a | 7A | 160 | 160 | 160 | 150 |
| miR-30e | 4 (120 Mb) | 7A | 4.6 | 5.4 | 4.5 | 4.5 |
| miR-31 | 4 (89 Mb) | 7A | 0.8 | 0.8 | 0.8 | 0.6 |
| miR-187 | 18 (25 Mb) | 7A | 3.4 | 3.2 | 2.4 | 2.7 |
| miR-203 | 12 (113 Mb) | 7B | 0.5 | 0.1 | 0.1 | 0.1 |
| miR-451 | 11 (78 Mb) | 7B | 2.3 | 4.4 | 1.1 | 0.9 |
| miR-212 | 11 (75 Mb) | 8 | 3.1 | 3.2 | 2.7 | 1.9 |
| miR-323 | 12 (111 Mb) | 8 | 0.8 | 1.0 | 0.7 | 0.6 |
Mean relative expression across strain for the 20 miRNAs assayed in individual animals (n = 8 for all strains). Expression was normalised to the geometric mean of RNU6B, RNU19, and miR-16
aThere is more than one chromosomal location for the given miRNA: miR-26a at 9 (119 Mb) and 10 (126 Mb) and miR-9 at 3 (88 Mb), 7 (87 Mb), and 13 (84 Mb)
Fig. 1.
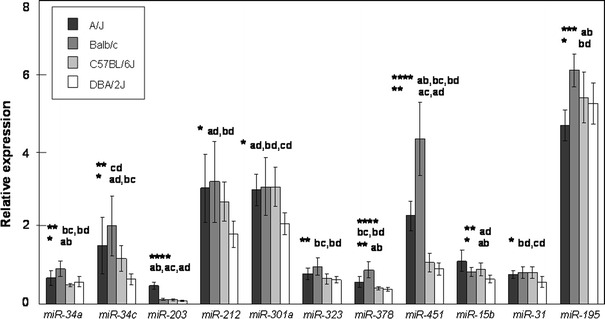
Relative expression for the 11 miRNA assays with a significant main effect of strain. Error bars: 95% confidence intervals. Significant differences (Fisher LSD post-hoc test) are represented by the first two letters of the name of the differing strains (e.g., ab for A/J vs. BALB/cJ). * p < 0.05, ** p < 0.005, *** p < 0.0005, **** p < 0.00005
Sex differences
Only miR-212 expression had a nominally significant main effect of sex (p = 0.03), which did not survive either the FDR or Bonferonni multiple-testing correction approaches. Females had higher miR-212 expression than the males.
Strain and sex interactions
Only miR-203 expression had a significant interaction between strain and sex (Table 1) that survived both the Bonferonni (p < 0.05) and FDR (q < 0.05) multiple-testing approaches. The female and male A/J animals significantly differed from the females and the males of the other three strains, as well as differed from each other (Fisher LSD test, p < 0.00005) (Fig. 2).
Fig. 2.
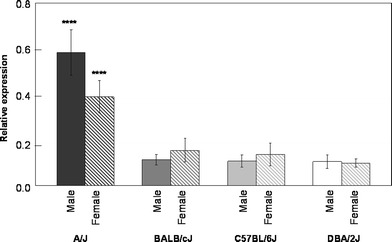
Relative expression of miR-203 has a significant interaction of strain and sex ([F(3,24) = 10.7, p = 1e-4], q < 0.1). Error bars depict 95% confidence intervals. Fisher LSD post-hoc tests indicate that the A/J females and A/J males were both significantly different from all the other groups (**** p < 0.00005)
Correlations of miRNA expression with phenotype
There were 144 correlations of miRNA relative expression and phenotype that were nominally significant (p < 0.05) (these data are summarized in Supplementary Table 2). Of these 144 correlations only two survived a low-stringency FDR multiple-testing correction (q < 0.20): miR-34c expression and total white blood cell count (r = −0.999, p = 6e-4), and miR-301a expression and percentage of monocytes in the total number of leukocytes (r = −0.999, p = 6e-4).
Table 3 summarizes the number of nominally significant correlations and the phenotypic categories (those with at least three significant correlations) for each miRNA. In particular, miR-195 expression has multiple correlations with measures of bone mineral content and density; miR-34a, miR-323, miR-378, and miR-451 expressions correlate to behavioural measures for exploration on the elevated plus maze task; miR-34c and miR-323 expressions correlate with behavioural measures for anxiety on the elevated plus maze task; miR-34c, miR-323, miR-378, and miR-451 expressions correlate with learning and memory measures; and miR-212 expression correlates with grooming duration on a series of anxiety tasks (elevated plus maze, light-dark box, and open field test).
Table 3.
Summary of correlations between miRNA expression and phenotype
| MiRNA | Correlated traits | Phenotypic categories (with more than 3 significant correlations) | |
|---|---|---|---|
| p < 0.05 | FDR q < 0.2 | ||
| miR-15b | 3 | 0 | None |
| miR-34a | 16 | 0 | Behaviour—anxiety (4) |
| Behaviour—exploratory (3) | |||
| Cardiovascular (4) | |||
| miR-195 | 9 | 0 | Bone (5) |
| miR-34c | 14 | 1 | Behaviour—anxiety (3) |
| Behaviour—learning and memory (3) | |||
| Blood haematology (4) | |||
| miR-301a | 16 | 1 | Blood haematology (6) |
| miR-378 | 16 | 0 | Behaviour—exploratory (4) |
| Behaviour-learning and memory (4) | |||
| miR-31 | 14 | 0 | Blood haematology (4) |
| miR-203 | 10 | 0 | None |
| miR-451 | 16 | 0 | Behaviour—exploratory (3) |
| Behaviour—learning and memory (4) | |||
| miR-212 | 10 | 0 | Behavior—anxiety (3) |
| miR-323 | 20 | 0 | Behaviour—anxiety (5) |
| Behaviour—exploratory (3) | |||
| Behaviour—learning and memory (4) | |||
| Blood haematology (3) | |||
Summary of the significant correlations of the strain means of miRNA relative expression and phenotypes available from the Mouse Phenome Database. The phenotypic categories are listed if there were at least three significantly correlated phenotypes falling within that category. The number of significant phenotypic categories is given in parentheses
Correlations of miRNA expression and mRNA expression
There were 292 correlations of miRNA relative expression and mRNA expression for those genes with predicted miRNA binding sites that were nominally significant (p < 0.05), of which 153 were negative correlations. Only two correlations survived a low-stringency FDR multiple-testing correction (q < 0.20): miR-195 and unc93b1 expression (r = 1.0, p = 8.9e-5) and miR-212 and mppe1 expression (r = 1.0, p = 5.3e-5).
Table 4 summarizes the number of nominally significant correlations for each miRNA, for all correlations and for only the negative correlations. Table 4 also summarizes the results of the KEGG pathway analysis. The gene classes with at least two nominally significant correlations are listed, as are the constituent genes for this group.
Table 4.
Summary of miRNA and mRNA correlations and KEGG gene class
| MiRNA | Correlations | Gene class | |||
|---|---|---|---|---|---|
| p < 0.05 | q < 0.2 | r < −0.95 | Correlations (all, negative) | Genes | |
| miR-15b | 34 | 0 | 22 | Purine metabolism (2,0) | polr2h, rpo1–4 |
| Pyrimidine metabolism (2,0) | polr2h, rpo1–4 | ||||
| Neuroactive ligand-receptor interaction (2,1) | glp1r (neg), pmch | ||||
| RNA polymerase (2,0) | polr2h, rpo1-4 | ||||
| miR-34a | 10 | 0 | 6 | None | None |
| miR-195 | 39 | 1 | 22 | MAPK signaling pathway (2,0) | fgf23, prkaca |
| Insulin signaling pathway (2,1) | lipe (neg), prkaca | ||||
| Regulation of actin cytoskeleton (2,0) | fgf23, wasf1 | ||||
| Calcium signaling pathway (2,1) | htr6 (neg), prkaca | ||||
| miR-34c | 31 | 0 | 16 | GnRH signaling pathway (2,1) | itpr3, pla2g10 (neg) |
| Long-term depression (2,1) | itpr3, pla2g10 (neg) | ||||
| Neuroactive ligand-receptor interaction (2,2) | grik4 (neg), p2ry2 (neg) | ||||
| miR-301a | 26 | 0 | 10 | None | None |
| miR-378 | 10 | 0 | 6 | None | None |
| miR-31 | 21 | 0 | 12 | Glutathione metabolism (2,2) | gclc (neg), idh1 (neg) |
| Regulation of actin cytoskeleton (2,0) | diap3, itgb6 | ||||
| miR-203 | 59 | 0 | 31 | Cell adhesion molecules (4,3) | cd276 (neg), cldn (neg), cldn5 (neg), glg1 |
| Glycan structures - biosynthesis (2,0) | alg12, fut8 | ||||
| Leukocyte transendothelial migration (2,2) | cldn3 (neg), cldn5 (neg) | ||||
| Tight junction (2,2) | cldn3 (neg), cldn5 (neg) | ||||
| N-glycan biosynthesis (2,0) | alg12, fut8 | ||||
| miR-451 | 24 | 0 | 12 | None | None |
| miR-212 | 24 | 1 | 8 | None | None |
| miR-323 | 14 | 0 | 8 | None | None |
Summary of the correlations between miRNA and mRNA (from Nadler et al. 2006) and KEGG pathway analysis. The gene classes are shown for those classes with at least two nominally significant correlations. Those genes which had a negative correlation are denoted by (neg)
Discussion
Recently, numerous studies have taken advantage of the phenotypic and genetic variations across inbred strains of mice to better understand complex behaviour and disease (Svenson et al. 2007). Part of the strength of such an approach comes from the potential to use cumulative data for these strains, which is catalogued in various web-based databases for phenotype [such as the Mouse Phenome Project for phenotypic data (Bogue et al. 2006)] and gene expression data [such as the Gene Expression Omnibus for gene expression data (Barrett and Edgar 2006)]. The diversity of phenotypes available for these strains has led to successful investigation into interactions of genetic variation and genetic expression with various phenotypes (Grupe et al. 2001; Guo et al. 2006, 2007; Hillebrandt et al. 2005; Hovatta et al. 2005; Liu et al. 2007; Nadler et al. 2006; Pletcher et al. 2004). Such approaches have highlighted genes that play a significant role in behaviour and disease pathology (Guo et al. 2006, 2007; Hillebrandt et al. 2005; Hovatta et al. 2005; Liu et al. 2007). In this study we used this approach to investigate the effects that variation in miRNA expression across common inbred strains have on various phenotypes, including mRNA expression. This analysis allowed us to better characterise the effects of miRNA expression on phenotypes and gene expression, with the aim of a better understanding of the functional consequences of the miRNAs.
Correlations of the differentially expressed miRNAs with the traits from the Mouse Phenome Database revealed a number of potential functions for these miRNAs. The expression of a number of these miRNAs was significantly correlated with behavioural measures for exploration (miR-34a, miR-378, miR-451, and miR-323), anxiety (miR-34c and miR-323), and learning and memory (miR-34c, miR-323, miR-378, and miR-451). While miRNAs have previously been implicated in synaptic development (Fiore and Schratt 2007), this is the first indication of miRNA involvement in anxiety, learning and memory, and exploration measures. Although these findings are preliminary and would require a much larger number of genotypes to be conclusive, they are in line with the findings of miRNA involvement in various mental disorders, including schizophrenia (Beveridge et al. 2008), DiGeorge syndrome (Landthaler et al. 2004), mental retardation (Lin et al. 2006), and Tourette’s syndrome (Chou et al. 2007).
The functions of a number of these differentially expressed miRNAs have been implicated in numerous biological processes, determined either from functional studies or inferred from expression studies. Both miR-15b and miR-195 are highly expressed in regenerating pancreas and they inhibit neurogenin 3 (NGN3) protein expression, which is known to be inhibited during normal pancreatic regeneration (Joglekar et al. 2007). miR-195 has also been shown to be important for cardiac function as shown in mice overexpressing miR-195 (van Rooij et al. 2006), while miR-15b also has altered expression in T-cell differentiation (Wu et al. 2007). miR-378 promotes cell survival by targeting SUFU and FUS1 expression (Lee et al. 2007). miR-451 accumulates in both infected and healthy red blood cell lines (Rathjen et al. 2006).
In addition, some of these miRNAs have been implicated in disease pathology. Alcohol increases the expression of miR-212 and is correlated with induced gut leakiness, a key factor in alcoholic liver disease (Tang et al. 2008). miR-203 has altered expression in chronic inflammatory diseases (Sonkoly et al. 2008), while miR-31 expression is altered in colorectal cancer (Slaby et al. 2007). miR-34a has altered expression in tumor-suppressive conditions (Tong and Nemunaitis 2008). miR-195 and miR-34a both are highly expressed in chronic lymphocytic leukaemia (Zanette et al. 2007). Expression of miR-34a and miR34c is increased by DNA damage and oncogenic stress in a p53-dependent method (He et al. 2007). miR-203 expression was downregulated in oral squamous cell carcinoma (Kozaki et al. 2008) and has altered expression in chronic inflammatory diseases (Sonkoly et al. 2008). miR-378 and miR-451 have upregulated expression in the late stages of erythropoiesis (Bruchova et al. 2007).
Traditionally, miRNAs have been thought to cause post-transcriptional gene silencing by base pairing with target sites on specific mRNAs, resulting in either mRNA cleavage or translational repression (Kim 2005). More recently, new evidence suggests that miRNAs can also enhance mRNA expression (Check 2008; Place et al. 2008). We attempted to validate potential miRNA binding sites by correlating miRNA expression with mRNA expression for those genes with miRNA binding sites. This approach would be able to detect only miRNAs that cause mRNA cleavage or enhancement of mRNA expression. Only half of the nominally significant correlations had negative correlational coefficients (52%). While this suggests that miRNA is as likely to enhance mRNA expression as it is to inhibit its expression, these findings should be interpreted cautiously because it is likely that many of our nominally significant correlations are due to chance.
KEGG pathway analysis was conducted for the nominally significant miRNA and mRNA expression correlations with the hope of revealing downstream pathways affected by these miRNAs. One of the stronger findings is that miR-203 may regulate four genes that code for cell adhesion molecules. Cell adhesion molecules are important in the regulation of inflammation (Smith 2008) and miR-203 has previously been implicated in inflammatory diseases (Sonkoly et al. 2008). We also found that miR-34c may regulate genes involved in long-term depression and neuroactive ligand receptors, which is interesting as our phenotypic analysis revealed that miR-34c is a candidate gene for anxiety and learning and memory measures, both of which are influenced by genes from these gene classes.
Finding differences in miRNA expression across strains opens the door for further studies on various mouse genetic reference populations, including recombinant inbred mice, consomics, and outbred populations. Mouse genetic reference populations such as C57BL/6J × DBA/2J recombinant inbred mice (BXD RI) have been integral to the understanding of complex biological systems (Bystrykh et al. 2005; Wang et al. 2003) and a key tool in systems genetics approaches (Churchill 2007). The use of such populations has been particularly good for investigating the underlying genetics of various phenotypes, including mRNA expression, with the existence of numerous references describing the successful use of this approach (Belknap and Crabbe 1992; de Haan and Williams 2005; Hsu et al. 2005). Now that significant differences in miRNA expression have been detected within the progenitor strains of these populations, this approach similarly can be used to determine the effects of miRNA expression on various phenotypes, from behavioural phenotypes to mRNA expression (Chesler et al. 2004).
We found that the hippocampal expression of nearly 7% of all the screened miRNAs was nominally significant across strains. All 11 of these nominally significant assays survived multiple testing by controlling the FDR to q < 0.10; thus, only one of these 11 nominally significant findings is likely due to chance. When we lowered the FDR rate to q < 0.05, eight survived, which was still approximately 5% of all miRNAs investigated. At this FDR level it is unlikely that any of these eight assays are due to chance. Thus, depending on the stringency level, between 5 and 7% of all miRNAs investigated were differentially expressed across strains. It should be noted that we searched the primer sequences for the assays we investigated using the perlegen sequence data and found no SNPs within the miRNAs for the genes we investigated.
Previous microarray experiments conducted within our group have found that the total percentage of mRNAs that have significant differences in hippocampal expression across strains vary from 2% (Fernandes et al. 2004) to an upper limit of 20% (unpublished observation). This is similar to the 2–30% seen in other studies (Nadler et al. 2006; Sandberg et al. 2000). In our current study, we found that 5–7% of all tested miRNAs significantly differ in hippocampal expression, thus falling well within the wide range seen for mRNAs in previous studies. It should be noted that direct comparisons of these studies are difficult due to differences in the techniques, statistical power, and the stringency of the multiple-testing approaches used.
A recent study calculated the number of samples needed to detect gene expression fold changes using microarrays for varying power levels, false detection rates, and expression variablities (Wei et al. 2004). For inbred mice they found that for the genes whose variation of expression levels falls into the lower half of the distribution, you need 7 animals per group to have 90% power to detect a 1.5-fold or greater change in expression at α = 0.01. For more variable genes a much greater sample number is required to obtain a similar power. The miRNAs that we failed to replicate in individual animals generally had higher variation than those that we were able to replicate. If we assume that the power calculations conducted for microarrays are relevant for TaqMan RT-PCR, then the failure to replicate these findings in individual animals is likely due to a lack of sufficient power for these highly variable genes, rather than them not being real effects.
While we did not find any main effects of sex that survived multiple-testing correction, we did find a significant interaction between strain and sex for miR-203 expression (p = 1e-4). The underlying cause of this interaction is unknown, although it may relate to sex-specific regulation of miRNA expression (Zhang et al. 2007a). The failure to find many effects of sex on miRNA expression may be accounted for by the fact that we were specifically screening for strain differences.
In conclusion, this is the first observation of miRNA expression differences across inbred mouse strains, with approximately 5–7% of all investigated miRNAs differentially expressed across strains. By correlating the expression of these differentially expressed miRNAs with in silico phenotype data and mRNA expression, we were able to better characterise the effects of these miRNAs on both phenotype and the regulation of mRNA expression. This approach has allowed us to nominate miRNAs that have potential roles in anxiety, exploration, and learning and memory. In addition, the finding of these differentially expressed miRNAs opens the door for further studies on various mouse genetic reference populations, including recombinant inbred mice, consomics, and outbred populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Contributor Information
Michael J. Parsons, Email: m.parsons@har.mrc.ac.uk
Leonard C. Schalkwyk, Email: l.schalkwyk@iop.kcl.ac.uk
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, et al. Sequence variants in SLITRK1 are associated with Tourette’s Syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC. Chromosome mapping of gene loci affecting morphine and amphetamine responses in BXD recombinant inbred mice. Ann N Y Acad Sci. 1992;654:311–323. doi: 10.1111/j.1749-6632.1992.tb25977.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. microRNA target predictions: the microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Bogue MA, Grubb SC, Maddatu TP, Bult CJ. Mouse Phenome Database (MPD) Nucleic Acids Res. 2006;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35:1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Check E. RNA interference: hitting the on switch. Nature. 2007;448:855–858. doi: 10.1038/448855a. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7(5):485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Chou IC, Wan L, Liu SC, Tsai CH, Tsai FJ. Association of the Slit and Trk-like 1 gene in Taiwanese patients with Tourette syndrome. Pediatr Neurol. 2007;37:404–406. doi: 10.1016/j.pediatrneurol.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Recombinant inbred strain panels: a tool for systems genetics. Physiol Genomics. 2007;31:174–175. doi: 10.1152/physiolgenomics.00181.2007. [DOI] [PubMed] [Google Scholar]
- de Haan G, Williams RW. A genetic and genomic approach to identify longevity genes in mice. Mech Ageing Dev. 2005;126:133–138. doi: 10.1016/j.mad.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Fernandes C, Paya-Cano JL, Sluyter F, D’Souza U, Plomin R, et al. Hippocampal gene expression profiling across eight mouse inbred strains towards understanding the molecular basis for behaviour. Eur J Neurosci. 2004;19:2576–2582. doi: 10.1111/j.0953-816X.2004.03358.x. [DOI] [PubMed] [Google Scholar]
- Fiore R, Schratt G. MicroRNAs in synapse development: tiny molecules to remember. Expert Opin Biol Ther. 2007;7:1823–1831. doi: 10.1517/14712598.7.12.1823. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, et al. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Guo Y, Weller P, Farrell E, Cheung P, Fitch B, et al. In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol. 2006;24:531–536. doi: 10.1038/nbt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, et al. In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci USA. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- Hillebrandt S, Wasmuth HE, Weiskirchen R, Hellerbrand C, Keppeler H, et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat Genet. 2005;37:835–843. doi: 10.1038/ng1599. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Tennant RS, Helton R, Marr RA, Singer O, et al. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- Hsu HC, Li L, Zhang HG, Mountz JD. Genetic regulation of thymic involution. Mech Ageing Dev. 2005;126:87–97. doi: 10.1016/j.mad.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–612. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang SJ, Ying SY. First in vivo evidence of microRNA-induced fragile X mental retardation syndrome. Mol Psychiatry. 2006;11:616–617. doi: 10.1038/sj.mp.4001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE. 2007;2:e651. doi: 10.1371/journal.pone.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler JJ, Zou F, Huang H, Moy SS, Lauder J, et al. Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype. Genetics. 2006;174:1229–1236. doi: 10.1534/genetics.106.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, et al. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- Smith CW. Adhesion molecules and receptors. J Allergy Clin Immunol. 2008;121:S375–S379. doi: 10.1016/j.jaci.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Svenson K, Smith RV, Magnani P, Suetin H, Paigen B, et al. Multiple trait measurements in 43 inbred mouse strains captures the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center responsse by microRNA-55. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Wei Q, Liu S, Huang J, Mao X, Chu X, et al. Comparison of hybridization behavior between double and single strands of targets and the application of asymmetric PCR targets in cDNA microarray. J Biochem Mol Biol. 2004;37:439–444. doi: 10.5483/bmbrep.2004.37.4.439. [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


