Abstract
The introduction of modified or labeled nucleotides into RNA is a powerful RNA engineering tool as it enables us to investigate how native RNA modifications affect RNA function and structure. It also helps in the structural analysis of RNA. A modified nucleotide can be introduced into a specific position of RNA by the method of two-step enzymatic ligation of RNA fragments. However, this method requires a complicated purification step between the two ligation steps that results in low yields of the ligation product. Here we have developed a new ligation technique employing periodate oxide that eliminates this purification step. This increases the total yield of the ligation product and makes it a faster procedure.
INTRODUCTION
To introduce point mutations in RNAs, the in vitro RNA transcription method using a DNA template and T7 RNA polymerase is widely employed (1). However, there are many RNA molecules that bear modified nucleosides that cannot be generated by this method. Such modified nucleosides play important roles in RNA functions in many cases (2) and thus the site-specific introduction or deletion of the modification in a particular RNA molecule would be helpful in elucidating the effect of the modified nucleoside. However, this requires a method other than the simple in vitro transcription method. This is also true for the site-specific stable-isotopic labeling of RNA for NMR analysis (3), the site-specific introduction of photoreactive nucleotide analogs for cross-linking experiments (4), and other situations.
In such cases, RNA molecules including a modified or labeled residue can be constructed with RNA ligase. This method has been used to introduce modified nucleosides in cases where an appropriate modification enzyme is not available (5), and for NMR measurements that require a stable-isotopic labeled nucleoside (6,7). This method demands two-step ligation. After the first ligation (ligation of a modified or labeled nucleotide to an RNA fragment that is the 5′ part of the target RNA molecule), the products must be purified by high-performance liquid chromatography (HPLC) or polyacrylamide gel electrophoresis (PAGE) to prevent side reactions in the second step. However, it is not easy to separate the (n)mer substrate from the (n + 1)mer product that results from ligating a mononucleotide to long RNA. These difficulties have discouraged RNA researchers from using this two-step ligation method.
In this study, we sought to simplify the two-step ligation method by using periodate oxidation to inactivate the unligated substrate in the first step and thereby block side reactions in the second step. The rationale for using periodate oxidation, which has been used to investigate the various roles the 3′ terminus plays in RNA function (8–11), is that it only cleaves the 3′-end cis-diol bond of RNA. This method would eliminate the PAGE/HPLC purification step between the first and the second ligation steps and therefore would simplify the overall strategy.
Recently, we found two taurine-containing modified uridines [5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U)] in human and bovine mitochondrial tRNAs (12). Yasukawa et al. have shown that these modifications are related to mitochondrial diseases (13,14). At present, we are interested in the function and property of various modified uridines (15), including τm5U and τm5s2U, which occur at the anticodon first position of tRNAs that correspond to two-codon families ending in purine. Here we demonstrate the utility of our quick two-step RNA-ligation method in introducing modified uridines into tRNAs. As our model system, we incorporated an unmodified uridine into the anticodon first position of the transcribed Escherichia coli tRNALeu.
MATERIALS AND METHODS
Preparation of E.coli tRNALeuUAA
The primers 5′-GATCCGGGTAATACGACTCACTATAGGCCGGATGGTGGAATCGGTAGACACAAGGGATTTAAAATCCCTCGGCGTTCGCGCTGTGCGGGTTCAAGTC CCGCTCCGGCTAG-3′ and 5′-AATTCTAGCCGGAGCGGGACTTGAACCCGCACAGCGCGAACGCCGAGGGATTTTAAATCCCTTGTGTCTACCGATTCCACCATCCGG CCTATAGTGAGTCGTATTACCCG-3′ that encompass the E.coli tRNALeuUAA gene sequence were hybridized to each other and the resultant DNA was inserted into the multi-cloning site of the pUC19 vector (TOYOBO) by the method of Sampson and Uhlenbeck (16). The nucleotide sequence of the plasmid was confirmed by the dideoxy-termination method (17) using a 3100 Genetic Analyzer (Applied Biosystems). The template DNA was prepared from E.coli JM109 cells cultivated on a large scale and completely digested with EcoRI. It was then transcribed with T7 RNA polymerase (1,18). The reaction mixture contained 200 nM DNA template, 40 mM Tris–HCl (pH 8.0), 24 mM magnesium chloride, 2 mM spermidine, 5 mM dithiothreitol (DTT), 0.01% Triton, 2 mM each of ATP, CTP, GTP, and UTP, 10 mM GMP and 57 µg/ml T7 RNA polymerase. After incubation for 4 h at 37°C, the sample was extracted with phenol and dialyzed against water, then recovered by ethanol precipitation. The transcribed RNA was then incubated for 30 min at 37°C in 50 mM Tris–HCl (pH 8.0), 10 mM magnesium chloride, 150 mM potassium chloride, 1 mM DTT, 1 mM each of ATP and CTP and 10 µg/ml E.coli CCA enzyme to repair the CCA of the 3′ terminus. The sample was purified by 10% denaturing PAGE.
Preparation of the 5′- and 3′-half fragments of E.coli tRNALeuUAA
The hammerhead ribozyme (19) used to cleave the tRNA was transcribed and purified as described above except for the CCA repair. The template DNA for the transcription of the hammerhead ribozyme was prepared by using the primers 5′-GATCCGGGTAATACGACTCACTATAGGGATTTTCTGATGAGCCGAAAGGCGAAAATCCCTTGTGTCTAG-3′ and 5′-AATTCTAGACACAAGGGATTTTCGCCTTTCGGCTCATCAGAAAATCCCTATAGTGAGTCGTATTACCCG-3′. The tRNA substrate was then specifically cleaved with the ribozyme at the 3′ side of U34 (Fig. S1). The reaction mixture contained 50 mM Tris–HCl (pH 8.0), 25 mM magnesium chloride, 0.02 A260 units of tRNA, 0.012 A260 units of ribozyme, and 0.01 A260 units of DNA (5′-CGATTCCACCATCCGGCC-3′ and 5′-TGGTAGCCGGAGCGGG-3′). After incubation for 2 h at 37°C, the 5′- and 3′-half fragments were separated and purified by 10% denaturing PAGE. The 5′ and 3′ fragments were processed as below (see Fig. S1).
The 5′ fragment was treated with 0.1 M HCl for 4 h at 0°C to hydrolyze the 2′,3′-cyclic phosphates resulting in the 3′-phosphate. The sample was diluted with 2 vol of H2O, followed by recovery using ethanol precipitation. The 3′-phosphate and the U34 nucleoside of the 5′ fragment were removed by E.coli alkaline phophatase (BAP) treatment (6), followed by deprivation of the 3′ nucleoside using NaIO4 and lysine–HCl (20). This was followed by a second BAP treatment.
The 3′ fragment was treated with T4 polynucleotide kinase as described by Ohtsuki et al. (20) to generate a 5′-phosphate end and a 3′-OH end.
3′(2′),5′-Diphosphorylation of uridine
The 3′(2′),5′-diphosphorylation of uridine was performed according to Barrio et al. (21). In this method, the 3′,5′- and 2′,5′-diphosphates cannot be separated, but the mixture can be used directly with T4 RNA ligase since the 3′,5′ component of the mixture is the substrate and the 2′,5′ component is neither a substrate nor an inhibitor (22). A mixture of 0.3 mmol of uridine and 3 mmol of pyrophosphoryl chloride was stirred at –10 to –15°C. After 4–5 h, the reaction was quenched by the rapid addition of ice and a chilled solution of 2 M triethylammonium bicarbonate (TEAB) (pH 8.0) to adjust the final concentration to 0.05 M. The sample was then purified on a column of DEAE–Sephadex Fast Flow (Amersham Pharmacia Biotech) with a linear gradient ranging from 0.05 to 1 M TEAB at 4°C. The fractions containing the uridine diphosphate (pUp) were pooled and evaporated to dryness under vacuum to remove the excess TEAB.
The first ligation
The 5′ fragment (34mer; 34OH) and pUp were ligated using T4 RNA ligase. The reaction was performed at 11°C for 0.5–16 h in the reaction mixture containing 50 mM Tris–HCl (pH 7.6), 15 mM magnesium chloride, 3.5 mM DTT, 15 µg/ml bovine serum albumin (BSA), 5% polyethylene glycol, 300 µM ATP, 1.8 mM pUp, 90 µM 5′ fragment and 1.6 U/µl T4 RNA ligase. After phenolization, the RNA was recovered by ethanol precipitation.
Periodate oxidation of the first ligation product
The cis-diol group (RNA 3′ end without phosphate) of the ligation products was cleaved with periodic acid to generate 2′,3′-dialdehydes. The reaction was carried out for 40 min at 0°C in the dark in a total volume of 100 µl containing the first ligation products and 10 mM NaIO4. Since only the unligated substrate (34OH) still bears a cis-diol group at its 3′ end, it was selectively destroyed at its 3′ end. NaIO4 was removed from the RNAs by ethanol precipitation. The 3′-phosphate of the 5′ fragment was then removed by BAP treatment (6).
The second ligation
The 5′ fragment (the mixture of the first ligation resultants oxidized with NaIO4) and the 3′ fragment (90 µM each) were heated at 65°C for 7 min and annealed at room temperature for 1 h in 50 mM Tris–HCl (pH 7.6) and 15 mM magnesium chloride. This RNA complex was incubated at 37°C for 1 h in a buffer containing 60 mM Tris–HCl (pH 7.6), 17.5 mM magnesium chloride, 3.5 mM DTT, 10 µg/ml BSA, 300 µM ATP and 1.6 U/µl T4 RNA ligase. The sample was purified by 10% denaturing PAGE.
Mass spectrometry
An LCQ ion-trap mass spectrometer (ThermoFinnigan) equipped with an electrospray ionization source and a HP1100 liquid chromatography system (Agilent) were used to analyze RNase T1-digested RNA. Oligonucleotides produced by RNase T1 digestion were analyzed as the negative ion form by LC/MS according to the literature (23) with a slight modification. The digests were placed directly onto a Spercosil LC-DB C18 column, 300 × 1.0 mm (Sperco). The solvent system consisted of 0.4 M 1,1,1,3,3,3-hexafluoro-2-propanol (pH 7.0, adjusted with triethylamine) in H2O (A) and 50% methanol (B). These solvents were used as follows: 5–80% B for 0–33 min, 80% B for 33–35 min, 80–5% B for 35–40 min. The chromatographic effluents (70 µl/ml) were conducted into the ion source without prior splitting. Negative ions were scanned over m/z ranging from 620 to 2000 throughout the separation under the following conditions: flow rate of sheath gas, 55 arb; capillary temperature, 235°C; spray voltage, 5 kV.
Aminoacylation of tRNAs
Escherichia coli leucyl-tRNA synthetase (LeuRS) was prepared as described (24). Aminoacylation of 0.03 A260 units of tRNAs was carried out at 37°C in 30 µl of a reaction mixture containing 100 mM Tris–HCl (pH 7.6), 5 mM magnesium acetate, 2 mM ATP, 20 mM potassium chloride, 1 mM DTT, 20% dimethylsulfoxide, 100 µM [14C]leucine and 1 ng/µl LeuRS. At appropriate times, 9 µl aliquots were withdrawn from the reaction mixture and placed onto dry Whatman 3MM filters. The filters were washed three times with 5% (w/v) trichloroacetic acid and once with ethanol, and then dried, and measured by a liquid scintillation counter (ALOKA).
RESULTS
Two-step ligation using periodate oxidation
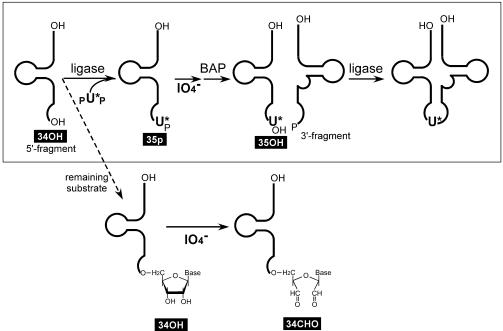
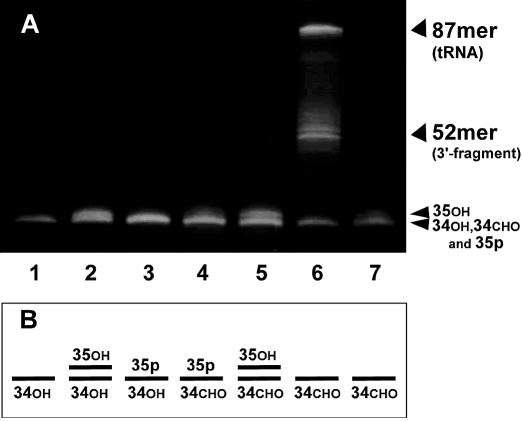
The Materials and Methods describes our successful generation of the 5′- and 3′-half fragments of tRNALeuUAA and pUp. Figure 1 shows the overall strategy that we use to introduce a modified or unmodified uridine into the tRNA. In the first ligation, pUp was added at much higher concentrations than the 5′ fragment because it was obtained more abundantly than the 5′ fragment and because it consists of a mixture of nucleoside 3′,5′- and 2′,5′-diphosphates. Although the efficiency of the first ligation is over 80% after 4–16 h of incubation, the first ligation used here was performed for only 30 min so that only half of the 5′ fragments were ligated, which would help to visualize the effect of periodate oxidation clearly by gel electrophoresis (Fig. 2). On the gel shown in Figure 2, the mobility of the 5′ fragment ligated to pUp (35p; lane 3) or oxidized with NaIO4 (34CHO; lane 7) was the same as that of the original 5′ fragment (34OH; lane 1). The ligation product that was dephosphorylated with BAP (35OH) separated slightly from 34OH (lane 2) and 34CHO (lane 5). Lanes 2 and 5 indicate that the ligation efficiency was ∼40%. Lane 6 shows the result of the second ligation using the 3′ fragment and the 5′ substrate mixture shown in lane 5. Comparison of lanes 5 and 6 indicates that almost all (over 80%) of the 35OH was ligated to the 3′ fragment whereas 34CHO was not.
Figure 1.
The scheme used to introduce a modified uridine (U*) into the wobble position of tRNALeuUAA. The substrate lacking the modified uridine (34OH) is inactivated prior to the second ligation by periodate oxidation.
Figure 2.
(A) Analysis of ligated RNAs by 10% denaturing PAGE. Lane 1, the 5′ fragment (34OH); lane 2, the 5′ fragment after the first ligation followed by BAP treatment (35OH); lane 3, the 5′ fragment after the first ligation (35p); lane 4, the 5′ fragment after the first ligation followed by BAP treatment and periodate oxidation; lane 5, the 5′ fragment after the first ligation followed by BAP treatment, periodate oxidation and BAP treatment (35OH); lane 6, the second ligation between the 5′ and 3′ fragments; lane 7, the 5′ fragment after periodate oxidation (34CHO). (B) The lower part of (A) summarized in a simple depiction.
This quick two-step ligation method was also performed to confirm the overall yield. It took us ∼9 h to complete the operation shown in Figure 1, namely, the first ligation (4 h), the phenol treatment and ethanol precipitation (30 min), the NaIO4 treatment and ethanol precipitation (1 h), the BAP treatment (30 min), the phenol treatment and ethanol precipitation (30 min), and the second ligation with the annealing (2 h). The product was purified by PAGE. Using 1.3 nmol of the 5′ and the 3′ fragments (0.3 and 0.46 A260 units, respectively), we obtained 0.29 nmol of the full-length tRNA.
Confirmation of the tRNA sequence by mass spectrometry
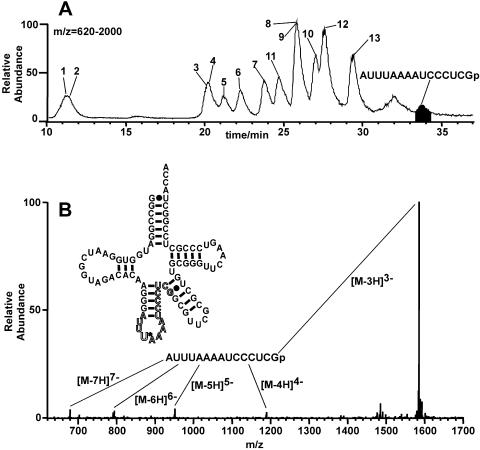
To confirm the sequence of the tRNA obtained as described above, it was digested with RNase T1 and the resultant fragments were analyzed by mass spectrometry. The sequence of the RNase T1 fragment that includes U34 is AUUUAA AAUCCCUCGp, a 15mer, whereas RNase T1 digestion of the tRNA without U34 would result in the appearance of the 14mer AUUAAAAUCCCUCGp. Figure 3 shows that the 15mer fragment, not the 14mer, was detected, which indicates that almost all of the 34OH was inactivated by NaIO4 before the second ligation.
Figure 3.
Mass spectrometric analysis of oligonucleotides derived from the tRNALeuUAA obtained by the ligation method. (A) Mass chromatograms of oligonucleotides obtained by RNase T1 digestion of the tRNALeuUAA. Base peaks in the mass chromatogram of m/z = 620–2000, within which each oligonucleotide was detected. Peak numbers represent RNA fragments derived from tRNALeuUAA: 1, CGp; 2, UGp; 3, CCGp; 4, CUGp; 5, AUGp; 6, UAGp; 7, UUCGp; 8, UCCCGp; 9, CUCCGp; 10, AAUCGp; 11, CUACCA (3′ terminal); 12, UUCAAGp; 13, ACACAAGp. (B) Mass spectrum in the range 33.51–34.16 min. [M-3H]3–, [M-4H]4–, [M-5H]5–, [M-6H]6– and [M-7H]7– ions were detected.
Biological activity of the tRNA constructed by the quick two-step ligation method
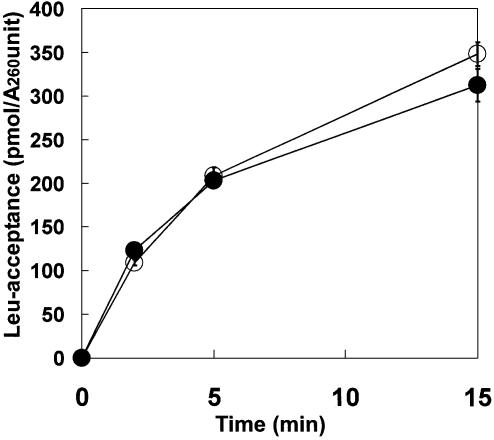
The constructed and the transcribed E.coli tRNALeuUAA molecules were aminoacylated by E.coli LeuRS. As shown in Figure 4, the leucine-accepting activity of the altered tRNA was almost identical to that of the transcribed counterpart.
Figure 4.
Leucylation analysis of the transcribed and constructed tRNAs (open and closed circles, respectively). The reaction conditions used are described in the Materials and Methods.
DISCUSSION
Advantages of the quick two-step RNA ligation method
This method is easier and takes less time than the standard ligation method that is used for mononucleotide introduction into RNA (5–7). The efficiencies of the ligation and gel-purification steps are very important for the overall yield of the RNA molecular surgery because the other operations (phosphorylation, dephosphorylation, 3′-nucleoside deprivation, etc.) can be performed with no loss. In the protocol without the step of periodate oxidation (5–7), purification after the first ligation is the most troublesome step because the ligation product has to be separated from the substrate RNA, which is only one base shorter than the product. Such purification requires the use of quite a long (e.g. 40 cm) polyacrylamide gel, which significantly lowers the yield, especially if small substrate amounts are used. In contrast, the quick two-step ligation method can employ comparatively small amounts of the substrate. In fact, we obtained 0.29 nmol of the final product from 1.3 nmol each of the 5′ and the 3′ fragments. Our method demands only a single gel-purification step with a short gel (e.g. 10 cm) because the final RNA product is much longer than the 5′ and the 3′ fragments.
Preparation of materials
In this study, the 5′ and the 3′ fragments were prepared by digestion of the tRNA transcript with the hammerhead ribozyme. It is actually easier to prepare RNA fragments by T7 transcription or chemical synthesis than to prepare them by RNA digestion. However, this study aimed to demonstrate that a specific position in an RNA molecule could be cleaved and that a mononucleotide could be introduced into this position. This strategy can thus also be used to mutagenize RNA bearing modifications, while the in vitro RNA transcription method can mutagenize only unmodified RNA.
A nucleoside 3′,5′-diphosphate can be obtained from a nucleoside by the method described above. Polynucleotide kinase lacking 3′-dephosphorylation activity is also useful to produce nucleoside 3′,5′-diphosphate if the nucleoside 3′-phosphate is available (6).
Application to site-specific labeling and modification
Site-specific stable-isotopic labeling of RNAs followed by NMR analysis generates information about the local conformation around the labeled site. However, isotopic labeling of RNA by automated chemical synthesis is not common because labeled RNA monomers for chemical synthesis are not commercially available. Several methods using enzymes to site-specifically label RNAs have been devised. These include segmental labeling by ligation of RNase H-cleaved fragments with DNA ligase (25) or ribozyme-cleaved fragments with RNA ligase (26), and mono- or dinucleotide labeling by ligation of synthetic RNA fragments and labeled nucleotide with RNA ligase (6,7,27). The method described here improved the latter technique. It should be noted that this method can be applied to mono- or dinucleotide labeling but not to trinucleotide (or longer segment) labeling because of the substrate specificity of T4 RNA ligase. Mono- or dinucleotide can only be the ‘donor’ substrate of T4 RNA ligase, however, trinucleotide can be both the ‘donor’ substrate and the ‘acceptor’ substrate. Thus, in our protocol (Fig. 1), BAP treatment of the first ligation product generates undesired ‘acceptor’ substrate if the labeled insert is longer than trinucleotide.
Modified nucleosides sometimes play important roles in RNA functions (2). Our method is useful for introducing modified nucleosides into specific RNA positions, thus allowing us to investigate the effect of the modification. Among other applications, this method also permits the introduction of (i) photoreactive nucleotide analogs for cross-linking experiments (4), (ii) nucleoside analogs for the analysis of local structure (28), (iii) unnatural nucleosides for the generation of an artificial genetic code (29), and so on. We have ourselves used this method to introduce τm5U and 5-carboxymethylaminomethyluridine (cmnm5U) into the anticodon first position of E.coli tRNALeuUAA (data not shown). This has permitted us to investigate the translation activity and fidelity of the variant tRNAs in reading the UUR (R = A or G) codon. We will report the properties of these modifications in the tRNA wobble position in the near future.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Grants-in-Aid from the Ministry of Culture, Sports, Science and Technology of Japan to K.W. and T.O.
REFERENCES
- 1.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane B.G. (1998) Historical perspectives on RNA nucleoside modifications. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 1–20. [Google Scholar]
- 3.Varani G., Aboul-ela,F. and Allain,H.-T. (1996) NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc., 29, 51–127. [Google Scholar]
- 4.Hanna M.M., Bentsen,L., Lucido,M. and Sapre,A. (1999) RNA-protein crosslinking with photoreactive nucleotide analogs. Methods Mol. Biol., 118, 21–33. [DOI] [PubMed] [Google Scholar]
- 5.Helm M., Giege,R. and Florentz,C. (1999) A Watson–Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry, 38, 13338–13346. [DOI] [PubMed] [Google Scholar]
- 6.Ohtsuki T., Kawai,G. and Watanabe,K. (1998) Stable isotope-edited NMR analysis of Ascaris suum mitochondrial tRNAMet having a TV-replacement loop. J. Biochem., 124, 28–34. [DOI] [PubMed] [Google Scholar]
- 7.Ohtsuki T., Kawai,G. and Watanabe,K. (2002) The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNASerUCU having a short T arm and lacking the entire D arm. FEBS Lett., 514, 37–43. [DOI] [PubMed] [Google Scholar]
- 8.Neu H.C. and Heppel,L.A. (1964) Nucleotide sequence analysis of polyribonucleotides by means of periodate oxidation by cleavage with an amine. J. Biol. Chem., 239, 2927–2934. [PubMed] [Google Scholar]
- 9.Whitfeld P.R. (1965) Application of the periodate method for the analysis of nucleotide sequence to tobacco mosaic virus RNA. Biochim. Biophys Acta, 108, 202–210. [DOI] [PubMed] [Google Scholar]
- 10.Steinschneider A. and Fraenkel-Conrat,H. (1966) Studies of nucleotide sequences in tobacco mosaic virus ribonucleic acid. 3. Periodate oxidation and semicarbazone formation. Biochemistry, 5, 2729–2734. [DOI] [PubMed] [Google Scholar]
- 11.Tanner N.K. and Cech,T.R. (1987) Guanosine binding required for cyclization of the self-splicing intervening sequence ribonucleic acid from Tetrahymena thermophila. Biochemistry, 26, 3330–3340. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T., Suzuki,T., Wada,T., Saigo,K. and Watanabe,K. (2002) Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J., 21, 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasukawa T., Suzuki,T., Ueda,T., Ohta,S. and Watanabe,K. (2000) Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. J. Biol. Chem., 275, 4251–4257. [DOI] [PubMed] [Google Scholar]
- 14.Yasukawa T., Suzuki,T., Ishii,N., Ueda,T., Ohta,S. and Watanabe,K. (2000) Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett., 467, 175–178. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama S. and Nishimura,S. (1995) Modified nucleosides and codon recognition. In Soll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynsthesis and Function. American Society for Microbiology, Washington, DC, pp. 207–223. [Google Scholar]
- 16.Sampson J.R. and Uhlenbeck,O.C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA, 85, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messing J. (1983) New M13 vectors for cloning. Methods Enzymol., 101, 20–78. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham P.R. and Ofengand,J. (1990) Use of inorganic pyrophosphatase to improve the yield of in vitro transcription reactions catalyzed by T7 RNA polymerase. Biotechniques, 9, 713–714. [PubMed] [Google Scholar]
- 19.Blount K.F. and Uhlenbeck,O.C. (2002) The hammerhead ribozyme. Biochem. Soc. Trans., 30, 1119–1122. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsuki T., Kawai,G., Watanabe,Y., Kita,K., Nishikawa,K. and Watanabe,K. (1996) Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res., 24, 662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrio J.R., Barrio,M.C., Leonard,N.J., England,T.E. and Uhlenbeck,O.C. (1978) Synthesis of modified nucleoside 3′,5′-bisphosphates and their incorporation into oligonucleotides with T4 RNA ligase. Biochemistry, 17, 2077–2081. [DOI] [PubMed] [Google Scholar]
- 22.England T.E. and Uhlenbeck,O.C. (1978) Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry, 17, 2069–2076. [DOI] [PubMed] [Google Scholar]
- 23.Apffel A., Chakel,J.A., Fischer,S., Lichtenwalter,K. and Hancock,W.S. (1997) Analysis of oligonucleotides by HPLC–electrospray ionization mass spectrometry. Anal. Chem., 69, 1320–1325. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu Y., Inoue,A., Tomari,Y., Suzuki,T., Yokogawa,T., Nishikawa,K. and Ueda,T. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol., 19, 751–755. [DOI] [PubMed] [Google Scholar]
- 25.Xu J., Lapham,J. and Crothers,D.M. (1996) Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc. Natl Acad. Sci. USA, 93, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim I., Lukavsky,P.J. and Puglisi,J.D. (2002) NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc., 124, 9338–9339. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura A., Muto,Y., Watanabe,S., Kim,I., Ito,T., Nishiya,Y., Sakamoto,K., Ohtsuki,T., Kawai,G., Watanabe,K. et al. (2002) Solution structure of an RNA fragment with the P7/P9.0 region and the 3′-terminal guanosine of the tetrahymena group I intron. RNA, 8, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strobel S.A. (1999) A chemogenetic approach to RNA function/structure analysis. Curr. Opin. Struct. Biol., 9, 346–352. [DOI] [PubMed] [Google Scholar]
- 29.Hirao I., Ohtsuki,T., Fujiwara,T., Mitsui,T., Yokogawa,T., Okuni,T., Nakayama,H., Takio,K., Yabuki,T., Kigawa,T. et al. (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol., 20, 177–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.