The research group of Professor Andrey Belozersky with whom I started my academic career in 1955 consisted of two parts: one was located at the Department of Plant Biochemistry, Moscow State University, and the other at the A. N. Bach Institute of Biochemistry, Academy of Sciences of the USSR. This biochemical group was one of the most creative in the country. It was world-renowned because of several important discoveries in the field of nucleic acid studies. In the thirties of the last century, it succeeded in settling the question of the universal occurrence of two known types of nucleic acids, ribonucleic acid (RNA) and deoxyribonucleic acid (DNA), in living matter. At that time, many biochemists believed that RNA is a characteristic component of plants and fungi, whereas DNA (designated as “thymonucleic acid” or “animal nucleic acid”) belongs to the animal kingdom. The presence of DNA in plant cells raised doubts, as the positive cytochemical Feulgen reaction in plant cell nuclei was the only indirect evidence. Belozersky and colleagues were the first to isolate thymine and then DNA (thymonucleic acid) from higher plants (1, 2), thus proving the universal occurrence of DNA. The next series of studies was carried out on bacteria (3) and demonstrated that both RNA and DNA were present there, again confirming the idea of the universality of the occurrence of both types of nucleic acids in organisms of different phylogenetic kingdoms. At the same time, the studies on bacteria showed that these organisms were deserving of special attention because of the high content of nucleic acids in their cells. During the years from 1939 to 1947, the systematic studies of the content of nucleic acids in bacteria of various taxonomic families, of different ages, and under different physiological conditions were performed in both subgroups headed by Belozersky (4). The high level of nucleic acids in cells was postulated to be in direct relation to their biological activities, growth rate, and cell proliferation.
I joined the group in 1954 as a graduate student, formally at the Institute of Biochemistry of the Academy of Sciences, but the place of my experimental work was in the well equipped new building of the Biological Faculty at the Moscow State University. By that time, the Journal of Biological Chemistry had published a series of papers by Chargaff and colleagues in which the first convincing results that the base composition of nucleic acids can vary in different organisms were presented (5–8). Crick and Watson had just published their famous papers on DNA structure and its implications for gene duplication and transcription into RNA (9, 10). The following questions had arisen. What is the range of variations of base compositions of DNA and RNA in different organisms? Does the total RNA just copy the total DNA of the cell, thus repeating its base composition, or do DNA-independent fractions of RNA exist? In 1956, I started work on testing the idea of the presumable correlation between the base compositions of RNA and DNA. The result was unexpected: the total DNA base composition manifested wide species variations, whereas the RNA composition was found to be surprisingly conserved (11). At the same time, statistical analysis of the data showed that a positive correlation of the base compositions of total RNAs of different species with their DNA compositions does exist, although at a low regression value (12). The data were interpreted in such a way that a relatively small fraction of species-specific (i.e. gene-specific) RNA exists at the background of the main mass of evolutionarily conserved (presumably “non-genetic” or non-coding) RNA, which consists for the most part of ribosomal RNA. These results and interpretations were widely discussed in the literature and at conferences, in particular by F. H. Crick (13), F. Jacob and J. Monod (14), C. Levinthal (15), S. Spiegelman (16), and M. Yčas (17). To illustrate the situation in those years, two citations are given below.
The coding problem has so far passed through three phases. In the first, the vague phase, various suggestions were made, but none was sufficiently precise to admit disproof. The second phase, the optimistic phase, was initiated by Gamov in 1954, who was rash enough to suggest a fairly precise code. This stimulated a number of workers to show that his suggestions must be incorrect and, in doing so, increased somewhat the precision of thinking in this field. The third phase, the confused phase, was initiated by the paper of Belozersky and Spirin in 1958… The evidence presented there showed that our ideas were in some important respects too simple. (cited from Ref. 13, p. 35)
It has long been believed that structural information was transferred from the genes to stable templates, such as ribosomal RNA, copied along the genes and maintaining in the cytoplasm the information necessary for protein synthesis. Every gene was supposed to determine the production of a particular type of ribosomal particles which in turn ensured the synthesis of a particular protein (see Crick, 1958 (18)). In recent years, however, this hypothesis has encountered several difficulties. 1. The diversity of base composition found in the DNA of different bacterial species is not reflected in the base composition of ribosomal RNA (Belozersky and Spirin, 1960). (cited from Ref. 14, p. 195)
Thus, Belozersky and I found ourselves among the pioneers of messenger RNA studies. Our results were the first indications that only a small fraction of total RNA of normal (non-infected) cells copies DNA (genes) and, hence, could be supposed to play the role of messenger (as such an RNA was named by Jacob and Monod (19)) from DNA to proteins (i.e. to be a coding RNA). In 1962, Belozersky retired from his position as head of the laboratory at the Institute of Biochemistry, which I inherited. Together with my colleagues, we decided to move our investigations to the study of mRNA in eukaryotic (animal) cells. Using a new object, fish embryos, we made another discovery, that mRNA in eukaryotic cells does not exist in a free form, but, even when it is not engaged in translation, it is present in the form of messenger ribonucleoproteins (RNP particles) with a characteristic protein/RNA ratio of ∼3:1 (20, 21). These messenger RNP particles were named informosomes. In oogenesis and early embryogenesis, the RNP particles were proposed to be a masked form of mRNA (22). Many years later, N. Standart, T. Hunt, and associates presented one of the most elegant experimental proofs of this proposal (23, 24).
Major Non-coding RNA That Forms Ribosome Structure
Yet as the great bulk of the cellular RNA was implied to be a non-coding RNA (11, 12), my interest was shifting to the structural and functional characteristics of this substance. As the RNA of ribosomes was already known to comprise at least 80% of the total RNA of a bacterial cell, it was quite evident that the major non-coding RNA should be ribosomal RNA. This expectation was confirmed by the analyses of base compositions of RNA-containing fractions of bacterial cells conducted by several groups (25–27). Our first contribution to the understanding of ribosomal RNA was the demonstration that its high-molecular-weight molecules are constituted of a single covalently continuous polyribonucleotide chain each (28–31) but are not composed of smaller RNA subunits, as had been assumed previously (32–35).
Self-folding of Ribosomal RNA into Specific Compact Particles
The discovery of the self-folding of the high-polymer polyribonucleotide chains into specific compact globular bodies was the principal achievement that attracted us to further studies of ribosomes. First, it was demonstrated that the conformation of a high-polymer RNA can change from the state of an unfolded flexible chain in the absence of Mg2+ at low ionic strength to the state of more compact rod-like particles, still flexible but possessing a developed secondary structure in the presence of Mg2+ at moderate ionic strength, and further to the state of well shaped compact globules at elevated Mg2+ concentrations and ionic strengths (Refs. 36–38; see also Refs. 30 and 31). Later, my colleague V. D. Vasiliev and associates showed that electron microscopy images of two species of isolated ribosomal RNA (16 S and 23 S) in the compactly folded state are different in their shapes and strongly resemble the images of isolated 30 S and 50 S ribosomal subunits, respectively (39, 40). This led us to boldly assert that the specific shape and gross structure of ribosomal particles are determined by self-folding of their high-polymer ribosomal RNAs (41). More recently, this assertion was confirmed by direct x-ray structural analyses of ribosomes (“The shape [of the 30S ribosomal particle] is largely determined by the RNA component; none of the gross morphological features is all protein.” (cited from Ref. 42)). Thus, ribosomal RNA could be considered as the structural core of ribosomal particles.
Conformational Mobility of Ribosomal RNA and Ribosomes
In addition to the structure-forming capacity of ribosomal RNA, the high conformational mobility of the folded RNA depending on ionic conditions, temperature, and the presence of some solutes seemed to be an intriguing property of the RNA in light of its possible functional role in the ribosome (43). When compact ribosomal particles were exposed to the same physical and chemical conditions that were used in the RNA studies, they exhibited a similar conformational response. Depletion of Mg2+ caused stepwise unfolding of ribosomal subunits through several discrete intermediate states without loss of ribosomal proteins, thus demonstrating the scaffold role of ribosomal RNA in the ribosome structure, on one hand, and the possibility of conformational mobility of ribosomal particles without their destruction, on the other (44, 45).
Self-assembly of Ribosomal Proteins on Ribosomal RNA
Another type of reversible structural transformation of ribosomal particles in vitro was shown upon their exposure to high ionic strength in the presence of Mg2+ (46, 47). Under these conditions, ribosomal proteins dissociated from ribosomal RNA in a stepwise manner while the compactness of the particles and their gross morphology remained the same (see also Ref. 41). This stepwise disassembly of compact ribosomal particles was found to be reversible, with restoration of their biological activity (47, 48). The experiments on successful reassembly (reconstitution) of biologically active ribosomal particles were simultaneously published by the groups of M. Nomura (49) and M. Meselson (50). (It is noteworthy that 3 years before, preliminary results on the in vitro assembly of ribosome-like particles from ribosomal RNA-containing “CM-particles” and cell lysate proteins were obtained and reported at the Cold Spring Harbor Symposium (51).)
Conformational Movements in Translating Ribosome
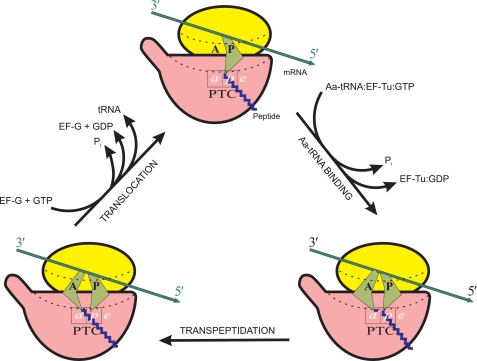
The function of the ribosome is to translate the genetic information encoded in the nucleotide sequences of mRNA into amino acid sequences of polypeptide chains of proteins. During the process of translation, the ribosome performs the unidirectional driving of tRNA macromolecules through itself and the coupled drawing of the mRNA chain from its 5′- to 3′-end. In the course of translation, the free energies of the transpeptidation reaction and the GTP hydrolysis reaction are consumed (Fig. 1). Thus, the translating ribosome can be considered as a conveying molecular machine, simultaneously being a “technological” protein-synthesizing machine. Obviously, the ribosome as a conveying machine must be capable of performing its own mechanical movements. More than 2 decades ago, it was proposed that the functional movements of the translating ribosome are based on the overall construction of the ribosome, allowing certain anisotropic motions generated by thermal Brownian movements of large blocks of the ribosome and the ribosomal subunits (52). These ideas were further developed in subsequent publications (53–55).
FIGURE 1.
General schematic representation of the elongation cycle of a translating ribosome. Each cycle results in (i) elongation of a growing peptide chain by one amino acid residue (formation of one peptide bond), (ii) hydrolysis of two GTP molecules, (iii) entry of one molecule of aminoacyl (Aa)-tRNA into intersubunit channel, (iv) exit of one molecule of deacylated tRNA from the intersubunit channel, and (v) movement of mRNA chain by three nucleotides toward the 3′-end. Here, as well as in Figs. 3–5, ribosomal particles are shown in the orientation in which the small subunit (yellow) is on the top and the large subunit (red) is on the bottom. The head of the small subunit and the central protuberance of the large subunit are facing the viewer, with the L7/L12 stalk of the large subunit directed to the left. In this orientation, the bound L-like tRNAs must face the viewer by their external angles (“elbows”), as in reality they are facing the head of the small subunit; however, here, as well as in Figs. 3–5, for the sake of better discerning between the A and P site tRNAs, the external angles of their symbolical depictions (green) are shown to be drawn apart. The intersubunit channel accommodating the mRNA and tRNAs is traced by dotted lines. A and P are the tRNA-binding sites on the small subunit, and a, p, and e are the binding sites for their 3′ termini, either acylated or deacylated, on the large subunit. (For the sake of better clarity, the subsites on the small subunit are designated by capital letters, A and P, as originally proposed and usually accepted (59, 94), whereas the large subunit subsites, which are localized within a small area of the PTC and nearby, are designated by lowercase italicized letters, a and p, as proposed elsewhere (92).)
Intersubunit Movements
The fact that ribosomes are universally built from two loosely associated and easily separable subunits in all living beings is one of the most fascinating properties of the translation machinery. The two subunits (Fig. 2), the small one (the so-called 30 S in prokaryotes or 40 S in eukaryotes) and the large one (50 S in prokaryotes or 60 S in eukaryotes), have different functions: the small subunit is responsible for the “genetic” functions of the ribosome, such as binding of mRNA and decoding of genetic information, whereas the large subunit acts as its “catalytic” partner, being responsible for the formation of peptide bonds and the attraction of protein catalysts for GTP hydrolysis. Thus, a clear division of labor exists between the two ribosomal subunits. It is remarkable that none of the subunits alone is capable of performing the coupled unidirectional movement of mRNA and tRNA, the conveying function designated as translocation. On the basis of the above knowledge, I proposed that (i) the main functional purpose of the two-subunit construction of the ribosome is the organization of the translocation mechanism of the ribosome, (ii) translocation requires mutual mobility of the ribosomal subunits, and (iii) translocation proceeds through an intermediate state when the products of the transpeptidation reaction (peptidyl-tRNA and deacylated tRNA) occupy positions with shifted 3′-ends of tRNAs on the large subunit but yet non-shifted codon-anticodon duplexes on the small subunit (56–58). Similar ideas were published at the same time by M. S. Bretscher (59), although the two models differed in detail.
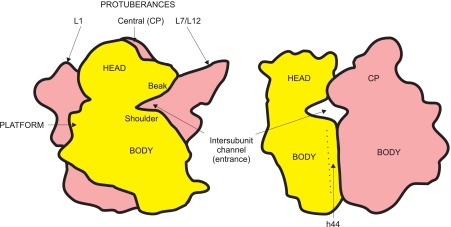
FIGURE 2.
Contours of two ribosomal subunits, the small 30 S (yellow) and the large 50 S (red), associated in the full 70 S ribosome, with designations of some morphological features. Left, the so-called overlap projection when the 30 S subunit is facing the viewer and covers part of the 50 S subunit; right, the lateral projection viewed from the side of the L7/L12 stalk. CP, central protuberance.
The mechanistic principle of my model was based upon the idea that the associated subunits of the translating ribosome pass through the stage of an “unlocked” (open) ribosome. Thus, the ribosome was considered as a particle oscillating between “locked” (closed) and unlocked (open) conformations. The unlocked states were proposed to be required both at the aminoacyl-tRNA binding step to allow the large substrate (aminoacyl-tRNA) to enter into the intersubunit space of the ribosome and at the translocation step to facilitate the products' movement inside the ribosome (peptidyl-tRNA) and exit from the ribosome (deacylated tRNA). Experimental testing of the hypothesis was delayed, however, for many following years because of the lack of adequate methodologies to study the dynamics of macromolecular complexes.
Nevertheless, the attempts to detect macroconformational changes within ribosomes during translation were undertaken from time to time. The first experimental evidence in favor of intraribosomal conformational mobility of the translating ribosome came from comparison of the compactness of the particles before and after translocation. It was shown by sedimentation analysis that the sedimentation coefficient of the post-translocation ribosome is somewhat less than that of the pre-translocation ribosome (the difference was ∼1 S) (60). However, this fact could be explained either by changing the composition of the ribosome as a result of translocation (loss of the deacylated tRNA molecule) or by a conformational change, such as some “swelling” of one of the ribosomal subunits or of the ribosome as a whole (the latter could result from a widening of the intersubunit space). Neutron scattering in various mixtures of H2O and D2O allowed these alternatives to be distinguished. The point is that the RNA component of the ribosome becomes contrast-matched, i.e. “invisible” for neutrons, in a solvent with a proper proportion of light and heavy water (70% D2O). This enabled the measurement of compactness (radius of gyration (Rg)) of only the protein component of the ribosomal particle, irrespective of the number of bound tRNA molecules. It was found that the Rg of the protein component of the post-translocation ribosome was somewhat greater than that of the pre-translocation ribosome (ΔRg = 1–3 Å) (61, 62). In other words, translocation made the whole ribosome slightly less compact. These experiments were the first physical evidence of a conformational change in the translating ribosome as a result of translocation. However, they did not answer the question of whether the slight decrease in ribosome compactness upon translocation reflects an intersubunit change or a conformational alteration within one of the subunits. Further neutron-scattering experiments, including those with selectively deuterated ribosomal subunits (either 30 S or 50 S), led to the conclusion that conformational changes within the small subunit made a major contribution to the effect of the increase in the Rg of the full ribosome upon translocation (see “Intrasubunit Large-block Mobility”) (61, 63).
Recent developments in the cryoelectron microscopy technique allowed J. Frank and colleagues to demonstrate a real intersubunit movement coupled with translocation: they detected a rotational shift of one ribosomal subunit relative to the other around an axis perpendicular to the subunit interface (64, 65). This rotation of the small subunit relative to the large subunit was estimated to be ∼6° counterclockwise if viewed from the small subunit. The rotation was accompanied by a widening of the intersubunit mRNA channel (64). The observation of such a rotation was confirmed in the studies by H. F. Noller's group using a cross-linking technique, fluorescence resonance energy transfer (FRET) methodology, and “translation-libration-screw” (TLS) crystallographic refinement (66–68). The ribosome was shown to be fixed (locked) in the rotated form upon binding of elongation factor (EF)-G, the catalyst of translocation, until EF-G and deacylated tRNA were released from the ribosome (64, 69). More recently, using single-molecule FRET methodology, it was found that ribosomes undergo spontaneous intersubunit movements oscillating between the original (“classical”) and rotated forms, with the equilibrium shifted toward either the original or rotated forms depending on the functional state of the ribosome (70). The following conclusions can be made from the recent FRET data. (i) Vacant ribosomes thermally oscillate between the original and rotated forms with relatively low forward and reverse rotation rates; the equilibrium is somewhat shifted toward the original form (the proportion of the two forms in the equilibrium mixture is ∼3:2). (ii) The binding of N-acylated aminoacyl-tRNA to the P site somewhat reduces the forward rotation rate and correspondingly slightly shifts the equilibrium toward the original (non-rotated) form, but still the proportion of the rotated form in the equilibrium mixture may be significant (up to one-third). (iii) The occupancy of the A site by N-acylated aminoacyl-tRNA and the P site by deacylated tRNA, which models the translating ribosome after transpeptidation, induces rapid oscillation of ribosomes between the original and rotated forms, with the equilibrium shifted to the rotated form; this state should correspond to the pre-translocation state ribosome. (iv) The binding of the translocation catalyst EF-G with a non-hydrolysable GTP analogue, when deacylated tRNA still remains in the P site (the situation that simulates the first step of translocation), fixes the rotated form of the ribosome. (v) The transition to the final post-translocation state, after GTP hydrolysis and the release of EF-G and deacylated tRNA, when peptidyl-tRNA occupies the P site and the A site becomes vacant, to some extent restores the situation mentioned in Conclusion (ii), but with somewhat higher rates of both the forward and reverse reactions. Thus, both pre-translocation and post-translocation state ribosomes in the absence of elongation factors oscillate between the original (classical) and rotated forms. The presence of deacylated tRNA in the P site after transpeptidation strongly stimulates the rates of both forward and, to a less extent, reverse rotational shifts of ribosomal subunits, shifting the equilibrium toward the rotated form. The binding of EF-G fixes the rotated form.
The properties of the rotated form of the ribosome, such as the high rate of oscillation between the alternative conformations (in the absence of EF-G), the permissibility of translocational intraribosomal shifts of peptidyl-tRNA and deacylated tRNA, and the competence to accept EF-G as a translocation catalyst (64, 65, 69), as well as the widening of the intersubunit mRNA channel (64), imply that the rotated form is equivalent to the unlocked state proposed earlier for the hypothetical intermediate in ribosomal translocation (56–58). It is noteworthy that more than 2 decades ago, based on general considerations of a number of facts concerning translocation, the following was written (cited from Ref. 52): “An equilibrium exists between the locked and unlocked states of the ribosome: the ribosome fluctuates between two states,” and “Attachment of EF-G with GTP fixes a certain ”unlocked“ state of the ribosomal complex where a greater freedom is allowed for diffusional movements of tRNA ligands, within the limit assigned by the construction of the ribosome.”
Intrasubunit Large-block Mobility
More than 2 decades ago, the unique conformations of two specifically folded high-polymer ribosomal RNAs were proposed to underlie the specific anisotropic motility of structural blocks of the ribosomal subunits, this being the structural basis for functional intrasubunit movements (52). This mostly intuitive statement was presented and further developed in a number of subsequent publications (53–56). Recently, the analysis of structural dynamics in the ribosome by TLS crystallographic refinement directly confirmed the assertion (68). In accordance with the dense mutual packing and interpenetration of the RNA domains in 23 S ribosomal RNA of the large ribosomal subunit (71), as compared with the loosely arranged and weakly interacting RNA domains of 16 S RNA of the small subunit (42), the two subunits were found to manifest very different levels of thermal mobility of their blocks. The main body of the large subunit proved to be almost monolithic, with low levels of structural fluctuations inside. At the same time, two peripheral protuberances of the large subunit, the block of helices H43-H44 with proteins L11-L10-(L7/L12)4 (the so-called L7/L12 stalk) and the block of helices H76-H77-H78 with protein L1 (Fig. 2, left), demonstrated extremely high levels of thermal mobility (68). As for the small ribosomal subunit, high levels of thermal mobility were recorded in all domains of 16 S RNA (see below).
The first experimental demonstration of the independent high mobility of the L7/L12 stalk of the large ribosomal subunit was made in 1982 by my colleague A. T. Gudkov and associates, who studied isolated large ribosomal subunits and whole 70 S ribosomes using NMR (72). As a matter of fact, that was the first case of experimental detection of the large-block mobility in the ribosome. The functional significance of the mobility of the L7/L12 stalk followed from the fact that the attachment of EF-G, the catalyst of translocation, to the ribosome resulted in immobilization of the stalk (73). Parallel immunoelectron microscopy studies of cross-linked functional complexes of elongation factors (EF-G and EF-Tu) with ribosomes performed by my colleagues A. S. Girshovich and V. D. Vasiliev and associates visualized the catalyst proteins at the L7/L12 stalk and its base on the large ribosomal subunit (74, 75). The following footprinting analyses made by H. F. Noller and colleagues revealed the regions of 23 S ribosomal RNA that were protected by bound EF-G; they proved to be helices H43-H44 of the 23 S RNA, which serve as the base of the L7/L12 stalk (76). Thus, the movable L7/L12 stalk was found to be involved in binding of elongation factors to the ribosome, leading to immobilization of the stalk. Cryoelectron microscopy observations demonstrated that the GTP-induced binding of EF-G, as well as EF-Tu, to the ribosome is accompanied by positioning of the L7/L12 stalk closer to the central protuberance of the 50 S ribosomal subunit; this movement was proposed to be part of a general mechanism of loading translation factors into the ribosome's factor-binding site (77, 78).
The mobility of the other side protuberance of the large ribosomal subunit, the L1-H76-H77-H78 block, was revealed from cryoelectron microscopy reconstructions and x-ray crystallographic studies of the ribosomes in different functional states and first proposed and then proved to be involved in the displacement and exit of deacylated tRNA during the final stage of translocation (65, 78–82). Most recently, in elegant experiments using single-molecule FRET, the real-time dynamics of the L1 protuberance was followed, and its movement relative to the body of the large ribosomal subunit was demonstrated (83); three distinct conformational states, open, half-closed, and fully closed, were observed.
As already mentioned, the first experimental evidence for a functional intrasubunit conformational change in the small (30 S) ribosomal subunit came from the neutron-scattering experiments of Serdyuk et al. (Ref. 63; see also Ref. 61). It was proposed that “the movement of the head [of the small ribosomal subunit] relative to the passive 50S subunit is the main mechanical act of translocation” (63). Recently, two different conformations of bacterial 70 S ribosomes (designated I and II) were revealed by x-ray crystallographic analysis; the main difference was that the head of the small subunit in the type II ribosome compared with the type I ribosome was rotated as a rigid block around the neck in the direction of the mRNA- and tRNA-conveying path during translocation, i.e. counterclockwise if viewed from the top of the head (82). The rotation was estimated to be up to 12° or ∼20 Å at the subunit interface. Independently, cryoelectron microscopy reconstructions of eukaryotic 80 S ribosomes demonstrated the rotational movement of the head relative to the body of the small ribosomal subunit upon binding of eEF2 in the rotated (“ratcheted”) form of the ribosome (78, 84). Thus, rotation of the head of the small ribosomal subunit was really shown to be coupled with translocation.
The analysis of the dynamics of thermal structural movements in the small ribosomal subunit by the TLS crystallographic refinement method (68) showed that the most movable region of this subunit is the block of helices h30–h34 of domain III of 16 S RNA, which forms the so-called “bill” or “beak,” the prominent part of the head at the entrance into the intersubunit channel (Fig. 2). As was demonstrated by x-ray crystallographic analysis, the anisotropic displacement of this structural block is realized in the process of aminoacyl-tRNA binding to the A site of the ribosome: upon binding of the anticodon hairpin of tRNA in the A site, this block moves toward the “shoulder,” the part of the body at the other side of the entrance into the intersubunit channel (85, 86). As a result, the anticodon hairpin is found to be occluded (locked) in the A site.
Another highly movable part of the small ribosomal subunit is the minor 3′-terminal domain of 16 S RNA (helices h44-h45). Helix H44, the longest hairpin of the subunit that ranges from the head to the end of the body and forms a number of important contacts with the large subunit, displays a tendency for rotational motions around an axis approximately parallel to its long axis and located at the subunit interface (68). This rotational movement may play a pivotal role in the mutual mobility of the ribosomal subunits and in translocation (65, 81, 82, 87).
A significant mobility of the side lobe formed by domain II of 16 S RNA, the so-called platform, relative to the rest of the small subunit was also shown (68, 88). The functional role of this motion may be associated with the processes of mutual movements of the ribosomal particle and mRNA during initiation of translation (89, 90).
Locking-Unlocking Principle (Closed and Open Conformations)
The principle of most functional conformational movements seems to be simple: movements are based on thermal anisotropic fluctuations, where the anisotropy is determined by the structure of a movable body and its environment, and ligand binding induces fixation of one of the alternative conformations in a less movable state. This can be called “locking,” “induced fit,” or “maximization of non-covalent bonds” between a ligand and surrounding groups, and it is usually accompanied by closing of a binding pocket around a ligand. The binding of the codon-cognate anticodon helix (anticodon stem-loop) to the A site on the 30 S ribosomal subunit described by Ogle et al. (85, 86) in terms of transition from an open to a closed form is a remarkable example of such a locking. Induced fit allowing maximization of contacts is considered there as a physical mechanism of the selectivity of codon-directed binding of a cognate tRNA in the tRNA-binding pocket. The open (unlocked) form should be more relaxed (fluctuating), whereas the closed (locked) form seems to be more rigid. The situation may also be considered as an oscillation between two (or more) alternative local conformations, with the equilibrium shifted toward the open form when a ligand is absent, whereas the presence of the proper ligand (cognate anticodon stem-loop) shifts the equilibrium to the closed form and reduces the rate of the reverse reaction.
The same principle can be applied to all functional movements in the translating ribosome. The ribosome (in the absence of bound EF-G) oscillates between locked (classical, closed) and unlocked (rotated, open) conformations, with the equilibrium positions and the forward and reverse reaction rates being dependent on its functional state (70). In the post-translocation state, the oscillation rate is not high but is essential, and the equilibrium is shifted, to a greater or lesser extent, toward the locked (closed or non-rotated) conformation. In the pre-translocation state, the oscillation rate is much higher, and the equilibrium is shifted to the unlocked (open or rotated) conformation.
In all cases, the binding of a GTP-bound translation factor results in immobilization of the L7/L12 stalk and formation of a closed pocket with participation of the adjacent tRNA entrance region (induced fit, locked local conformation), which results in the selection and fixation of an unlocked conformation of the ribosome. Hydrolytic cleavage of the factor-bound GTP leads to relaxation of the closed conformation of the factor (unlocking of its domains), the loss of its high affinity for the L7/L12 pocket, and thus a local unlocking event (relaxation of the pocket), allowing some movements at the subunit interface (see below). However, the temporary presence of the factor in the relaxed GDP form may still prevent the return to the original (locked) conformation of the ribosome. The spontaneous release of the factor with GDP from the ribosome allows returning to the equilibrium situation with the prevailing locked conformation of the translating ribosome.
It is logical to infer that the binding of aminoacyl-tRNA to the A site of the post-translocation ribosome should occur with an unlocked conformation of the post-translocation ribosome and lead to fixation of a locked conformation, first of all, because of formation of an additional strong bridge between the two ribosomal subunits. On the other hand, it seems evident that the transpeptidation reaction requires the fixed locked conformation to firmly position the aminoacyl residue in the immediate vicinity of the peptidyl group of the P site peptidyl-tRNA. Thus, transpeptidation should occur in the locked state ribosome. As a result of transpeptidation, two strong intersubunit bridges become disrupted: the P site tRNA is now deacylated (Fig. 1) and cannot be further retained in the donor p-site of the peptidyltransferase center (PTC) on the large ribosomal subunit, whereas the A site tRNA has lost the affinity for the acceptor a-site of the PTC. Then, the ribosome is allowed to be unlocked.
Stepwise Conveyance of tRNA and mRNA through the Translating Ribosome
In considering the ribosomal translocation phenomenon, the process of passing of mRNA through the translating ribosome is usually viewed as a passive driven movement, whereas tRNA translocation is regarded as an active driving act. Indeed, triplet-by-triplet movement of mRNA is fully determined by unidirectional movements of the tRNA anticodons bound to their cognate codons in the ribosome. However, it is rather thermal Brownian motions that provide a translating ribosome with all driving forces so that the “driving” function of tRNA should be understood conditionally: codon-anticodon duplexes move as whole units, but it is tRNA residues that are fully responsible for successive step-by-step fixation along the ribosomal conveyer path.
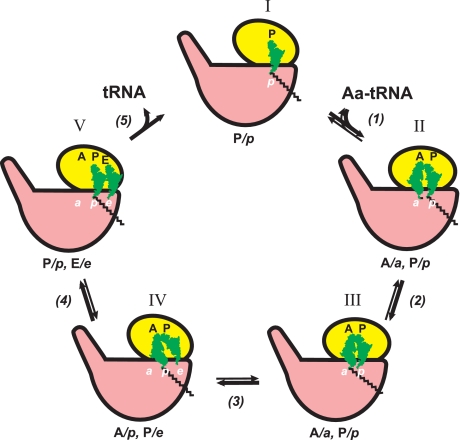
Whereas the mRNA chain extends along the mRNA- binding groove exclusively on the small ribosomal subunit, the tRNA molecules, including aminoacyl-tRNA, peptidyl-tRNA, and deacylated tRNA, occupy discrete sites, each of them being shared between two ribosomal subunits. Hence, each tRNA-binding site is subdivided into two subsites: the small subunit subsites accommodate anticodon arms, whereas the large ribosomal subunit interacts with the acceptor ends (Fig. 1). The positions of the aminoacyl-tRNA and peptidyl-tRNA prior to transpeptidation can therefore be designated by two letters, such as A/A and P/P (91) and A/a and P/p (92), respectively (Fig. 1, lower right). In both models of translocation proposed 4 decades ago (56–59), intersubunit movement was considered as a mechanism required for translocation of peptidyl-tRNA and deacylated tRNA, and an intermediate state in the process of the movement of the peptidyl-tRNA from the A/a to P/p position was postulated. The intermediate state was assumed to be the result of the high affinity of the newly formed CCA-aminoacyl-CO- grouping for the p-site of the PTC (58): “Having a high affinity for the corresponding neighboring site of the peptidyl-transferase center, it [the grouping] can spontaneously pass to this site (peptidyl translocation) and get firmly hooked there.” In other words, the spontaneous transition of the product peptidyl-tRNA from the A/a position to the intermediate A/p position was supposed and considered to be a prerequisite for subsequent EF-G/GTP-driven translocation of the rest of the tRNA (56–58). The fact that translocation of tRNA molecules does proceed stepwise through a discrete intermediate state was established in a series of chemical footprinting studies by D. Moazed and H. F. Noller (91, 93, 94). The main finding was that, after the transpeptidation reaction between aminoacyl-tRNA in the A site and peptidyl-tRNA bound in the P site, the acceptor ends of the products (elongated peptidyl-tRNA and deacylated tRNA) are not firmly retained in their previous positions on the large subunit, a and p, respectively (Fig. 1, lower left), but tend to spontaneously migrate from a to p and from p to e, respectively (see Fig. 3, step 1). As a result, the products of the transpeptidation reaction are found in the so-called “hybrid” positions, A/p and P/e. (The term was introduced earlier in the model proposed by M. S. Bretscher (59).)
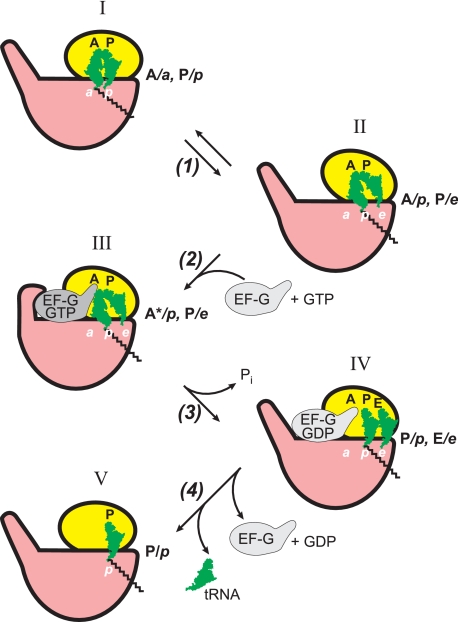
FIGURE 3.
Proposed sequence of events during EF-G/GTP-promoted translocation in terms of tRNA ligand positions and the ribosomal locking-unlocking (closing-opening) concept. The ribosomes in the unlocked form (where in reality the small subunit is rotated relative to the large subunit around the axis perpendicular to the subunit interface) are conditionally depicted with the small subunits somewhat rotated relative to the large subunits in the plane of the figure (positions II–IV).
The process of EF-G/GTP-catalyzed translocation including intermediate hybrid state and ribosome locking-unlocking is schematically presented in Fig. 3. As mentioned above, transpeptidation results in the disruption of chemical groups responsible for firm retention of the ribosomal subunits in the locked state so that such a destabilization should lead to restoration of the equilibrium between the locked (“non-rotated”) and unlocked (“rotated”) conformations of the ribosome. Thus, it seems that the spontaneous shift of the acceptor ends of the product tRNA residues and the appearance of the hybrid state after transpeptidation are allowed because of establishment of the locking-unlocking equilibrium (step 1). Indeed, cryoelectron microscopy studies and chemical footprinting and FRET analyses showed that the hybrid situation correlates with the rotated state of the ribosome (65, 67, 69, 70). It is remarkable that binding of EF-G with a non-cleavable GTP analogue (that is EF-G with GTP prior to GTP hydrolysis) was shown to fix the rotated state of the ribosome and the hybrid positions of peptidyl-tRNA and deacylated tRNA (“locking of the unlocked state of the ribosome”) (step 2, position III). (The asterisk with A in position III indicates that the binding state of the tRNA residue in the A site of the small subunit is somehow distorted by the intervention of the elongated domain IV of EF-G.) The hydrolysis of GTP by the ribosome-bound EF-G (step 3) seemingly does not reverse the ribosome to the original (non-rotated) form until EF-G is released from the ribosome. At the same time, the hydrolysis of GTP and the following release of orthophosphate from EF-G lead to relaxation of the rigid domain structure of EF-G and to interdomain rearrangements, allowing slippage of the two codon-anticodon duplexes from the A and P sites to the P and E sites on the small ribosomal subunit and thus establishing the P/p-E/e situation in the unlocked (rotated) form of the ribosome (position IV) (95, 96). Subsequent release of the weakly bound EF-G with GDP (step 4) permits the reverse transition of the ribosome into the original (non-rotated) form with peptidyl-tRNA and deacylated tRNA in the P/p and E/e positions, respectively, and then the spontaneous release of deacylated tRNA, establishing the final post-translocation state (position V).
It should be mentioned that the scheme in Fig. 3 is simply first approximations, and it is likely that the processes involved are more complex. In particular, the passing through intermediate positions and unlocked (rotated) conformations of the translating ribosome (Fig. 3, steps 2 and 3) may include additional short-lived intermediates and transition states (see, for example, Refs. 97 and 98).
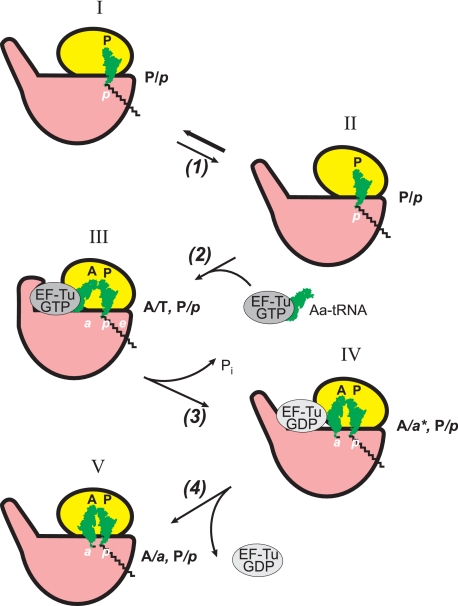
An analogous sequence of events, including passing through an intermediate hybrid state, seems to be realized during the process of EF-Tu-promoted entry of aminoacyl-tRNA into the empty A site of the post-translocation state ribosome (Fig. 4) (Refs. 77 and 91; see also Ref. 92 for review). However, the rotated form of the unlocked ribosome seems to be not involved in the process of aminoacyl-tRNA binding: the cross-link between protein S6 (at the platform of the small ribosomal subunit) and protein L2 (at the nearby L1 side of the large subunit; see Fig. 2) that blocks the rotational movement between the subunits does not prevent either EF-Tu-dependent binding of aminoacyl-tRNA or its factor-free binding (66). An unlocked form of the ribosome that is supposedly required for the entry of aminoacyl-tRNA into the intersubunit space (56–58) may be realized because of the open (unlocked) state of the empty A site of the small ribosomal subunit, when the beak and shoulder at the entrance of the intersubunit channel (Fig. 2) are slightly drawn apart (85, 86). Also, the involvement of the rotational movement of the small subunit head in unlocking the intersubunit channel cannot be excluded. In any case, a codon-cognate aminoacyl-tRNA·EF-Tu·GTP complex is selected such that its anticodon arm interacts with mRNA in the A site of the small ribosomal subunit, and the protein moiety binds in the factor-binding pocket at the base of L7/L12 stalk of the large subunit (Fig. 4, step 2). This leads to closing (locking) of the pocket around EF-Tu and setting of aminoacyl-tRNA in an intermediate A/T hybrid state (position III). Next, the ribosome-induced hydrolysis of GTP on EF-Tu (step 3) results in the loss of contacts of EF-Tu with the acceptor arm of aminoacyl-tRNA and weakening of its interactions with the large ribosomal subunit. The released aminoacylated CCA end of aminoacyl-tRNA moves away and approaches the a-site of the PTC of the large subunit (position IV). (The asterisk with a in position IV indicates that the binding state of the aminoacylated end in the a-site of the large subunit is not yet competent for the reaction with the donor substrate, peptidyl-tRNA.) The release of the weakly bound EF-Tu·GDP complex (step 4) leads to the A/a-P/p state in the locked ribosome, with tightly closed reacting groups. Now the ribosome is ready to catalyze the transpeptidation reaction between the properly settled substrates (position V).
FIGURE 4.
Proposed sequence of events during EF-Tu/GTP-promoted binding of aminoacyl-tRNA in terms of tRNA ligand positions and the ribosome locking-unlocking (closing-opening) concept. Aa, aminoacyl.
GTP-dependent Catalysis of Conformational Transitions
The discovery by Y. Nishizuka and F. Lipmann in 1966 of EF-G as a ribosome-dependent GTPase catalyzing translocation (99, 100) led to the hypothesis that translocation as an act of mechanical work used for movement of mRNA and tRNAs is driven by the free energy from the GTP hydrolysis reaction (101). Translocation was considered as useful work in a thermodynamically uphill process. The hypothesis had a strong influence on the scientific community and was widely accepted. However, very soon, several reports appeared in which translocation was observed in cell-free translation systems in the absence of EF-G and GTP (102–105). Nevertheless, some doubts about the purity of the ribosomes and/or other constituents of the translation mixtures remained. The breakthrough, quite unexpected, was made in my laboratory when the sulfhydryl group reagent p-chloromercuribenzoate (pCMB) was added to the translation mixture to inactivate possible traces of EF-G: it was found that the presence of the reagent strongly stimulated spontaneous translocation (106). Moreover, pretreatment of bacterial ribosomes or just small ribosomal subunits with pCMB produced the same stimulation effect (107). Furthermore, it was demonstrated that the stimulation of the factor-free translocation was caused by pCMB modification of ribosomal protein S12 (108) and that the same effect could be achieved by removal of protein S12 from the ribosome (109). Since then, the phenomenon of factor-free, GTP-independent translocation has been fully acknowledged.
Based on the discovery of factor-free translocation and the previous knowledge of factor-free binding of aminoacyl-tRNA to the mRNA-programmed ribosome, several variants of factor-free and one factor-promoted (either EF-Tu-promoted or EF-G-promoted) translation systems were invented and used in laboratory practice (110–113). On the other hand, theoretical considerations led to principal conclusions concerning the mechanism and energetics of translocation (Refs. 114 and 115; see also Refs. 52 and 53). First, translocation in the absence of EF-G clearly showed that the molecular mechanism of translocation is intrinsic to the ribosome itself and not introduced by EF-G. Second, translocation without GTP as an energy substrate provided evidence that translocation is a thermodynamically spontaneous (downhill) process and, hence, principally does not require energy to be performed. Third, the realization of the full elongation cycle without both elongation factors and any other energy source except aminoacyl-tRNA proved that the transpeptidation reaction in the ribosome must be the only source of energy to drive the elongation cycle. The latter implied that the free energy of the transpeptidation reaction is accumulated in the pre-translocation state ribosome (the products are not released yet!), which makes this state thermodynamically unstable.
At the same time, the elongation factors with GTP strongly increase the elongation rate, and EF-G with GTP specifically accelerates translocation. Also, EF-Tu makes codon-dependent binding of aminoacyl-tRNA much faster compared with the slow, factor-free binding of the substrate. Thus, both elongation factors can be considered as enzyme-like catalysts of thermodynamically allowed, spontaneous processes (114, 115). However, there are two important peculiarities of the catalytic action of the elongation factors (as well as other GTP-dependent translation factors): (i) catalysis is coupled with GTP hydrolysis, and (ii) the processes catalyzed are not chemical reactions of covalent transformations but are instead the acts of conformational transitions.
Conformational flexibility and large-block mobility can provide conditions for association-dissociation (attachment-detachment) processes to pass through intermediate states with partially formed or, respectively, disrupted contacts between partner macromolecules. It is likely that intermediate states are required, first of all, to avoid a kinetic blockade in the case of extensive multicenter interactions between macromolecules. From this assertion, the possibility of catalysis of conformational rearrangements and transitions can be directly deduced. Similar to the enzymatic catalysis of covalent chemical reactions, which is based on the affinity of an enzyme for the transition state of the reaction, the catalysis of conformational rearrangements is made possible by virtue of the affinity of a protein (a catalyst of the rearrangement) for an intermediate conformational state. Elongation factors EF-Tu and EF-G, as well as other GTP-dependent translation factors (IF2 and RF3), can be considered to be such catalysts of conformational rearrangements of the ribosome.
The requirement for nucleoside triphosphates and their hydrolysis in the processes of conformational catalysis could be inferred from the following consideration. In the case of enzymatic catalysis of a covalent reaction, formation of the complex between an enzyme and a transition state intermediate is followed by decay of the intermediate and formation of the reaction products spontaneously released from the enzyme (due either to low affinity for the enzyme or to low concentration of the products in the medium). Hence, liberation of the enzyme upon completion of the reaction is paid for by the change in free energy of the catalyzed covalent reaction itself. In the case of catalysis of a conformational transition, the catalyzed process is usually not a reaction accompanied by a significant decrease of free energy of the system. This circumstance can be overcome by coupling the catalysis act with an exergonic chemical reaction, such as hydrolysis of a nucleoside triphosphate (54). Thus, when an elongation factor with GTP has an affinity for a conformational intermediate and binds to it, the detachment of the factor or its displacement will be required to complete the conformational transition. This will demand energy compensation at the expense of an exergonic process that would be capable of sufficiently lowering the free energy of the system. It is the coupled chemical covalent reaction of GTP hydrolysis that can provide energy for the factor detachment from a conformational intermediate through the change in its affinity.
In light of what is stated above, it is meaningful that translocation can be catalyzed by EF-G in vitro without GTP cleavage. In our early experiments, EF-G with a non-cleavable GTP analogue interacted with pre-translocation state ribosomes, and the subsequent removal of EF-G from the ribosomes by a physical washing-off procedure resulted in the appearance of post-translocation state ribosomes capable of continuing the elongation cycle (“translocation by attachment-detachment of EF-G”) (116, 117). From this observation, the main role of GTP cleavage in the process of translocation was suggested to be destruction of a ligand (GTP) that imparted affinity of EF-G for the ribosome and thus the removal of the factor from its complex with a translocational intermediate.
Coupling of conformational catalysis to hydrolysis of nucleoside triphosphates is not a phenomenon uniquely characteristic of only translation processes. Similar GTP- or ATP-dependent catalysis occurs in various processes in which acceleration of non-covalent macromolecular rearrangements by proteins with GTPase or ATPase activities, as in the case of chaperonins, DNA topoisomerases, RNA helicases, G-protein interactions, etc., is observed. These catalytic proteins can be considered as a special class of enzymes called “energases” (118). Also, ATP- or GTP-dependent catalysis of molecular displacements through alternating attachment and detachment of a moving part of a substrate takes place in molecular movement systems, such as myosin locomotion on actin filaments, kinesin and dynein locomotion on microtubules, and active transmembrane transports.
Brownian Motion, a Conformational Ratchet Wheel, and Energy-dependent Pawls in the Elongation Cycle of the Translating Ribosome
Peculiarities of Molecular Machines
The manifestations of physical laws in a micro-world can strongly differ from those in the macro-world. In particular, a number of principles that underlie the work of power-stroke macro-machines, such as the internal-combustion engine or the electric motor, cannot be realized at the molecular level. Three main features of molecular machines should be mentioned. (i) The first is the small masses of macromolecules and their complexes. From this fact, it follows that the structural blocks of molecular machines are practically inertialess and incapable of providing momentum conservation for longer than a fraction of a nanosecond (119). Indeed, flyweight wheels, pendulums, rectilinear inertial motions, and other inertia energy storage systems are not used in molecular devices. (ii) The second feature is the conformational flexibility of structural blocks and joints. Molecular bodies, such as proteins, nucleic acids, and their complexes, are made of flexible polymers with movable side groups, so they can hardly satisfy the requirements of mechanical accuracy. That is why it is unlikely that molecular machines can use rigid levers, cranks, hooks, axles, wheels, and other mechanical constrictions for force transmission from an engine to a mover. (iii) The third feature is Brownian motions and internal thermal self-oscillations of all parts of a molecular machine. As a result, the structural elements of molecular machines are not strictly fixed in space but rather undergo permanent conformational fluctuations. Therefore, all the work of molecular machines should have stochastic rather than mechanically determined character.
Hence, because neither mechanical energy nor high-precision mechanics can be realized at the molecular level, molecular machines, including machines of the conveying type, must be considered as constructions moving without mechanical engines, mechanical transmissions, and mechanical movers. They are based on quite different principles deduced from the above-mentioned features of molecular systems. Here, all of the following considerations will be based on Feynman's thermal ratchet model, in which the driving force for molecular directional movements is essentially Brownian motion but biased in a certain direction by using free energy released from chemical exergonic reactions (120–123).
Indeed, the engines of most molecular machines are fuelled by so-called high-energy compounds, usually ATP or GTP (or the product of high-energy group transfers from ATP, as in the case of aminoacyl-tRNA). As a rule, binding of a high-energy substrate to a specific site of the engine induces a transition from a fluctuating loose (unlocked, open) conformation of the site into a fixed (locked, closed) conformation due to formation of non-covalent bonds between the site and the substrate (induced fit). This conformational change is the first stroke of the engine. Next, the bound high-energy substrate is catalytically hydrolyzed (or chemically transformed in another way) at the binding pocket, the affinity of the reaction products for the pocket decreases, and the conformation again becomes mobilized (unlocked, open). This is the second conformational stroke of the engine. It is followed by the release of the reaction products (or their displacement to neighboring sites). All the displacements displayed require no special energy-dependent motive forces, such as directional pulling or pushing, but are randomly generated by Brownian motions and properly fixed by binding affinities.
As a paradigmatic example of the two-stroke action of a molecular engine, the locking-unlocking cycle of elongation factor EF-Tu (124–127) can be considered. Its globular molecule is composed of two blocks of approximately equal masses (domain I and domains II + III) connected by a flexible strand. In the free state or in complex with GDP, contact between the two halves is weak, so it exists in a relaxed conformation, probably oscillating between open (with the halves drawn somewhat apart) and closed forms, which are in equilibrium shifted toward the open form. Specific binding of GTP with domain I leads to formation of a GTP-binding pocket around the ligand and particularly around its γ-phosphate group (induced fit), resulting in local rearrangements in domain I and the appearance of new groups on the domain interface that have an affinity for the surface of the other domain. Thus, the two halves firmly stick together, i.e. the conformation becomes closed and fixed in this state (stroke 1). Now, this closed and locked conformation is competent for binding of aminoacyl-tRNA and attachment to the ribosome. Binding of the ternary aminoacyl-tRNA·EF-Tu·GTP complex to the ribosome induces the hydrolytic cleavage of GTP with the following release of the split-off phosphate, leading to a reverse local rearrangement and consequent unlocking (opening) of the overall conformation of EF-Tu (stroke 2). This event causes loss of EF-Tu affinity for aminoacyl-tRNA and the ribosome and its release from the ribosome. In this example, the fuel is GTP, which is combusted in the chemical reaction of hydrolysis; the engine is domain I, where the catalytic center for hydrolysis is located; and the transmission is the coupling between local conformational changes around the energy substrate in response to GTP binding and GTP decay, on one hand, and gross conformational movements of the locking-unlocking type, on the other. It is in such a way that the shuttle delivery of aminoacyl-tRNA into the translating ribosome is accomplished. Thus, EF-Tu works as a molecular machine, yet its shuttle function is relatively simple and cannot be considered as a true conveying function.
Main Engine of the Ribosome as a Molecular Machine
The translating ribosome is a true conveying machine that directionally passes compact tRNA molecules and the tRNA-bound mRNA chain through itself (Fig. 1). The ribosome was found to be capable of performing this function in the absence of GTP and elongation factors, when aminoacyl-tRNA is the sole high-energy substrate present in the medium (110, 111). Hence, the main fuel for the translating ribosome as a molecular machine must be the molecules of aminoacylated tRNA, and the main engine must be the PTC catalyzing the exergonic reaction of transpeptidation. The PTC is organized by domain V of the compactly folded ribosomal RNA of the large ribosomal subunit (71). It is localized in the middle of the large subunit at the subunit interface and can be subdivided into the acceptor substrate site (designated as A or a) and the donor substrate site (designated as P, p, or d) (128). Below, the cyclic process of factor-free (“non-enzymatic”) translation in which PTC as an engine moves all the working cycle is considered.
It is convenient to begin the consideration from the post-translocation state ribosome with peptidyl-tRNA in the P site (Fig. 5, position I). In this state, the ribosome must be competent for accepting aminoacyl-tRNA and thus someway unlocked. In any case, aminoacyl-tRNA is allowed to enter the A site of the ribosome and binds, first to a cognate codon at the A site of the small subunit and then with the PTC a-site, which has an affinity for the aminoacyl-adenosyl residue (stroke 1). The addition of the intersubunit connection may lead to fixation of a locked form of the ribosome (position II).
FIGURE 5.
Factor-free elongation cycle and main functional states of the translating ribosome. The ribosomes in the unlocked forms (positions I, IV, and V) are shown with the small subunits somewhat rotated relative to the large subunits in the plane of the figure. In the case of the factor-promoted working cycle, step 1 is catalyzed by EF-Tu with GTP (see Fig. 4), and step 4 involves EF-G with GTP (see Fig. 3). Aa, aminoacyl.
Now, both substrates of the transpeptidation reaction are positioned side-by-side in the PTC of the locked ribosome, and their reacting groups are tightly drawn together (Fig. 5, position II). The subsequent transpeptidation reaction (stroke 2) results in replacement of the free amino group of aminoacyl-RNA in the a-site of the PTC by the amide group connecting the aminoacyl-tRNA residue with the peptidyl residue, as well as in the appearance of deacylated tRNA in the p-site of the PTC (position III): aminoacyl-tRNA″ + peptidyl-tRNA′ → peptidyl-aminoacyl-tRNA″ + tRNA′.
As a result, the chemical situation in the PTC has been strongly changed: the a- and p-sites of the PTC accommodate reaction products that have no strong affinity for the sites. This implies that the previous interactions have disappeared and that the intersubunit situation is destabilized. In such a situation, the ribosome starts to oscillate between closed (non-rotated) and open (rotated) forms at a high rate, with the equilibrium shifted toward the open form (70). Simultaneously, the products should tend to leave their previous sites in the PTC. Under conditions of Brownian motions, the weakly bound groups become dissociated and then caught by sites with higher affinities for them (stroke 3). As a result, the newly formed peptide group with its ester group at the CCA terminus of tRNA″ will be reset from the a-site of the PTC to the p-site of the PTC, whereas the deacylated terminus of tRNA′ will be positioned in the e-site nearby (the “weak to strong binding state transition,” cited from Ref. 121). Thus, the so-called hybrid state (A/p-P/p) is established when the ribosome is in the open (rotated) form (position IV).
The state established is thermodynamically unstable: the free energy of the transpeptidation reaction is found to be stored, at least partly, in the pre-translocation state ribosome (Fig. 5, position IV), probably in sterically strained conformations of the acceptor arms of tRNA residues and distorted intersubunit contacts. In the absence of the translocation catalyst EF-G, a high kinetic barrier prevents a fast downhill transition. However, thermal motion can provide slow spontaneous dissociation of tRNA residues complexed with mRNA codons from the A and P sites on the small subunit and subsequent reassociation with the P and E sites, respectively (stroke 4), where their strained conformations become relaxed, with simultaneous closing of the ribosome and thus establishing the original (non-rotated) form in the P/p-E/e state (position V). Spontaneous release of deacylated tRNA from the E site (stroke 5) completes the factor-free translocation step and makes the completed cycle irreversible, and the next cycle can start from position I.
Additional Molecular Engines of the Translating Ribosome
In the case of the factor-promoted elongation cycle, the additional energy of GTP is expended in overcoming the high kinetic barriers in the processes of aminoacyl-tRNA binding and translocation. These GTP-dependent catalytic acts are performed by two subsidiary engines assembled for a time from an elongation factor (EF-Tu or EF-G) and the side protuberance (the so-called L7/L12 stalk) of the large ribosomal subunit at the entrance to the intersubunit channel. In the process of aminoacyl-tRNA binding (Fig. 4), a ternary aminoacyl-tRNA·EF-Tu·GTP complex interacts with the L7/L12 stalk and its base, thus leading to immobilization of the flexible stalk, formation of the factor-binding pocket around EF-Tu (induced fit), and, presumably, fixation of the ribosome in an unidentified open form. The open ribosome admits the aminoacyl-tRNA moiety of the complex into the intersubunit entrance for codon-anticodon recognition and binding to the A site of the small ribosomal subunit. Thus, after this step, the aminoacyl-tRNA is set in the intermediate hybrid A/T position. Next, GTP is hydrolyzed on the bound EF-Tu, the EF-Tu·GDP complex loses its affinity for the aminoacylated acceptor arm of aminoacyl-tRNA, and the released aminoacylated CCA terminus is caught by the a-site of the PTC. The aminoacyl-tRNA becomes fixed in the final A/a position, and the ribosome is closed. The EF-Tu·GDP complex is now weakly retained in the pocket at the L7/L12 stalk and spontaneously leaves it.
Likewise, in the process of translocation, EF-G and GTP interact with the L7/L12 stalk and its base on the pre-translocation ribosome oscillating between the two forms (Fig. 3). This leads to immobilization of the flexible stalk, formation of the factor-binding pocket around EF-G (induced fit), and fixation of the ribosome in the open (rotated) form. The open (rotated) state allows the tRNA residue of the peptidyl-tRNA and deacylated tRNA to move within the intersubunit space and thus be caught by the P site and E site, respectively, of the small ribosomal subunit. Hence, the products of the transpeptidation reaction are now found in post-translocation positions P/p and E/e. The EF-G·GDP complex, weakly bound at the L7/L12 stalk, is then spontaneously released.
Transmission and Mover
In all the cases considered above, an energy substrate that feeds a molecular engine is “combusted” (i.e. contributes to the decrease of the thermodynamic potential of the system) in three steps: first, when it binds to a binding site and thus forms non-covalent bonds in a binding pocket; second, when an exergonic reaction of its chemical transformation occurs; and third, when the product of the reaction is released. All steps lead to conformational changes: the first induces conformational shifts because of attraction of flexible groups of the binding site (induced fit), the second changes the bound ligand and thus abolishes a part of the previous contacts, and the third finally liberates an engine. The local conformational changes within an engine may change contacts between more distant groups and larger blocks and thus be coupled with gross conformational transitions in a molecular machine (refer to the example of EF-Tu described above). In any case, the coupling between primary conformational changes in response to binding of the energy substrate and its subsequent decay and more distant rearrangements will result in changes of affinities of binding sites for the conveyed substrates. This is the way that transmission from an engine to a mover is principally organized in molecular machines, including the ribosome. In the case of the ribosome, binding of the aminoacylated terminus of aminoacyl-tRNA to the a-site of the PTC and the subsequent change of the aminoacyl-tRNA into peptidyl-tRNA induce, through conformational transmission mechanisms of the ribosome, such global rearrangements as mutual rotation of the ribosomal subunits, locking-unlocking effects, and, possibly, shifts of other structural blocks (such as the L1 protuberance).
Again, it should be mentioned that movements within transmission mechanisms of molecular machines are generated mainly by thermal motions. The motions are anisotropic, as they are spatially limited by construction of a machine. Mutual affinities of blocks and other structural elements determine interactions between them and fixation of certain connections and rearrangements. In other words, conformational changes during the transmission process are the events of relaxation, thermal motion, and temporary fixation of certain conformational states.
Concerning the movers of molecular machines, the same considerations can be applied. The conveyed ligands are moved by thermal energy, but their diffusional path may be limited by a channel and, most importantly, determined by alternating changing affinities of binding sites, from weakening of ligand retention at its site to a temporary fixation of a ligand at the next position. This provides unidirectional transitions from a weak to a strong binding state along the conveying path. In the case of translating ribosomes (see Figs. 1 and 5), an aminoacyl-tRNA selected by the A site codon firmly binds to both its binding site on the small subunit (A) and the binding site in the PTC of the large subunit (a). After transpeptidation, the affinity of the acylated terminal group for the a-site is abolished, and it dissociates from the a-site and, because of flexibility of the terminal CCA sequence, may randomly (but within sterically allowed limits) move until it becomes fixed at the new affinity p-site. At the same time, because the deacylated terminus of the tRNA has lost high affinity for the p-site of the PTC, the flexible CCA end sequence becomes dissociated and caught by the e-site in the vicinity of the PTC. These events induce instability at the subunit interface and at the A and P sites on the small subunit. The destabilized and unlocked intersubunit state, as well as possibly the strained tRNA conformations, results in weakening of retention of the tRNA residues in their A and P sites on the small ribosomal subunit. Because of the unlocked state of the ribosome, the tRNA residues are allowed to move from the A and P sites and reassociate with the stronger nearby binding sites, P and E, respectively (tRNA residue translocation). At the final step of translocation, the deacylated tRNA spontaneously dissociates from the E site and e-site and quits the translating ribosome. It is in this way that the tRNA residues are directionally conveyed through the intersubunit channel, from the entrance hole at the small subunit beak and L7/L12 protuberance toward the exit hole and L1 protuberance, using thermal motions as immediate moving impulses for the conveyed ligands. Thus, the mover of the translating ribosome as a conveying machine is the sequence of tRNA-binding sites A, P, E: their alternating changes of affinities for tRNA residues provide the unidirectional “weak to strong binding state transitions” of the conveyed molecules.
Ratchet and Pawl
As a matter of fact, thermal motions, including Brownian movement and intrinsic thermal conformational fluctuations in an isothermal medium and anisotropic system, provide random impulses that are continually displacing structural blocks of a molecular machine and its ligands, thereby creating a field of sterically allowed conformations and positions at each stage of the transition from one state to another. The selection of the proper conformation and position is determined by thermodynamic parameters. Alternating events of attachment-detachment and fixation-relaxation, rather than mechanically directed shifts by means of rigid couplings and mechanical energy, seem to be the most likely basis of the working cycles of molecular machines, including the ribosome. In this model, the binding of a ligand, the chemical transformations of a bound ligand, and its (or its product) release are the energy contributions to molecular and ligand movements, not as a direct feed to a mechanical shift but as a means to rectify random fluctuations by selection and fixation of the conformations and positions that serve to create a unidirectional process. In other words, in terms of the considered model, the binding of a ligand and its chemical transformations can be viewed as the energy needed to enable operation of the “pawls.”
In Feynman's ratchet machine, the ratchet wheel rotates only in one direction because of a pawl (which is here equivalent to Maxwell's Demon) that allows a forward step and prevents a backward step. In a molecular machine, this function is realized because of the principle of the weak to strong binding state transitions (121). In the translating ribosome, the sequential unidirectional shifts of two moieties of tRNA ligands, a → p, A → P, p → e, P → E (Figs. 3 and 5), obey this principle. The corresponding changes in affinities of the binding sites along this path are provided by energy contributions to the translating ribosome, first and foremost by the intraribosomal transpeptidation reaction enabled by aminoacyl-tRNA input and deacylated tRNA output. Thus, the working cycle of the translating ribosome (for the sake of simplicity, one may look at the factor-free elongation cycle in Fig. 5) can be considered as a Feynman's ratchet wheel, with pawls appearing along the conveying path in response to energy-dependent conformational shifts. On the other hand, if one were to focus on the irreversibility (unidirectionality) of the cycle as a whole, it is clear that the only practically irreversible step of the cycle is the release of deacylated tRNA into the medium after translocation.
Acknowledgments
I am very grateful to all the past and present members of my laboratory and to other colleagues mentioned in this paper for their contributions to our experimental work as well as for their theoretical discussions on the topics considered here. I wish to express my special gratitude to my former co-workers Lydia Gavrilova and Nadezhda Belitsina, with whom most experiments on translocation were carried out, and to Alexander Chetverin, Alexey Finkelstein, and Oleg Ptitsyn for help in formulating statements on the energetics and molecular physics of ribosomes. I am also very obliged to Alexey Finkelstein, Harry Noller, Loren Runnels, and Alexey Ryazanov for critical reading of the manuscript, helpful comments, and editing of the text.
REFERENCES
- 1.Kiesel A., Beloserskii A. (1934) Hoppe-Seyler's Z. Physiol. Chem. 229, 160–166 [Google Scholar]
- 2.Belozersky A. N., Dubrovskaya I. I. (1936) Biokhimiya 1, 665–675 [Google Scholar]
- 3.Belozersky A. N. (1940) Mikrobiologiya 9, 107–113 [Google Scholar]
- 4.Belozersky A. N. (1947) Cold Spring Harbor Symp. Quant. Biol. 12, 1–6 [Google Scholar]
- 5.Vischer E., Chargaff E. (1948) J. Biol. Chem. 176, 703–714, 715–734 [PubMed] [Google Scholar]
- 6.Chargaff E., Vischer E., Doniger R., Green C., Misani F. (1949) J. Biol. Chem. 177, 405–416 [PubMed] [Google Scholar]
- 7.Vischer E., Zamenhof S., Chargaff E. (1949) J. Biol. Chem. 177, 429–438 [PubMed] [Google Scholar]
- 8.Chargaff E., Magasanik B., Vischer E., Green C., Doniger R., Elson D. (1950) J. Biol. Chem. 186, 51–67 [PubMed] [Google Scholar]
- 9.Watson J. D., Crick F. H. (1953) Nature 171, 737–738 [DOI] [PubMed] [Google Scholar]
- 10.Watson J. D., Crick F. H. (1953) Nature 171, 964–967 [DOI] [PubMed] [Google Scholar]
- 11.Spirin A. S., Belozersky A. N., Shugaeva N. V., Vanyushin B. F. (1957) Biokhimiya 22, 744–754 [PubMed] [Google Scholar]
- 12.Belozersky A. N., Spirin A. S. (1958) Nature 182, 111–112 [DOI] [PubMed] [Google Scholar]
- 13.Crick F. H. (1959) Brookhaven Symp. Biol. 12, 35–39 [PubMed] [Google Scholar]
- 14.Jacob F., Monod J. (1961) Cold Spring Harbor Symp. Quant. Biol. 26, 193–209 [DOI] [PubMed] [Google Scholar]
- 15.Signer E. R., Torriani A., Levinthal C. (1961) Cold Spring Harbor Symp. Quant. Biol. 26, 31–34 [DOI] [PubMed] [Google Scholar]
- 16.Spiegelman S. (1961) Cold Spring Harbor Symp. Quant. Biol. 26, 75–90 [DOI] [PubMed] [Google Scholar]
- 17.Yčas M. (1969) The Biological Code, North-Holland Publishing Co., Amsterdam [Google Scholar]
- 18.Crick F. H. (1958) Symp. Soc. Exp. Biol. 12, 138–163 [PubMed] [Google Scholar]
- 19.Jacob F., Monod J. (1961) J. Mol. Biol. 3, 318–356 [DOI] [PubMed] [Google Scholar]
- 20.Spirin A. S., Belitsina N. V., Ajtkhozhin M. A. (1964) J. Gen. Biol. 25, 321–338 (English Translation (1965) Fed. Proc.24, T907–T915) [PubMed] [Google Scholar]
- 21.Spirin A. S. (1969) Eur. J. Biochem. 10, 20–35 [DOI] [PubMed] [Google Scholar]
- 22.Spirin A. S. (1966) Curr. Topics Dev. Biol. 1, 1–38 [PubMed] [Google Scholar]
- 23.Standart N., Dale M., Stewart E., Hunt T. (1990) Genes Dev. 4, 2157–2168 [DOI] [PubMed] [Google Scholar]
- 24.Standart N., Hunt T. (1990) Enzyme 44, 106–119 [DOI] [PubMed] [Google Scholar]
- 25.Woese C. R. (1961) Nature 189, 920–921 [DOI] [PubMed] [Google Scholar]
- 26.Miura K. I. (1962) Biochim. Biophys. Acta 55, 62–70 [DOI] [PubMed] [Google Scholar]
- 27.Midgley J. E. (1962) Biochim. Biophys. Acta 61, 513–525 [DOI] [PubMed] [Google Scholar]
- 28.Spirin A. S., Milman L. S. (1960) Dok. Akad. Nauk SSSR 134, 717–720 [Google Scholar]
- 29.Spirin A. S. (1961) Biokhimiya 26, 511–522 [Google Scholar]
- 30.Spirin A. S. (1962) in Acides Ribonucléiques et Polyphosphates: Structure, Synthèse et Fonctions, Colloques Intrernationaux CNRS No. 106, Strasbourg, 6–12 Juillet 1961 (Ebel J.-P., Grunberg-Manago M. eds) pp. 75–87, Édition du CNRS, Paris [Google Scholar]
- 31.Bogdanova E. S., Gavrilova L. P., Dvorkin G. A., Kisselev N. A., Spirin A. S. (1962) Biokhimiya 27, 387–402 [PubMed] [Google Scholar]
- 32.Hall B. D., Doty P. (1959) J. Mol. Biol. 1, 111–126 [Google Scholar]
- 33.Takanami M. (1960) Biochim. Biophys. Acta 39, 152–154 [DOI] [PubMed] [Google Scholar]
- 34.Brown R. A., Ellem K. A., Colter J. S. (1960) Nature 187, 509–511 [Google Scholar]
- 35.Aronson A. I., McCarthy B. J. (1961) Biophys. J. 1, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spirin A. S. (1960) J. Mol. Biol. 2, 436–446 [Google Scholar]
- 37.Kisselev N. A., Gavrilova L. P., Spirin A. S. (1961) J. Mol. Biol. 3, 778–783 [DOI] [PubMed] [Google Scholar]
- 38.Spirin A. S. (1963) Prog. Nucleic Acid Res. 1, 301–345 [Google Scholar]
- 39.Vasiliev V. D., Selivanova O. M., Koteliansky V. E. (1978) FEBS Lett. 95, 273–276 [DOI] [PubMed] [Google Scholar]
- 40.Vasiliev V. D., Zalite O. M. (1980) FEBS Lett. 121, 101–104 [DOI] [PubMed] [Google Scholar]
- 41.Vasiliev V. D., Serdyuk I. N., Gudkov A. T., Spirin A. S. (1986) in Structure, Function, and Genetics of Ribosomes (Hardesty B., Kramer G. eds) pp. 128–142, Springer-Verlag New York Inc., New York [Google Scholar]
- 42.Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Nature 407, 327–339 [DOI] [PubMed] [Google Scholar]
- 43.Spirin A. S. (1964) Macromolecular Structure of Ribonucleic Acids, Reinhold Publishing Corp., New York [Google Scholar]
- 44.Spirin A. S., Kiselev N. A., Shakulov R. S., Bogdanov A. A. (1963) Biokhimiya 28, 920–930 [PubMed] [Google Scholar]
- 45.Gavrilova L. P., Ivanov D. A., Spirin A. S. (1966) J. Mol. Biol. 16, 473–489 [DOI] [PubMed] [Google Scholar]
- 46.Spirin A. S., Belitsina N. V., Lerman M. I. (1965) J. Mol. Biol. 14, 611–615 [DOI] [PubMed] [Google Scholar]
- 47.Lerman M. I., Spirin A. S., Gavrilova L. P., Golov V. F. (1966) J. Mol. Biol. 15, 268–281 [DOI] [PubMed] [Google Scholar]
- 48.Spirin A. S., Belitsina N. V. (1966) J. Mol. Biol. 15, 282–283 [DOI] [PubMed] [Google Scholar]
- 49.Hosokawa K., Fujimura R. K., Nomura M. (1966) Proc. Natl. Acad. Sci. U.S.A. 55, 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staehelin T., Meselson M. (1966) J. Mol. Biol. 16, 245–249 [DOI] [PubMed] [Google Scholar]
- 51.Spirin A. S. (1963) Cold Spring Harbor Symp. Quant. Biol. 28, 267–268 [Google Scholar]
- 52.Spirin A. S. (1985) Prog. Nucleic Acids Res. Mol. Biol. 32, 75–114 [DOI] [PubMed] [Google Scholar]
- 53.Spirin A. S. (1988) in The Roots of Modern Biochemistry (Kleinkauf H., von Dören H., Jaenicke R. eds) pp. 511–533, Walter de Gruyter & Co., Berlin [Google Scholar]
- 54.Spirin A. S. (2002) FEBS Lett. 514, 2–10 [DOI] [PubMed] [Google Scholar]
- 55.Spirin A. S. (2004) RNA Biol. 1, 3–9 [PubMed] [Google Scholar]
- 56.Spirin A. S. (1968) Dok. Akad. Nauk SSSR 179, 1467–1470 [PubMed] [Google Scholar]
- 57.Spirin A. S. (1968) Curr. Mod. Biol. 2, 115–127 [DOI] [PubMed] [Google Scholar]
- 58.Spirin A. S. (1969) Cold Spring Harbor Symp. Quant. Biol. 34, 197–207 [DOI] [PubMed] [Google Scholar]
- 59.Bretscher M. S. (1968) Nature 218, 675–677 [DOI] [PubMed] [Google Scholar]
- 60.Baranov V. I., Belitsina N. V., Spirin A. S. (1979) Methods Enzymol. 59, 382–397 [DOI] [PubMed] [Google Scholar]
- 61.Serdyuk I. N., Spirin A. S. (1986) in Structure, Function, and Genetics of Ribosomes (Hardesty B., Kramer G. eds) pp. 425–437, Springer-Verlag New York Inc., New York [Google Scholar]
- 62.Spirin A. S., Baranov V. I., Polubesov G. S., Serdyuk I. N., May R. P. (1987) J. Mol. Biol. 194, 119–126 [DOI] [PubMed] [Google Scholar]
- 63.Serdyuk I., Baranov V., Tsalkova T., Gulyamova D., Pavlov M., Spirin A., May R. (1992) Biochimie 74, 299–306 [DOI] [PubMed] [Google Scholar]
- 64.Frank J., Agrawal R. K. (2000) Nature 406, 318–322 [DOI] [PubMed] [Google Scholar]
- 65.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. (2003) Cell 114, 123–134 [DOI] [PubMed] [Google Scholar]
- 66.Horan L. H., Noller H. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4881–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ermolenko D. N., Majumdar Z. K., Hickerson R. P., Spiegel P. C., Clegg R. M., Noller H. F. (2007) J. Mol. Biol. 370, 530–540 [DOI] [PubMed] [Google Scholar]
- 68.Korostelev A., Noller H. F. (2007) J. Mol. Biol. 373, 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spiegel P. C., Ermolenko D. N., Noller H. F. (2007) RNA 13, 1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cornish P. V., Ermolenko D. N., Noller H. F., Ha T. (2008) Mol. Cell 30, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 72.Gudkov A. T., Gongadze G. M., Bushuev V. N., Okon M. S. (1982) FEBS Lett. 138, 229–232 [DOI] [PubMed] [Google Scholar]
- 73.Gongadze G. M., Gudkov A. T., Bushuev V. N., Sepetov N. F. (1984) Dok. Akad. Nauk SSSR 279, 230–232 [PubMed] [Google Scholar]
- 74.Girshovich A. S., Kurtskhalia T. V., Ovchinnikov Yu. A., Vasiliev V. D. (1981) FEBS Lett. 130, 54–59 [DOI] [PubMed] [Google Scholar]
- 75.Girshovich A. S., Bochkareva E. S., Vasiliev V. D. (1986) FEBS Lett. 197, 192–198 [DOI] [PubMed] [Google Scholar]
- 76.Moazed D., Robertson J. M., Noller H. F. (1988) Nature 334, 362–364 [DOI] [PubMed] [Google Scholar]
- 77.Valle M., Zavialov A., Li W., Stagg S. M., Sengupta J., Nielsen R. C., Nissen P., Harvey S. C., Ehrenberg M., Frank J. (2003) Nat. Struct. Biol. 10, 899–906 [DOI] [PubMed] [Google Scholar]
- 78.Spahn C. M., Gomez-Lorenzo M. G., Grassucci R. A., Jørgensen R., Andersen G. R., Beckmann R., Penczek P. A., Ballesta J. P., Frank J. (2004) EMBO J. 23, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Lorenzo M. G., Spahn C. M., Agrawal R. K., Grassucci R. A., Penczek P. A., Chakraburtty K., Ballesta J. P., Lavandera J. L., Garcia-Bustos J. F., Frank J. (2000) EMBO J. 19, 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. (2001) Cell 107, 679–688 [DOI] [PubMed] [Google Scholar]
- 81.Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. (2001) Science 292, 883–896 [DOI] [PubMed] [Google Scholar]
- 82.Schuwirth B. S., Borovinskaya M. A., Hau C. W., Zhang W., Vila-Sanjurjo A., Holton J. M., Cate J. H. (2005) Science 310, 827–834 [DOI] [PubMed] [Google Scholar]
- 83.Cornish P. V., Ermolenko D. N., Staple D. W., Hoang L., Hickerson R. P., Noller H. F., Ha T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor D. J., Nilsson J., Merrill A. R., Andersen G. R., Nissen P., Frank J. (2007) EMBO J. 26, 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ogle J. M., Brodersen D. E., Clemons W. M., Jr., Tarry M. J., Carter A. P., Ramakrishnan V. (2001) Science 292, 897–902 [DOI] [PubMed] [Google Scholar]
- 86.Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V. (2002) Cell 111, 721–732 [DOI] [PubMed] [Google Scholar]
- 87.Korostelev A., Trakhanov S., Laurberg M., Noller H. F. (2006) Cell 126, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 88.Gabashvili I. S., Agrawal R. K., Spahn C. M., Grassucci R. A., Svergun D. I., Frank J., Penczek P. (2000) Cell 100, 537–549 [DOI] [PubMed] [Google Scholar]
- 89.Yusupova G. Z., Yusupov M. M., Cate J. H., Noller H. F. (2001) Cell 106, 233–241 [DOI] [PubMed] [Google Scholar]
- 90.Yusupova G., Jenner L., Rees B., Moras D., Yusupov M. (2006) Nature 444, 391–394 [DOI] [PubMed] [Google Scholar]
- 91.Moazed D., Noller H. F. (1986) Cell 47, 985–994 [DOI] [PubMed] [Google Scholar]
- 92.Spirin A. S. (1999) Ribosomes, Kluwer Academic Publishers/Plenum Press, New York [Google Scholar]
- 93.Moazed D., Noller H. F. (1989) Cell 57, 585–597 [DOI] [PubMed] [Google Scholar]
- 94.Moazed D., Noller H. F. (1989) Nature 342, 142–148 [DOI] [PubMed] [Google Scholar]
- 95.Connell S. R., Takemoto C., Wilson D. N., Wang H., Murayama K., Terada T., Shirouzu M., Rost M., Schüler M., Giesebrecht J., Dabrowski M., Mielke T., Fucini P., Yokoyama S., Spahn C. M. (2007) Mol. Cell 25, 751–764 [DOI] [PubMed] [Google Scholar]
- 96.Frank J., Gao H., Sengupta J., Gao N., Taylor D. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19671–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Munro J. B., Altman R. B., O'Connor N., Blanchard S. C. (2007) Mol. Cell 25, 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan D., Kirillov S. V., Cooperman B. S. (2007) Mol. Cell 25, 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishizuka Y., Lipmann F. (1966) Proc. Natl. Acad. Sci. U.S.A. 55, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishizuka Y., Lipmann F. (1966) Arch. Biochem. Biophys. 116, 344–351 [DOI] [PubMed] [Google Scholar]
- 101.Lipmann F. (1969) Science 164, 1024–1031 [DOI] [PubMed] [Google Scholar]
- 102.Gordon J., Lipmann F. (1967) J. Mol. Biol. 23, 23–33 [Google Scholar]
- 103.Pestka S. (1968) J. Biol. Chem. 243, 2810–2820 [PubMed] [Google Scholar]
- 104.Pestka S. (1969) J. Biol. Chem. 244, 1533–1539 [PubMed] [Google Scholar]
- 105.Gavrilova L. P., Smolyaninov V. V. (1971) Molekul. Biol. 5, 883–891 [PubMed] [Google Scholar]
- 106.Gavrilova L. P., Spirin A. S. (1971) FEBS Lett. 17, 324–326 [DOI] [PubMed] [Google Scholar]
- 107.Gavrilova L. P., Spirin A. S. (1972) FEBS Lett. 22, 91–92 [DOI] [PubMed] [Google Scholar]
- 108.Gavrilova L. P., Spirin A. S. (1974) FEBS Lett. 39, 13–16 [DOI] [PubMed] [Google Scholar]
- 109.Gavrilova L. P., Koteliansky V. E., Spirin A. S. (1974) FEBS Lett. 45, 324–328 [DOI] [PubMed] [Google Scholar]
- 110.Gavrilova L. P., Spirin A. S. (1974) Methods Enzymol. 30, 452–462 [PubMed] [Google Scholar]
- 111.Gavrilova L. P., Kostiashkina O. E., Koteliansky V. E., Rutkevitch N. M., Spirin A. S. (1976) J. Mol. Biol. 101, 537–552 [DOI] [PubMed] [Google Scholar]
- 112.Spirin A. S., Kostiashkina O. E., Jonák J. (1976) J. Mol. Biol. 101, 553–562 [DOI] [PubMed] [Google Scholar]
- 113.Gavrilova L. P., Perminova I. N., Spirin A. S. (1981) J. Mol. Biol. 149, 69–78 [DOI] [PubMed] [Google Scholar]
- 114.Spirin A. S. (1978) Prog. Nucleic Acids Res. Mol. Biol. 21, 39–62 [DOI] [PubMed] [Google Scholar]
- 115.Chetverin A. B., Spirin A. S. (1982) Biochim. Biophys. Acta 683, 153–179 [DOI] [PubMed] [Google Scholar]
- 116.Belitsina N. V., Glukhova M. A., Spirin A. S. (1975) FEBS Lett. 54, 35–38 [DOI] [PubMed] [Google Scholar]
- 117.Belitsina N. V., Glukhova M. A., Spirin A. S. (1976) J. Mol. Biol. 108, 609–613 [DOI] [PubMed] [Google Scholar]
- 118.Purich D. L. (2001) Trends Biochem. Sci. 26, 417–421 [DOI] [PubMed] [Google Scholar]
- 119.Finkelstein A. V., Ptitsyn O. B. (2002) Protein Physics, Academic Press, London [Google Scholar]
- 120.Feynman R., Leighton R., Sands M. (1963) The Feynman Lectures on Physics, Addison-Wesley, Reading, MA [Google Scholar]
- 121.Vale R. D., Oosawa F. (1990) Adv. Biophys. 26, 97–134 [DOI] [PubMed] [Google Scholar]
- 122.Córdova N. J., Ermentrout B., Oster G. F. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ait-Haddou R., Herzog W. (2003) Cell Biochem. Biophys. 38, 191–214 [DOI] [PubMed] [Google Scholar]
- 124.Bertchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. (1993) Nature 365, 126–132 [DOI] [PubMed] [Google Scholar]
- 125.Kjeldgaard M., Nissen P., Thirup S., Nyborg J. (1993) Structure 1, 35–50 [DOI] [PubMed] [Google Scholar]
- 126.Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J. (1996) Structure 4, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 127.Abel K., Yoder M. D., Hilgenfeld R., Jurnak F. (1996) Structure 4, 1153–1159 [DOI] [PubMed] [Google Scholar]
- 128.Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) Science 289, 920–930 [DOI] [PubMed] [Google Scholar]