Abstract
The TP73 gene gives rise to transactivation domain-p73 isoforms (TAp73) as well as ΔNp73 variants with a truncated N terminus. Although TAp73α and -β proteins are capable of inducing cell cycle arrest, apoptosis, and differentiation, ΔNp73 acts in many cell types as a dominant-negative repressor of p53 and TAp73. It has been proposed that p73 is involved in myeloid differentiation, and its altered expression is involved in leukemic degeneration. However, there is little evidence as to which p73 variants (TA or ΔN) are expressed during differentiation and whether specific p73 isoforms have the capacity to induce, or hinder, this differentiation in leukemia cells. In this study we identify GATA1 as a direct transcriptional target of TAp73α. Furthermore, TAp73α induces GATA1 activity, and it is required for erythroid differentiation. Additionally, we describe a functional cooperation between TAp73 and ΔNp73 in the context of erythroid differentiation in human myeloid cells, K562 and UT-7. Moreover, the impaired expression of GATA1 and other erythroid genes in the liver of p73KO embryos, together with the moderated anemia observed in p73KO young mice, suggests a physiological role for TP73 in erythropoiesis.
The TP73 gene gives rise to multiple protein isoforms because of alternative mRNA splicing and promoter utilization as follows: TAp73 isoforms (containing the N-terminal transactivation domain) and TA-deficient ΔNp73 isoforms (1–5). The TP73 gene expresses at least seven C-terminal isoforms (α, β, γ, δ, ϵ, ζ, and η) that differ in their transcriptional activity and at least four alternatively spliced ΔN isoforms initiated at different ATG (6, 7). TAp73α and -β share many target genes with p53, inducing cell cycle arrest, apoptosis, and differentiation (8–11). Conversely, ΔNp73 isoforms can act as a dominant-negative suppressor of p53 and TAp73, abrogating their pro-apoptotic, differentiation, and growth suppression functions in diverse cell types (5, 12, 13). Nevertheless, several lines of evidence have revealed that ΔNp73 can also transactivate specific target genes, including neuronal pro-differentiation genes such as BTG2, and may either inhibit or stimulate p53 transcriptional activity, depending on the gene and the cellular context (14–18). These findings imply that ΔNp73α not only acts as an inhibitor of p53/TAp73 but can also cooperate with them. Furthermore, the recent observations that ΔNp73 acts as a pro-differentiation factor in acute promyelocytic leukemia cells and might be necessary for myeloid differentiation suggest that the functional role of ΔNp73 in the context of myeloid differentiation may differ from that in other cell types (19). However, the existence of a functional cooperation between ΔNp73α and TAp73 in myeloid cell differentiation remains unclear.
A model to explain p73 function in myeloid differentiation and leukemogenesis has been proposed based on the analysis of the p73 status in patients with advanced stages of chronic myeloid leukemia and acute myeloid leukemia. In this model, inappropriate expression of certain p73 isoforms could result in impaired cellular differentiation giving rise to leukemic cells (20, 21). However, there is little evidence as to which p73 variants (TA or ΔN) are expressed during differentiation and whether specific p73 isoforms have the capacity to induce, or hinder, this differentiation in leukemia cells.
In the process of addressing these questions, we found that constitutive expression of ΔNp73 in the human cell lines, K562 and UT-7, resulted in erythroid differentiation. Further analysis revealed that ΔNp73-induced erythroid differentiation required transcriptionally active TAp73. We demonstrated that TAp73 is capable of inducing GATA1 activity and is required for drug-induced GATA1 expression and differentiation. Consistent with an in vivo role for p73 in red blood cell differentiation, p73-deficient mice presented with moderate anemia as well as impaired expression of Gata1 and other erythroid related genes. Altogether, our data demonstrate a role of TP73 in erythroid differentiation and suggest a physiological role of this gene in developmental erythropoiesis.
EXPERIMENTAL PROCEDURES
Mice Husbandry and Dissection of Hematopoietic Tissue
Animal experimental procedures were conducted in agreement with European (Council Directive 86/609/CEE) and Spanish regulations (RD 1201/2005) for the protection of animals used for experimental and other scientific purposes. Mice heterozygous for TP73 on a mixed background C57BL/6 × 129/svJae were backcrossed, at least five times, to C57BL/6 to enrich for C57BL/6. Mice were genotyped as described before (4). Pregnant females at 14.5 days postcoitum (E14.5) were euthanized, and embryos were extracted and visually inspected independently by two different investigators. Fetal livers were dissected and processed to obtain RNA as described below.
Cell Culture
K562, HCT-116 p53−/−, and UT-7 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. UT-7 medium was supplemented with granulocyte-macrophage colony-stimulating factor 5 ng/ml.
Plasmids and Transfection
The cDNAs from human HA-TAp73α, TAp73α292, HA-ΔNp73α, and pcDNA3-HA-TAp73ϵ were described previously (22). The reporter plasmid GATA1-Luc (a gift from Dr. T. Nakano, Osaka University) contains a fragment of the GATA1 promoter, including three GATA1-binding sites (23). The cDNA from human HA-ΔNp73α was subcloned in the plasmid MT-CB6+ (a gift from Dr. Frank J Rauscher) in the KpnI/XbaI sites.
To generate K562-HA-ΔNp73α, -ΔNp73β, and -HA-TAp73ϵ cell lines, K562 cells (2 × 106) were electroporated (Bio-Rad Gene Pulser) in a serum-free medium with 10 μg of the following plasmids: pcDNA3-HA-ΔNp73α, pcDNA3-ΔNp73β, and pcDNA3-HA-TAp73ϵ. Twenty four h post-electroporation selection was applied (0.5 mg/ml G418), and individual clones were screened by Western blot assay. UT-7 cells were electroporated with 5 μg of the indicated plasmids in a serum-free medium.
Drugs and Erythroid Differentiation Analysis
To examine the extent of erythroid differentiation, cells were treated with either vehicle control, or with 1 μm AraC6 (Calbiochem), 40 μm hemin (Sigma), or 3 units/ml of human erythropoietin (EPO) (Sigma) in complete media. Hemoglobin-producing cells were scored by benzidine staining as described before (24). In each experiment at least 200 cells were counted in triplicate.
Western Blot Analysis
For protein analysis, cells were harvested after treatment with the indicated drug, processed as described before, resolved by SDS-PAGE, and detected by antibodies as indicated (25). The primary antibodies used were as follows: anti-p73 N-terminal (BL906 Abcam), anti-p73 C-terminal Ab-4 (NeoMarkers), anti-hemagglutinin (12CA5, Roche Applied Science), polyclonal anti-hemagglutinin (Y11, Santa Cruz Biotechnology), anti-p21 (C19, Santa Cruz Biotechnology), and anti-ERK2 (C14, Santa Cruz Biotechnology).
RNA Isolation, Semi-quantitative and Real Time RT-PCR Analysis
Total RNA was extracted with TriReagentTM (Ambion) following the manufacturer's protocols. cDNA was prepared using SuperScriptTM II First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Primer sequences used for PCR are described in supplemental Table 1. The cycling program for p73 was set for 35 cycles at 95 °C for 5 min, 95 °C for 1 min, 60 °C for 1 min, 72 °C for 2 min, 72 °C for 10 min; for ΔNp73: 39 cycles at 95 °C for 5 min, 95 °C for 1 min, 58 °C for 1 min, 72 °C for 2 min, 72 °C for 10 min; for GATA-1, ϵ-globin, glycophorin A, transferrin receptor II, and p21CIP1: 24 cycles at 95 °C for 5 min, 95 °C for 1 min, 56 °C for 1 min, 72 °C for 1.5 min, 72 °C for 10 min; and for GAPDH: 30 cycles at 95 °C for 5 min, 95 °C for 1 min, 60 °C for 1 min, 72 °C for 2 min, and 72 °C for 10 min. The expression of erythroid markers mRNA in mouse fetal livers was detected by real time quantitative RT-PCR in a StepOnePlusTM real time PCR system (Applied Biosystems) using an iQTM SYBR® Green SuperMix (Bio-Rad).
PCR was carried out at 50 °C for 2 min and 95 °C for 4 min, followed by 40 cycles of 95 °C for 1.5 min, 60 °C for 30 s, 72 °C for 30 s, and one cycle at 72 °C for 5 min. Primers sequences used for real time quantitative RT-PCR are described in supplemental Table 2. The expression level of erythroid markers mRNA were expressed as 2CtGAPDH− Cterythroid markers, Ct indicates the cycle threshold. Each assay was performed at least in duplicate.
Hematopoietic Indices
Two-week-old mice were bled by cardiac puncture, and hematopoietic indices were determined by counting in a hemocytometer using different solutions as follows: Hayem's solution (Sigma) for red blood cell count, Turk's solution (Sigma) for white cell count, and procaine solution (Sigma) for platelet count according to the manufacturer's instructions. Each experiment was counted in triplicate. The final values are the average of the values for all mice from a given genotype. Hematocrit was determined with the Drabkin reagent following the manufacturer's protocols (Sigma).
Flow Cytometry of Specific Erythroid Subpopulation
Percentage of erythroid precursors in each erythroid development stage was measured as described elsewhere (26). Briefly, to analyze the most related erythroid precursors obtained from bone marrow and spleen, 1 × 106 cells were stained with biotinylated murine anti-CD71 (Pharmingen) and TER-119-PE (Pharmingen) antibodies, washed, and incubated with streptavidin-tricolor (Caltag). A minimum number of 3 × 105 viable cells was acquired using an EPICS XL flow cytometer (Coulter Electronics).
Chromatin Immunoprecipitation (ChIP)
For each ChIP assay (3 × 107) cells, or the fetal livers, were fixed in 1% formaldehyde, lysed, and sonicated as described in the Upstate protocol. ChIP was done using Dynabeads-protein G (Invitrogen) coupled to the following antibodies: anti-p73 N-terminal mixture (BL906 Abcam, anti-p73 N-terminal (Imgenex) and Ab-5 (NeoMarkers)), anti-ΔNp73 IMG-313A (Imgenex) and polyclonal anti-p53 (FL-393, Santa Cruz Biotechnology). Dynabeads were incubated with lysates, and chromatin was eluted and purified as described before (27). Real time PCR of immunoprecipitated DNA was performed in an iQ5 apparatus (Bio-Rad) at 60 °C as the annealing temperature. Primers encompassing the p53-binding sites of the human p21 promoter were 5′-AGTTTGCAACCATGCACTTG and 5′-CTGATGCATGTGTGCTTGTG (amplicon, 180 bp).
An in silico prediction of p53-responsive elements (p53REs) within the human GATA1 locus was performed with the p53FamTaG data base. Primers encompassing the p53-binding sites of the human GATA1 promoter were 5′-GAAGAAATGGGGTGAGTCCA and 5′-GTGCTGAGTGGTTCCCTACCC (amplicon, 199 bp).
Luciferase Assay
Cells were electroporated with reporter plasmid GATA1-Luc (2 μg), 1 μg of pRLNull (Renilla luciferase), and 5 μg of the indicated expression vectors as follows: pcDNA-3, pcDNA3-TAp73α, pcDNA3-TAp73α292, pcDNA3-HA-ΔNp73α, pcDNA-ΔNp73β, or pcDNA3-HA-TAp73ϵ. Cellular extracts were prepared as described before (22). Measurements were taken in a Berthold's luminometer and normalized by the values of the Renilla luciferase of the same sample.
RNA Interference Analysis
The knockdown of p73 was performed using two anti-p73 siRNA oligos described previously by Dr. Meredith Irwin (28). To ablate ΔN-specific p73 isoforms, we used siRNA oligos described previously by Dr. A. Constanzo (29). All oligos used were synthesized by Dharmacon. Cells were electroporated with 100 nm siRNA oligos at 260 V and 1.05 millifarads (Bio-Rad Gene Pulser) in a serum-free medium and incubated for 48 h. Cells were treated with the indicated drug and incubated for 72 h; samples were collected at the indicated time and processed accordingly.
Preparation of Labeled cDNA and Hybridization of Microarrays
Total RNA from K562-V and K562-HA-ΔNp73α cells was isolated with a RNeasy mini kit (Qiagen) following the manufacturer's protocol. Biotinylated cDNAs were hybridized with human genome U133 Plus 2.0 array (Affymetrix) in the Genomic Facility of Centro de Investigaciones del Cancer (Salamanca, Spain) (30). Data analysis and a hierarchical cluster tree were generated using dChip software. Analysis of each cell line and/or condition was based on RNAs prepared from two independent experiments.
RESULTS
p73 Isoforms Are Differentially Regulated during Erythroid Differentiation in K562
To investigate the regulation and function of p73 variants (TA or ΔN) during hematopoiesis, we analyzed the p73 expression pattern in chemically induced differentiation in the p53-deficient K562 cells. These cells differentiated into the erythroid lineage upon treatment with cytosine arabinoside (AraC), or natural regulators of erythropoiesis like hemin and EPO (31, 32). We examined the expression pattern of the different p73 variants in K562 cells under either proliferating conditions (not treated) or after treatment with AraC, hemin, or EPO. Semi-quantitative RT-PCR using specific sets of primers that discriminate between the different C-terminal isoforms showed induction of p73α and -β mRNA upon treatment (Fig. 1, A–C, upper panels). This induction peaks at day 3, coinciding with the onset of differentiation (32). To determine whether these p73α and -β transcripts represented TA and/or ΔNp73 variants, we performed RT-PCR analysis with ΔN-specific primers (Fig. 1, A–C, lower panels) and Western blot analysis with an N-terminal (TA-specific) or a C-terminal (pan-p73) antibody (Fig. 1, D and E, respectively). We detected up-regulation of TAp73α protein with all the treatments analyzed (Fig. 1, D and E). On the other hand, ΔNp73 mRNA was up-regulated by AraC and hemin treatments (Fig. 1, A and B, lower panels), and consistently, we detected an up-regulation of ΔNp73α protein and to a minor extent ΔNp73β upon AraC treatment (Fig. 1E). However, we did not detect ΔNp73 up-regulation upon EPO treatment (Fig. 1C, lower panels).
FIGURE 1.
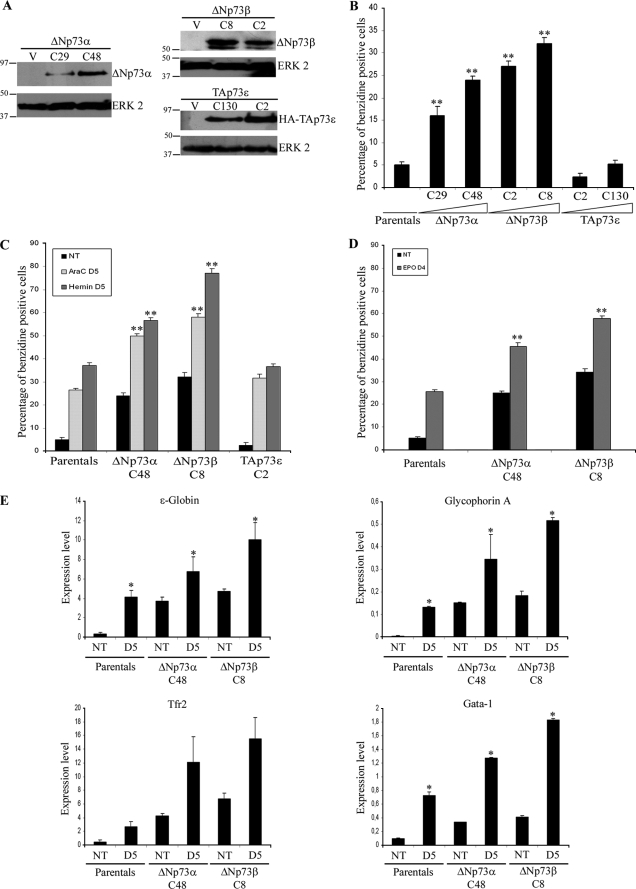
p73 isoforms are regulated during erythroid differentiation induced by AraC, hemin, and EPO in K562. A–C, expression of ΔN and TA p73 isoforms analyzed in K562 cells by semi-quantitative RT-PCR using specific primers for the different C-terminal p73 isoforms (upper panels) or the ΔN isoforms (lower panels). D–F, expression of TA and ΔNp73 isoforms in K562 (D and E) or UT-7 cells (F) was analyzed by Western blot analysis using anti-N-terminal, TA-specific antibody (D and F) or C-terminal pan-specific antibody (E). In all cases, samples were collected either under nontreated conditions (NT) or at different time points (days D1–D5) after the indicated treatment. Total ERK2 or GAPDH expression was assessed as loading control. The figures are representative of at least three independent experiments.
To examine whether p73 regulation during erythroid differentiation was also present in another human model of erythroid differentiation, we used UT-7 cells, a human megakaryoblastic leukemia cell line with erythroid potential only upon treatment with EPO (33). Consistent with a prevalent function in erythroid differentiation, TAp73α was up-regulated after EPO treatment in UT-7 cells (Fig. 1F).
Constitutive Expression of ΔNp73 Induces Erythroid Differentiation
The differential modulation of p73 isoforms (TAp73α and ΔNp73α and -β) during erythroid differentiation suggests a possible function for them in the process. We addressed this question via the generation of stable K562 cell lines expressing TAp73α, ΔNp73α, or ΔNp73β. We also transfected K562 cells with TAp73ϵ expression plasmid as a control, because of its lack of growth suppression function despite the presence of the TA domain (6). Although several clones expressing ΔNp73α, ΔNp73β, and TAp73ϵ were obtained, we repeatedly failed to get TAp73α-expressing clones. Therefore, we hypothesize that constitutive TAp73α expression was not compatible with long term culture in these cells. Two recombinant clones from each genotype were selected for further analysis (termed K562-ΔNp73α-C48 and -C29, K562-ΔNp73β-C8 and -C2, and K562-TAp73ϵ-C2 and -C130). These transfectant clones showed different levels of the transgene-encoded proteins (Fig. 2A). We next asked if the expression of these p73 isoforms affected K562 differentiation. To this end, K562 cells, as well as K562-ΔNp73α, -ΔNp73β, and -TAp73ϵ cells, were treated with AraC, hemin, or EPO. Differentiation was analyzed by a benzidine staining (Fig. 2, B–D) and expression analysis of erythroid-specific markers (ϵ-globin, glycophorin A, and GATA1) (Fig. 2E). Surprisingly, ΔNp73α and -β clones, but not TAp73ϵ, presented a significantly higher fraction of erythroid-differentiated cells than K562 under identical cell culture conditions in the absence of drugs (Fig. 2B). It is of note that the level of differentiation observed in each clone was proportional to the amount of ΔNp73 protein expressed, and even a moderated amount of ΔNp73 expression, like in clone C29 (Fig. 2A), resulted in a significant enhancement of erythroid differentiation (Fig. 2B). Furthermore, ΔNp73 clones presented a greater percentage of differentiated cells upon drug treatment than did the controls (Fig. 2, C and D). In sharp contrast with the effects of ΔNp73α and -β, TAp73ϵ expression neither induced differentiation nor affected the fraction of hemoglobinized cells in response to AraC (Fig. 2, B and C) indicating that the observed phenotype was specific, and not because of a mere overexpression of a p73 isoform. Consistent with the enhanced hemoglobin production of the ΔNp73 clones (Fig. 2B), real time RT-PCR analysis revealed that these clones expressed a higher level of the erythroid markers in nontreated conditions as well as with AraC than controls (Fig. 2E), thus confirming the pro-erythroid differentiation conditions induced by ΔNp73.
FIGURE 2.
Constitutive expression of ΔNp73 induces erythroid differentiation and enhances K562 differentiation potential. A, analysis of ΔNp73 or TAp73ϵ protein expression in stably transfected K562 cells. Western blot analysis of six of the obtained clones is as follows: ΔNp73αC29 and C48 (left panel), ΔNp73β C2 and C8 (upper right panel), and TAp73ϵ C2 and C130 (lower right panel). B–E, expression of ΔNp73 results in spontaneous erythroid differentiation and enhances AraC, hemin, and EPO induction of differentiation in K562. B–D, percentage of benzidine-positive cells was assessed in the indicated clones under nontreated culture conditions (B and NT), after 5 days of AraC or hemin treatment (C), or after 4 days with EPO (D). In all cases a minimum of 200 cells was counted in triplicate. The data are mean values from at least three independent experiments. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005). E, real time quantitative RT-PCR analysis of erythroid differentiation markers as follows: ϵ-globin, glycophorin A, and GATA1 of K562 control cells or representative clones expressing ΔNp73α (K562-ΔNp73αC48 and K562-ΔNp73C8 β). The experiments were performed at least three times, and similar results were obtained.
To further substantiate that the ΔNp73 clones were differentiating according to a normal erythroid program, a molecular characterization of ΔNp73-mediated differentiation was performed. For that purpose, a genome-wide analysis of gene expression through microarray hybridization was carried out in K562-ΔNp73α-C48, K562-ΔNp73β-C8, and K562-V cells. After filtering and statistical analysis, we found that ΔNp73α regulated 2805 genes with ≥3.45-fold changes and with a signal difference of ≥100, between both experimental conditions and p < 0.001(data not shown). As expected for an erythroid phenotype, a set of erythroid-specific genes (33 genes) was significantly up-regulated in the ΔNp73α cells (Table 1) (31, 34–70). These included erythroid markers (hemoglobins, glycophorins, transferring receptor, Rh-related antigens, or ALAS-2) as well as several erythroid-determining genes such as GATA1 and NFE2 (31, 34–70). Moreover, genes reported to be repressed during erythroid differentiation, like cyclin D1 and MEIS of FOSB, were down-regulated in ΔNp73α cells (Table 1) (68–70). Microarray analysis of the ΔNp73β clones revealed a similar profile to those of ΔNp73α (data not shown).
TABLE 1.
Data analysis of microarray, constitutive ΔNp73 expression induces regulation of erythroid differentiation-related genes
| Transcription factors | ID | -Fold change | Ref. | |
|---|---|---|---|---|
| Transcription factors | ||||
| ALAS2 | Aminolevulinate, δ-, synthase 2 (sideroblastic/hypochromic anemia) | 212 | 12.93 | 1, 2 |
| GATA1 | GATA-binding protein 1 (globin transcription factor 1) | 2623 | 2.68 | 3, 4 |
| KLF1 | Kruppel-like factor 1 (erythroid) | 10,661 | 2.45 | 5, 6 |
| NFE2 | Nuclear factor (erythroid-derived 2), 45 kDa | 4778 | 2.09 | 7, 8 |
| STAT5B | Signal transducer and activator of transcription 5B | 6777 | 1.89 | 9, 10 |
| TAL1 | T-cell acute lymphocytic leukemia 1 | 6886 | 2.41 | 11, 12 |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | 23,462 | −5.87 | 13 |
| Membrane and cytoplasmic proteins | ||||
| TFRC | Transferrin receptor (p90, CD71) | 7037 | 8.58 | 14 |
| BCL2L1 | BCL2-like I | 598 | 3.38 | 15, 16 |
| CD44 | CD44 molecule (Indian blood group) | 960 | 2 | 17, 18 |
| EPB41 | Erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | 2035 | 13.83 | 19, 20 |
| EPB49 | Erythrocyte membrane protein band 4.9 (dematin) | 2039 | 2.69 | 21, 22 |
| GYPA | Glycophorin A (MNS blood group) | 2993 | 10.58 | 23 |
| GYPB | Glycophorin B (MNS blood group) | 2994 | 9.06 | 22, 24 |
| GYPC | Glycophorin C (Gerbich blood group) | 2995 | 7.05 | 23 |
| GYPE | Glycophorin E | 2996 | 5.64 | 25 |
| HBA1 | Hemoglobin, α1 | 3039 | 3.19 | 26 |
| HBA2 | Hemoglobin, α2 | 3040 | 2.24 | 26 |
| HBB | Hemoglobin, β | 3043 | 2.71 | 26 |
| HBBP1 | Hemoglobin, β pseudogene 1 | 3044 | 3.55 | 26 |
| HBD | Hemoglobin, δ | 3045 | 4.24 | 26 |
| HBZ | Hemoglobin, ζ | 3050 | 6.37 | 26 |
| HMBS | Hydroxymethylbilane synthase | 3145 | 4.33 | 7, 27 |
| HMGN | Hemogen | 55,363 | 5.07 | 28 |
| MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 4170 | 50.02 | 29, 30 |
| RHCE | Rh blood group, CcEe antigens | 6006 | 5.49 | 31 |
| RHD | Rh blood group, D antigen | 6007 | 5.52 | 31 |
| RHAG | Rh-associated glycoprotein | 6005 | 4.88 | 32 |
| PTA1 | Spectrin, α, erythrocytic 1 (elliptocytosis 2) | 6708 | 10.77 | 33 |
| SPTB | Spectrin, β, erythrocytic (includes spherocytosis, clinical type I) | 6710 | 2.38 | 34 |
| CCND1 | Cyclin D1 | 595 | −7.94 | 35, 36 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 2354 | −83.21 | 37 |
| MEIS1 | Meis1, myeloid ecotropic viral integration site 1 homolog (mouse) | 4211 | −2.18 | 38 |
ΔNp73 Induces Accumulation of Transcriptionally Active TAp73, Which Is Necessary for the Erythroid Phenotype
We observed that some of the genes up-regulated in the K562-ΔNp73 transcriptome of the cell, like p21CIP1, BTG2, GDF15, or REELIN, were p53 and/or TAp73 target genes (Fig. 3A and data not shown). Because K562 cells lack p53, we hypothesized that the up-regulation of some of these genes could be p73-dependent. To address this question, we first investigated the status of endogenous TAp73 in these cells. In K562 cells we detected a low level of TAp73α protein, which rapidly increased upon AraC treatment (Fig. 1E and Fig. 3A). However, elevated levels of the TAp73α protein level were observed in nontreated ΔNp73 clones, comparable with those detected in treated K562 cells (Fig. 3A). This accumulation of TAp73α protein in ΔNp73-expressing cells is in agreement with previously reported stabilization of TAp73α by ΔNp73 (71). Furthermore, in all the clones analyzed TAp73 level was proportional to the amount of transgenic ΔNp73 expressed (data not shown), suggesting that ΔNp73 was responsible of the TAp73 accumulation. Moreover, a strong TAp73 induction, with a concomitant increase in hemoglobin-producing cells, was detected in K562 cells transiently transfected with an inducible ΔNp73 expression plasmid (Fig. 3, B and C), ruling out the possibility of an artifact due to the selection of a clone resistant to p73 growth inhibition. Consistently, constitutive expression of ΔNp73α and -β in UT-7 cells also resulted in an increase of endogenous TAp73α protein (supplemental Fig. 1A).
FIGURE 3.
ΔNp73 induces accumulation of transcriptionally active TAp73. A, Western blot analysis of endogenous TAp73α and p21Cip1 in nontreated conditions (NT) or following treatment with AraC (D1–D3) in K562 cells or in K562-ΔNp73α and -β clones without treatment. B and C, Western blot analysis of endogenous TAp73α and induced HA-ΔNp73 (B) or percentage of benzidine positive cells (C) in samples of K562 cells transiently transfected with an inducible HA-ΔNp73 expression plasmid or empty vector and collected at different time points upon activation with zinc.
The former results suggest that ΔNp73 results in elevated TAp73 protein, which in turn could regulate p73 target genes. To substantiate this hypothesis, we next investigated if TAp73 was transcriptionally regulating one of its classical targets, p21Cip1, in the ΔNp73-expressing cells (Fig. 3A). ChIP assays with TAp73-specific antibodies demonstrated that TAp73 was recruited to the p53-binding site of the p21CIP1 promoter upon treatment (Fig. 4A), suggesting a role for TAp73 in drug-induced p21CIP1 expression in K562.
FIGURE 4.
p73 is recruited to the p53 response element (RE) of the p21CIP1 (A) or the NOXA promoter (B and C) in a context-dependent manner. Chromatin immunoprecipitation assays (ChIP) of K562 untreated or treated for 3 days with AraC or K562-ΔNp73 α and β cells without treatment were performed using no antibody (H2O), anti-p53, anti-ΔNp73, or anti-TAp73 antibodies. Real time RT-PCR using primers to amplify the region encompassing p53RE of the human p21CIP1 or NOXA promoters was performed, and the data were normalized to input chromatin samples in each case. C and D, UT-7 or HCT116p53−/− cells were transfected with ΔNp73 expression plasmid, and 24 h later, ChIP assays were performed as before to analyze p73 recruitment to the p53RE of the p21Cip1 (C), and p21Cip1 expression levels were quantified by real time RT-PCR (D). The data are mean values from at least three independent experiments. NT, not treated. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005).
Analysis of the growth kinetics of the ΔNp73 clones demonstrated that these cells presented a slower growth rate than K562 controls even though cell cycle analysis did not reveal a significant accumulation of cells in either G1 or G2/M phase of the cycle (supplemental Fig. 2, A and B). Congruently, they expressed elevated levels of p21Cip1 under not treated conditions (Fig. 3A). Moreover, endogenous TAp73 was bound to the p21CIP1 promoter, despite the presence of high levels of ΔNp73 in these cells (Fig. 4A). To rule out the possibility that the results were due to the lack of specificity of the TAp73 antibody, we analyzed the binding of these proteins to the p53-response element of the pro-apoptotic NOXA promoter. No significant PCR product of this response element, or the p21CIP1, was obtained when we immunoprecipitated with the TAp73 antibody untreated K562 extracts (Fig. 4B). Furthermore, lack of significant PCR product with the anti-p53 antibody further demonstrated the specificity of the assay (Fig. 4, A and B).
ΔNp73 inhibitory function is exerted either by forming heterotetramers with TAp73 or by competing for binding to the same DNA target sequence (72, 73). However, despite the elevated levels of the ΔNp73 protein, we did not detect ΔNp73 binding to the p21CIP1 promoter in the ΔNp73 clones (Fig. 4A). Nevertheless, under the same experimental conditions we were able to detect both TAp73 and ΔNp73 binding to the NOXA promoter, indicating the presence of heterodimers in this promoter and suggesting that TA and ΔN isoforms were not binding to all the promoters in the same way (Fig. 4, A and B).
Therefore, we sought to investigate whether the binding of TA and ΔNp73 to a growth arrest promoter, like p21CIP1, would be different depending on the cellular context. For that purpose, we transfected ΔNp73α or -β into UT-7 or the human colon carcinoma cell line HCT116 p53−/−, both containing endogenous TAp73, under the same experimental conditions. ChIP assays showed that ΔNp73α or -β transfection in UT-7 cells resulted in preferential binding of endogenous TAp73 over ΔNp73 (TA > ΔN) to the p21CIP1 promoter with a concurrent p21Cip1 expression (Fig. 4, C and D). However, in HCT116p53−/− cells there was a ΔNp73 preferential binding over TAp73 to the promoter (ΔN ≥ TA), with no induction of p21Cip1 expression (Fig. 4, C and D). These results support the hypothesis in which the outcome of p73 isoforms interaction will depend on the cellular context.
Nevertheless, TAp73 recruitment to the p21CIP1 promoter might not necessarily indicate transcriptional activity. Thus, we next asked whether TAp73 was required for p21Cip1 expression in ΔNp73 cells. p73 knockdown was performed using two different siRNAs that inhibit expression of either TAp73 isoforms specifically (siRNA-p73-3) or all the p73 variants (siRNA-p73-4) (28). These siRNAs have been previously published and demonstrated to be efficacious siRNA constructs that are restricted to p73 and have no homology to p63 (74). Western blots were quantified by densitometric analysis and the values normalized to those of the total ERK values. We observed that the pan-p73 siRNA-p73-4 in K562-ΔNp73β cells repressed 33% of TAp73α and 61% of ΔNp73β expression (Fig. 5A). This inhibition resulted in an 80% decrease in p21Cip1 expression (Fig. 5A), denoting the requirement of functional p73 for p21Cip1 expression. However, in conditions in which TA-specific siRNA-p73-3 resulted in no significant reduction of ΔNp73β transgene expression (only −2.4%), but knocked down 28% of TAp73, a comparable repression of p21Cip1 (−75%) was obtained (Fig. 5A). Thus, a slight modulation of TAp73 expression, in the presence of sustained expression of ΔNp73β, was sufficient to abate p21Cip1 levels, in a similar manner to those obtained when all p73 forms were affected by the siRNA-p73-4. Consistently, a decrease in p21Cip1 level was observed by RT-PCR analysis with both sets of siRNA (Fig. 5C). Similar results were obtained when the experiments were performed in the ΔNp73α clone (Fig. 5, A and C). Furthermore, p73 knockdown alleviated the slower rate of proliferation of the ΔNp73-expressing cells (supplemental Fig. 2C). These data demonstrated that the endogenous TAp73 was required for p21Cip1 expression in ΔNp73 clones. Therefore, in K562 cells, ΔNp73 expression results in the accumulation of transcriptionally active TAp73.
FIGURE 5.
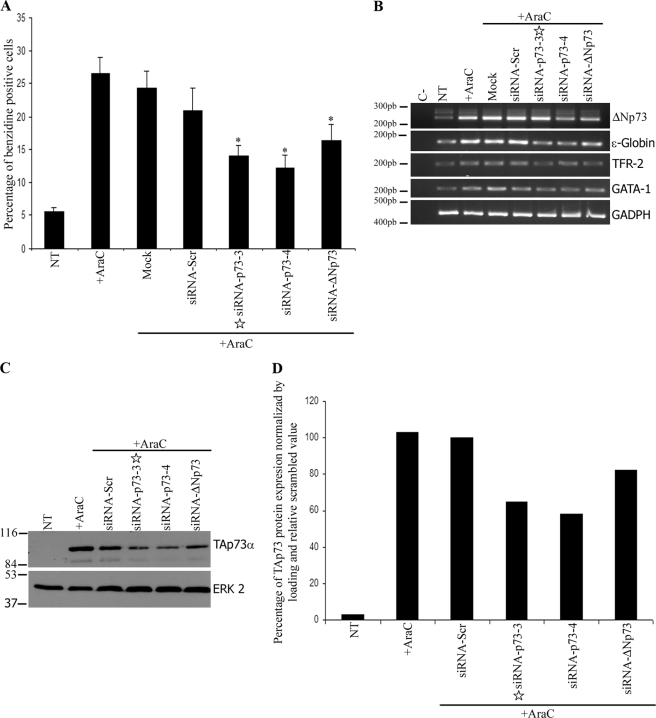
TAp73α functional inhibition results in suppression of p21Cip expression and attenuation of erythroid differentiation in the K562-ΔNp73 cells. A–D, K562-ΔNp73α and -β cells were transfected with annealing buffer only (mock) or siRNA against either all p73 isoforms (siRNA p73-4), TAp73 specific (siRNA p73-3 marked with a star), ΔNp73-specific (siRNA-ΔNp73), or scrambled control (siRNA-scr). After 72 h, samples were collected, and endogenous TAp73, transgenic ΔNp73, and p21Cip were analyzed by Western blot (A and D), benzidine assays (B), and RT-PCR of p21Cip and erythroid markers (C). For each ΔNp73 clone, the percentage of benzidine-positive cells obtained in the mock conditions was considered 100%, and the rest of the values were normalized to this one. The data are mean values from three independent experiments. Error bars indicate S.E. (**, p < 0.005). The asterisk in A refers to a nonspecific band.
We next asked whether TAp73 played a role in the erythroid phenotype of these clones. To that extent, we analyzed the effect of p73 knockdown in the erythroid differentiation of ΔNp73 clones. Partial inhibition of p73 with the pan-p73 siRNA resulted in a 42–44% decrease in the fraction of hemoglobinized cells (Fig. 5B, siRNA-p73-4) with a concomitant attenuation of expression of the erythroid differentiation markers (Fig. 5C), pointing to a relevant role of p73 in this process. Furthermore, 28–33% knockdown of TAp73 with the TA-specific siRNA-p73-3 resulted in a significant 34–38% reduction of differentiated cells in ΔNp73α and -β clones, respectively (Fig. 5, B and C), despite the remaining elevated levels (98%) of ΔNp73 (Fig. 5A). This result demonstrates that ΔNp73 is not sufficient to induce erythroid differentiation, and it requires the expression of TAp73 for this function. It is important to note that in our working conditions we do not knock down TAp73 completely, and hence, we still detect p21Cip1 as well as erythroid differentiation levels above those of nontreated K562 cells.
It is noteworthy that in ΔNp73 clones the pan-p73 siRNA-p73-4 only attained a partial TAp73 repression (33–44%), although in identical experimental conditions it silenced close to 90% of the AraC-induced TAp73 expression in parental cells (see Fig. 7, A and D, and Fig. 9A). This could be a reflection of the enhanced stability of TAp73 protein in the presence of ΔNp73. Furthermore, abatement of ΔNp73 expression in the ΔNp73 clones, using a previously validated ΔN-specific siRNA oligonucleotide with no sequence homology to TAp73 (29), resulted in a strong decrease of the TAp73α protein (Fig. 5D). Under the same experimental condition, these oligos did not affect TAp73ϵ protein levels in the TAp73ϵ-expressing clones, despite its sequence homology with TAp73α, ruling out the possibility that the siRNAs were directly affecting TAp73α. Neither affected the expression of the co-transfected green fluorescent protein vector under conditions in which ΔNp73β expression was repressed, excluding the possibility that the ΔN-specific siRNAs affected the viability of the transfected cells (supplemental Fig. 3), altogether suggesting that ΔNp73 is responsible of the accumulation of TAp73α.
FIGURE 7.
p73 functional inhibition attenuates drug-induced erythroid differentiation in K562 cells. A–F, p73 siRNA decreases the differentiation potential of AraC and hemin. siRNA was performed as before in K562 cells, and 48 h later the cells were treated with AraC (A–C) or hemin (D–F). Samples were collected 3 days after treatment to perform Western blot analysis (A and D), benzidine assays (B and E), and RT-PCR of erythroid-specific markers (C and F). The data are mean values from three independent experiments. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005). NT, not treated.
FIGURE 9.
p73 function is required for EPO-induced erythroid differentiation in K562 and UT-7 cells. A–F, siRNA was performed as before in K562 or UT-7 cells, and 48 h later the cells were treated with EPO. Samples were collected 3 days after treatment for Western blot analysis (A and D), benzidine assays (B and E), and RT-PCR of erythroid-specific markers (C and F).The data are mean values from at least three independent experiments. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005). NT, not treated.
Knockdown of ΔNp73, and therefore TAp73, also resulted in a significant repression of erythroid differentiation comparable with that obtained by the pan-p73 siRNAs (Fig. 5, B and D). Therefore, in K562 cells ΔNp73 expression brings about TAp73 protein accumulation, which is required for the erythroid differentiation.
GATA1 Is a Direct Target of TAp73, Which Is Required for Erythroid Differentiation in K562 Cells
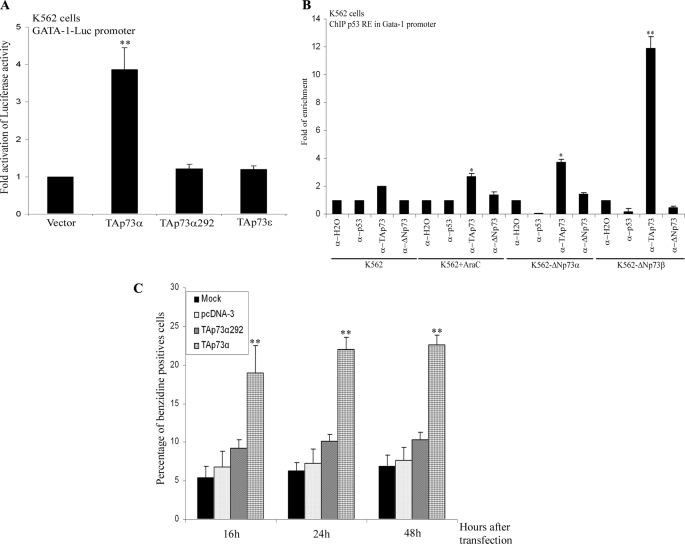
Our previous results indicate that TAp73 has a pro-erythroid differentiation function in K562-ΔNp73 cells. Furthermore, GATA1, a transcription factor determinant in erythropoiesis, together with some of its known targets (ALAS2, NFE2, EPOR, hemoglobins, and Bcl-XL) (37, 75) was found to be up-regulated in K562-ΔNp73 cells (Fig. 2E and Table 1). Hence, we hypothesized the existence of a functional link between p73 and GATA1. We first examined whether constitutive expression of TAp73 could induce GATA1 activity. We used a GATA1-luc reporter gene containing three GATA1-binding sites (23). Transient transfection of TAp73α, but not TAp73ϵ or TAp73α292, a DNA-binding incompetent mutant, significantly activated the GATA1-luc reporter (Fig. 6A).
FIGURE 6.
TAp73α induces GATA1 activity, binds to the GATA1 promoter, and induces erythroid differentiation. A, transcriptional analysis was performed in K562 cells transiently transfected with the GATA1-luciferase reporter promoter together with the indicated expression vectors. Luciferase activity was normalized by the Renilla activity of the same lysate. The data represent the mean of at least three experiments. B, endogenous TAp73 is recruited to the p53RE of the GATA1 promoter. ChIP of K562 control cells, untreated or with AraC for 3 days, or ΔNp73-K562 (α and β clones) under nontreated conditions were performed using no antibody (H2O), anti-p53, anti-ΔNp73, or anti-TAp73 antibodies. Real time RT-PCR using specific primers to amplify p53-binding sites of the human GATA1 promoter was performed, and the data were normalized to input chromatin samples of each case. C, K562 cells were transiently transfected with the indicated p73 expression vectors, and the percentage of benzidine-positive cells was assessed at different time points under normal culture conditions. The data are mean values from at least three independent experiments. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005).
To determine whether GATA1 was a direct transcriptional target of TAp73α, we performed an in silico prediction of p53-responsive elements within the human GATA1 locus using p53Family-TargetGenes data base (76). We identified a p53-response element within the human GATA1 promoter at position −3.326 kb (ACCCTTGTCTTAGCTCTTAGACATGCCTCGAGCCTGCCA). This sequence lies within a region considered a transcriptional control element located between −3.9 and −2.6 kb 5′ to the erythroid first exon that is sufficient to recapitulate GATA1 expression in the primitive and definitive erythroid lineages (77). ChIP assays with TAp73- and ΔNp73-specific antibodies demonstrated that TAp73 but not ΔNp73 was bound significantly to the GATA1 promoter in K562 parental cells treated with AraC for 3 days (Fig. 6B). In ΔNp73 clones TAp73, but not ΔNp73, was recruited to this p53-binding site (Fig. 6B). Consistently, transient expression of TAp73α, but not TAp73α292, induced erythroid differentiation in K562 cells (Fig. 6C).
The capacity of p73 to modulate GATA1 led us to propose that p73 could play a direct role in promoting erythroid maturation rather than a mere regulation of growth suppression through p21CIP1. To substantiate this claim, we first asked whether p73 was necessary for chemically induced erythroid differentiation using two compounds that triggered differentiation by different mechanisms. Even though p21Cip1 expression correlates with erythroid differentiation in many experimental models, it is not essential for all drug-induced differentiations (31). AraC inhibits cell proliferation inducing p21Cip1 expression, whereas hemin selectively activates the embryonic and fetal globin synthesis without inducing p21Cip1 or cell cycle arrest (31). p73 knockdown, with pan-p73 and TA-specific siRNA, in K562 cells treated with either AraC (Fig. 7, A–C) or hemin (Fig. 7, D–F) resulted in a significant reduction of hemoglobinized cells and a decrease in the expression of the erythroid markers. It is noteworthy that in both scenarios p73 functional inhibition decreases GATA1 expression (Fig. 7, C and F). These results confirm that the role of TP73 in erythroid differentiation is not dependent only on the induction of p21Cip1 or cell cycle arrest but corresponds to a common pathway of drug-induced differentiation shared by these drugs. Moreover, electroporation of K562 cells with the ΔNp73-si-RNA-specific oligos resulted in a comparable inhibition of AraC-induced erythroid differentiation (Fig. 8, A and B). Nevertheless, this inhibition was accompanied with a moderate (18%) reduction of AraC-induced TAp73α (Fig. 8, C and D) despite the demonstrated specificity of these oligos. However, even in conditions in which p73 knockdown was very effective, we still detected a percentage of hemoglobinized cells above the nontreated control, suggesting the existence of p73-independent mechanisms of drug-induced erythroid differentiation.
FIGURE 8.
ΔNp73 functional inhibition attenuates AraC-induced erythroid differentiation in K562 cells. siRNA was performed as before in K562 cells, and 48 h later the cells were treated with AraC. Samples were collected 3 days after treatment to perform benzidine assays (A), RT-PCR of erythroid-specific markers (B), or Western blot analysis (C and D). D, TAp73α protein expression from C was quantified by densitometry, normalized by the loading controls, and represented as relative value to the scrambled siRNA. The data are mean values from three independent experiments. Error bars indicate S.E. (*, p < 0.05; **, p < 0.005). NT, not treated.
p73α Function Is Required for EPO-induced Erythroid Differentiation in K562 and UT-7 Cell Lines and Plays a Role in Vivo in Erythropoiesis
To address whether p73 function was necessary during physiological erythropoiesis, we first analyzed p73 requirement in EPO-induced erythroid differentiation in K562 cells (Fig. 9, A–C). As a second model, we studied p73 involvement in EPO-induced differentiation in UT-7 cells (Fig. 9, D–F). We have previously demonstrated that in these cells TAp73α accumulation, induced by ectopic expression of ΔNp73α, was accompanied by a moderate increase in benzidine-positive cells (from 5% in vector-transfected cells to 13–15% in ΔNp73α- and -β transfected, respectively) (supplemental Fig. 1, A and B).
A partial p73 knockdown with siRNA-p73 in EPO-treated K562 cells resulted in an attenuation of hemoglobinized cells and in a sharp decrease in expression of erythroid markers (Fig. 9, A–C). Next we tested the effects of p73 silencing in UT-7 cells. Similar results were obtained with electroporation of 150–200 nm siRNA-p73-4 (Fig. 9, D–F). In these cells electroporation with 100 nm oligos did not achieve a significant repression of TAp73α expression (supplemental Fig. 4) and therefore did not significantly repress EPO-induced erythroid differentiation (Fig. 9E). Altogether these data indicate that TAp73 function is required, at least in part, for appropriate EPO-induced erythroid differentiation in human myeloid cells.
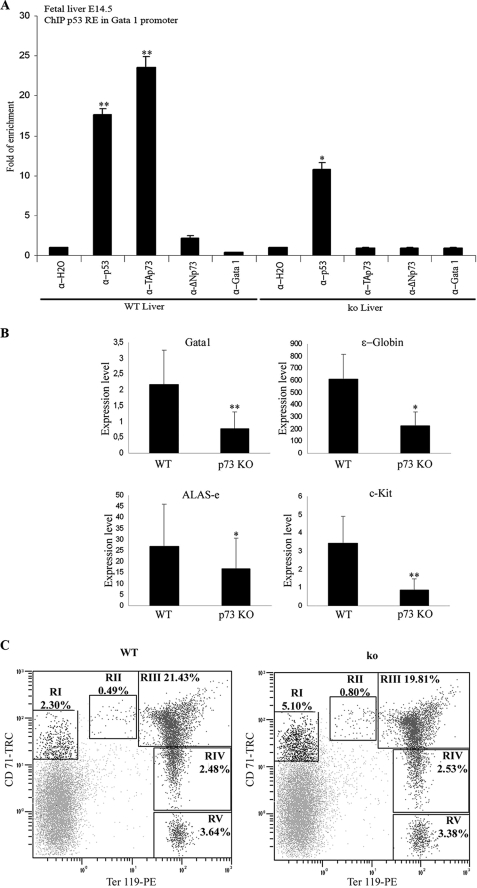
We next sought to investigate the possible role of p73 in physiological hematopoiesis in vivo. Because definitive hematopoiesis in the mouse embryo takes place in the fetal liver, we first analyzed whether p73 would bind to the p53RE within the Gata1 promoter in fetal livers of E14.5 embryos. ChIP analysis revealed that TAp73 and p53, but not ΔNp73, bind to this promoter in vivo (Fig. 10A), suggesting that Gata1 is an in vivo transcriptional target of p73. As a control, we performed ChIP analysis with fetal livers from p73KO E14.5 littermates (4) and did not detect any TAp73 binding (Fig. 10A). Because erythroid differentiation is dose-dependent with respect to GATA1 (37), we hypothesized that in the p73KO mice, lack of p73 could result in impaired GATA1 expression and therefore deficient erythropoiesis. We analyzed GATA1 expression in fetal livers of E14.5 p73KO embryos and control littermates. Real time quantitative RT-PCR analysis revealed significantly lower levels of GATA1 in p73KO when compared with control littermates (Fig. 10B). Consistently, expression of GATA1 target genes such as ALAS-e, ϵ-globin, or regulators like c-Kit, were also significantly diminished (Fig. 10B). Accordingly, macroscopic analysis revealed that 60% of p73KO embryos presented a pallor phenotype suggesting an anemic state (supplemental Fig. 5). Peripheral blood analysis of p73KO young mice (2 weeks old) confirmed a mild degree of anemia with 25% reduction in the red blood cells and 38% less hematocrit value than the control littermates (Table 2). Older animals were not analyzed because the half-life of p73KO mice is between 15 and 20 days (4). To further analyze the effect of p73 ablation on physiological hematopoiesis, we utilized a flow cytometric assay, with surface markers TER119\CD71 labeling, previously validated as an excellent method to quantify erythroid precursors and the maturation stage of differentiation in the hematopoietic tissue (78, 79). We observed a highly significant 2-fold accumulation (supplemental Fig. 5, p < 0.005) on the pre-committed erythroid progenitor(TER119low/CD71med) population in the bone marrow of P15 p73KO mice compared with control littermates (Fig. 10C). This accumulation could represent a blockage in the erythroid differentiation process before the commitment to pro-erythroblast. However, once the cells are committed to the erythroid differentiation, we did not detect any differences in the number of differentiated erythrocytes between wild type and p73KO bone marrow cells (Fig. 10C and supplemental Fig. 5). Altogether, these data support the role of p73 in physiological erythropoiesis.
FIGURE 10.
p73 plays a physiological role in developmental hematopoiesis. A, endogenous TAp73 is recruited to the p53RE of the Gata1 promoter in vivo. ChIP of E14.5 fetal livers extracts of wild type (WT) and p73KO (ko) embryos were performed using no antibody (H2O), anti-GATA1, anti-p53, anti-ΔNp73, or anti-TAp73 antibodies. Real time RT-PCR using specific primers to amplify p53-binding sites of the human GATA1 promoter was performed, and the data were normalized to input chromatin samples of each case. B, real time quantitative RT-PCR analysis of GATA1 and erythroid-related genes such as ALAS-e, ϵ-globin, and c-Kit in E14.5 p73KO livers compared with control littermates (WT). The data are mean values from at least three independent experiments from all the mice of each genotype (wild type, n = 7; p73KO, n = 6). Error bars indicate S.E. (*, p < 0.05; **, p < 0.005). C, representative example of the assessment of erythroid developmental stages in bone marrow of p73KO or wild type P15 mice. Cells at different stages were identified by double staining with anti-TER-119 and anti-CD71 antibodies. RI, TER119low/CD71med, pre-committed erythroid progenitor; RII, CD71high/TER-119low, proerythroblasts; RIII, CD71high/TER-119high, basophilic erythroblasts; RIV, CD71low-med/TER-119high, polychromatic and orthochromatic erythroblasts; RV, CD71−/TER-119high mature erythroid cells. The data are representative examples from all the mice of each genotype (wild type, n = 5; p73KO, n = 5).
TABLE 2.
Hematopoietic indices: p73KO mice showed moderated anemia in comparison with littermates
| Mouse type | Mouse number | Hemoglobin | Hematocrit | Red blood cell count | White blood cell count | Platelet count |
|---|---|---|---|---|---|---|
| g/dl | % | ×104/mm3 | ×103/mm3 | ×104/mm3 | ||
| Wild type | 13 | 7.2 ± 0.67 | 32.92 ± 1.07 | 327.92 ± 20.86 | 2.63 ± 0.39 | 37.15 ± 3.25 |
| p73KO | 11 | 4.7 ± 0.59a | 27.86 ± 0.64b | 241.85 ± 15.41b | 1.95 ± 0.33a | 34.57 ± 2.87 |
a p < 0.05.
b p < 0.005.
DISCUSSION
There is ample evidence indicating that leukemia, as well as other myeloproliferative disorders, result from alterations in the differentiation of hematopoietic cells (31). Therefore, investigation of the cellular and molecular processes involved in physiological, as well as abnormal, hematopoiesis could be very valuable in the understanding and treatment of these diseases.
Despite their sequence homology, the p53 family members possess both common as well as nonoverlapping functions (3). p73 maintains a specific role in cellular differentiation and development (22, 80, 81). The regulation of cellular differentiation has been described as a mechanism of tumor suppression (82). In this line, the role of p73 in leukemogenesis has been associated with its ability to regulate differentiation (19, 20, 83). In this study we provide a comprehensive analysis of the regulation and function of p73 variants during erythroid differentiation in K562 cells. Previous studies have shown that the K562 represents an important in vitro model for basic studies of hematopoietic differentiation (31). To investigate the regulation of p73 in hematopoietic cells, we examined the expression profile of p73 in the K562 cells under various culture conditions. We show that TAp73α was up-regulated by all the erythroid differentiation treatments analyzed, although ΔNp73 was up-regulated during AraC and hemin treatment. This regulation is also present in the EPO-induced differentiation of the human megakaryoblastic leukemia cell line UT-7. The modulation of specific p73 isoforms suggests a function for these isoforms in erythroid differentiation.
Although transient expression of TAp73 induced GATA1 activity and erythroid differentiation, we barely detected exogenous HA- TAp73 expression, or benzidine-positive cells, after 72 h post-transfection. This supports the hypothesis that constitutive expression of TAp73α at high levels, in the absence of ΔNp73 expression, is lethal in K562, making stable expression of TAp73α incompatible with long term cell culture. However, constitutive expression of ΔNp73 (α and β) results in enhanced erythroid differentiation in the two myeloid cell lines analyzed. Furthermore, in this work we have demonstrated that although ΔNp73 is necessary for the accumulation of the transcriptionally active TAp73, which is required for erythroid differentiation, it is not sufficient for the erythroid phenotype in the K562-ΔNp73 clones. Moreover, we have shown that attenuation of ΔNp73 expression hinders AraC-induced differentiation in K562 cells. This observation suggests that, in this context, ΔNp73 expression works as a positive regulator of erythroid differentiation, and it is consistent with the recent observation that ΔNp73 acts as a pro-differentiation factor in acute promyelocytic leukemia cells and might be necessary for myeloid differentiation (19).
This scenario would be in apparent contradiction with the trans-dominant function of ΔNp73. This inhibitory function is exerted at the oligomerization level and/or by competing for binding to the DNA target sequences. Furthermore, ΔNp73 can act as a transcription factor by binding to the promoter of its targets genes (14, 15, 18). However, it has been recently proposed that ΔNp73α could not only act as an inhibitor of p53/TAp73 functions but could also cooperate with p53 in the activation of certain genes in a cellular context-dependent manner (14, 15, 18). We have shown that TAp73 is present as homotetramers in the promoters in which it is active, like p21CIP1 or GATA1, because we do not detect ΔNp73 binding in the same experimental conditions in which we can detect binging to the NOXA promoter. However, ΔNp73 expression is required for the long term induction of erythroid differentiation by TAp73 in K562 cells. These results corroborate the need for ΔNp73 expression, along with TAp73α, to produce a stable erythroid phenotype in these cells. It is possible that ΔNp73 is acting as a survival factor that protects the cells from the apoptosis resulting from the enhanced TAp73 expression occurring during the differentiation process, without hindering TAp73 pro-differentiation function. From this perspective the lack of induction of ΔNp73 by EPO could be explained as the absence of a selective pressure for an additional anti-apoptotic mechanism, because EPO, by itself, promotes erythroid cell survival by regulating Bcl-XL expression through the JAK2/STAT5 pathway (84). ΔNp73 could exert this anti-apoptotic function by directly inducing the expression of anti-apoptotic genes like HSP70 (49), and/or by suppressing TAp73 transactivation of pro-apoptotic genes through the formation of heterotetramers.
We propose that the outcome of p73 isoform interactions will depend on the cellular context. Accordingly, in multipotent myeloid cells like K562 or UT-7, ΔNp73 will selectively block pro-apoptotic TAp73 target promoters (like NOXA) while allowing preferential TAp73 binding to, and transactivation of, the cell cycle arrest and pro-differentiation promoters (like p21CIP1 and GATA1), resulting in differentiation induction and survival of the differentiated cell. However, in a differentiation-incompetent epithelial cell line (HCT116 p53−/−), ΔNp73 will block binding of TAp73 to all the target genes inhibiting TAp73-induced growth arrest and apoptosis. In accordance with this hypothesis, we have demonstrated that in K562 or UT-7 cells there is a preferential binding of TAp73 versus ΔNp73 to promoters like p21CIP1 and GATA1. However, that is not the case in pro-apoptotic promoters like NOXA where a preferential ΔNp73 versus TAp73 binding is observed. Furthermore, while in UT-7 cells ΔNp73 transfection resulted in preferential endogenous TAp73 binding to the p21CIP1 promoter with a concomitant p21Cip expression, in HCT116p53−/− cells resulted in ΔNp73 preferential binding to the p21CIP1 promoter with no p21Cip expression. However, although our results support the hypothesis in which the outcome of p73 isoform interactions will depend on the cellular context, the molecular mechanism of this selective cooperation remains to be deciphered.
The hematopoietic system requires a continual replacement of cells through the life span of the organism. This is achieved through self-renewal of the hematopoietic stem cells and their sequential commitment to progenitor and precursor cells. The molecular mechanisms that determine these cell fate decisions are regulated by lineage-specific transcription factors (GATA1, KLF1, TAL1, SP1, PU.1, etc.) working in conjunction with general transcription factors (85). In our work, we have observed that in K562, a cell line with characteristics of multipotential progenitor cells, constitutive expression of ΔNp73 up-regulates some of these lineage-specific factors (GATA1, KLF1, and TAL1) probably through the up-regulation of TAp73α. Moreover, we have demonstrated for the first time that GATA1 is a direct target of TAp73α in vitro and in vivo. Furthermore, we have shown that reduction in p73 expression results in an impaired induction of GATA1 during drug-induced erythroid differentiation by EPO, hemin, and AraC in K562 and EPO-treated UT-7 cells.
GATA1 is a zinc finger transcription factor that plays a central role in erythroid development being a master regulator of terminal differentiation of erythroid precursors. Strikingly, analysis of p73KO embryos livers showed a significantly diminished expression in the fetal liver of Gata1 and other erythroid related genes, including globins. We also detected a significant decrease in the expression of c-Kit, known to be required for the optimal production of red cells in vivo through the collaboration with erythropoietin receptor and GATA1 (86). The altered levels of important regulators of erythroid differentiation in vivo in the p73KO embryos support the idea that TP73 fulfills a relevant function in this process. Moreover, peripheral blood analysis of p73KO 2-week-old mice confirmed moderated anemia in these animals, suggesting that p73 is required, at least in part, during erythropoiesis. In an attempt to state at which step of erythropoiesis p73 may play a role, we performed a flow cytometric assay that allows quantitative evaluation of erythroid differentiation in hematopoietic tissues (78, 79). We observed that lack of p73 resulted in an accumulation of the pre-committed erythroid progenitor population in the bone marrow of P15 p73KO mice. This accumulation could represent a blockage in the erythroid differentiation process before the commitment to pro-erythroblast. However, once the cells are committed to the erythroid differentiation, we do not detect any differences in the number of differentiated erythrocytes between wild type and p73KO bone marrow cells. An alternative explanation could be that the enhanced number of erythroid progenitors reflects a defect in the p73KO mice to increase their erythropoietic rate in response to stress (such as anemic state). Although further studies are needed to establish the requirement of TP73 in the development of hematopoietic precursors, our data point to a physiological role of the TP73 gene in erythropoiesis.
Supplementary Material
Acknowledgments
We thank all the people cited in the text for their generous gift of reagents. We are grateful to Dr. A. Fernández for helpful advice and discussion of the manuscript.
This work was supported by Grants SAF2006-10756, SAF2005-00461, SAF2008-1883, and RETIC-RD06/0020 from Spanish Ministerio de Educación y Ciencia (to M. C. M., J. L. S., and J. C. S.) and Grant LE030A07 from the Junta de Castilla y León (to M. M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
- AraC
- cytosine arabinoside
- RT
- reverse transcription
- EPO
- erythropoietin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ChIP
- chromatin immunoprecipitation
- oligo
- oligonucleotide
- siRNA
- small interfering RNA
- RE
- responsive element
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Bourdon J. C., Fernandes K., Murray-Zmijewski F., Liu G., Diot A., Xirodimas D. P., Saville M. K., Lane D. P. (2005) Genes Dev. 19, 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melino G., De Laurenzi V., Vousden K. H. (2002) Nat. Rev. Cancer 2, 605–615 [DOI] [PubMed] [Google Scholar]
- 3.Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006) Cell Death Differ. 13, 962–972 [DOI] [PubMed] [Google Scholar]
- 4.Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A., McKeon F., Caput D. (2000) Nature 404, 99–103 [DOI] [PubMed] [Google Scholar]
- 5.Ishimoto O., Kawahara C., Enjo K., Obinata M., Nukiwa T., Ikawa S. (2002) Cancer Res. 62, 636–641 [PubMed] [Google Scholar]
- 6.De Laurenzi V., Costanzo A., Barcaroli D., Terrinoni A., Falco M., Annicchiarico-Petruzzelli M., Levrero M., Melino G. (1998) J. Exp. Med. 188, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moll U. M., Slade N. (2004) Mol. Cancer Res. 2, 371–386 [PubMed] [Google Scholar]
- 8.Jost C. A., Marin M. C., Kaelin W. G., Jr. (1997) Nature 389, 191–194 [DOI] [PubMed] [Google Scholar]
- 9.Fang L., Lee S. W., Aaronson S. A. (1999) J. Cell Biol. 147, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung M. S., Yun J., Chae H. D., Kim J. M., Kim S. C., Choi T. S., Shin D. Y. (2001) Oncogene 20, 5818–5825 [DOI] [PubMed] [Google Scholar]
- 11.De Laurenzi V., Rossi A., Terrinoni A., Barcaroli D., Levrero M., Costanzo A., Knight R. A., Guerrieri P., Melino G. (2000) Biochem. Biophys. Res. Commun. 273, 342–346 [DOI] [PubMed] [Google Scholar]
- 12.Zaika A. I., Slade N., Erster S. H., Sansome C., Joseph T. W., Pearl M., Chalas E., Moll U. M. (2002) J. Exp. Med. 196, 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrenko O., Zaika A., Moll U. M. (2003) Mol. Cell. Biol. 23, 5540–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y., Kameoka M., Itaya A., Ota K., Yoshihara K. (2004) Biochem. Biophys. Res. Commun. 317, 865–872 [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y., Ota K., Kameoka M., Itaya A., Yoshihara K. (2006) Exp. Cell Res. 312, 1254–1264 [DOI] [PubMed] [Google Scholar]
- 16.Liu G., Nozell S., Xiao H., Chen X. (2004) Mol. Cell. Biol. 24, 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kartasheva N. N., Lenz-Bauer C., Hartmann O., Schäfer H., Eilers M., Dobbelstein M. (2003) Oncogene 22, 8246–8254 [DOI] [PubMed] [Google Scholar]
- 18.Goldschneider D., Million K., Meiller A., Haddada H., Puisieux A., Bénard J., May E., Douc-Rasy S. (2005) J. Cell Sci. 118, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 19.Mainardi S., Pelosi A., Palescandolo E., Riccioni R., Fontemaggi G., Diverio D., Testa U., Sacchi A., Grignani F., Lo-Coco F., Levrero M., Blandino G., Rizzo M. G. (2007) Cell Death Differ. 14, 1968–1971 [DOI] [PubMed] [Google Scholar]
- 20.Tschan M. P., Grob T. J., Peters U. R., Laurenzi V. D., Huegli B., Kreuzer K. A., Schmidt C. A., Melino G., Fey M. F., Tobler A., Cajot J. F. (2000) Biochem. Biophys. Res. Commun. 277, 62–65 [DOI] [PubMed] [Google Scholar]
- 21.Peters U. R., Tschan M. P., Kreuzer K. A., Baskaynak G., Lass U., Tobler A., Fey M. F., Schmidt C. A. (1999) Cancer Res. 59, 4233–4236 [PubMed] [Google Scholar]
- 22.Fernandez-Garcia B., Vaqué J. P., Herreros-Villanueva M., Marques-Garcia F., Castrillo F., Fernandez-Medarde A., León J., Marín M. C. (2007) Cell Death Differ. 14, 254–265 [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T., Suwabe N., Dai P., Yamamoto M., Ishii S., Nakano T. (2000) Oncogene 19, 134–140 [DOI] [PubMed] [Google Scholar]
- 24.Delgado M. D., Lerga A., Cañelles M., Gómez-Casares M. T., León J. (1995) Oncogene 10, 1659–1665 [PubMed] [Google Scholar]
- 25.Marin M. C., Jost C. A., Irwin M. S., DeCaprio J. A., Caput D., Kaelin W. G., Jr. (1998) Mol. Cell. Biol. 18, 6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meza N. W., Quintana-Bustamante O., Puyet A., Rio P., Navarro S., Diez A., Bueren J. A., Bautista J. M., Segovia J. C. (2007) Hum. Gene Ther. 18, 502–514 [DOI] [PubMed] [Google Scholar]
- 27.Wu S., Cetinkaya C., Munoz-Alonso M. J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L. G. (2003) Oncogene 22, 351–360 [DOI] [PubMed] [Google Scholar]
- 28.Irwin M. S., Kondo K., Marin M. C., Cheng L. S., Hahn W. C., Kaelin W. G., Jr. (2003) Cancer Cell 3, 403–410 [DOI] [PubMed] [Google Scholar]
- 29.Papoutsaki M., Lanza M., Marinari B., Nisticó S., Moretti F., Levrero M., Chimenti S., Costanzo A. (2004) J. Invest. Dermatol. 123, 1162–1168 [DOI] [PubMed] [Google Scholar]
- 30.Berwanger B., Hartmann O., Bergmann E., Bernard S., Nielsen D., Krause M., Kartal A., Flynn D., Wiedemeyer R., Schwab M., Schäfer H., Christiansen H., Eilers M. (2002) Cancer Cell 2, 377–386 [DOI] [PubMed] [Google Scholar]
- 31.Tsiftsoglou A. S., Pappas I. S., Vizirianakis I. S. (2003) Pharmacol. Ther. 100, 257–290 [DOI] [PubMed] [Google Scholar]
- 32.Neri L. M., Bortul R., Tabellini G., Borgatti P., Baldini G., Celeghini C., Capitani S., Martelli A. M. (2002) Cell. Signal. 14, 21–29 [DOI] [PubMed] [Google Scholar]
- 33.Jacobs-Helber S. M., Sawyer S. T. (2004) Blood 104, 696–703 [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T., Mitchell T., Sariban E., Sabbath K., Griffin J., Kufe D. (1985) Mol. Pharmacol. 27, 683–688 [PubMed] [Google Scholar]
- 35.Cox T. C., Bawden M. J., Martin A., May B. K. (1991) EMBO J. 10, 1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki N., Morimoto K., Tanimoto T., Hayakawa T. (1996) Arch. Biochem. Biophys. 328, 289–294 [DOI] [PubMed] [Google Scholar]
- 37.Ferreira R., Ohneda K., Yamamoto M., Philipsen S. (2005) Mol. Cell. Biol. 25, 1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 39.Basu P., Morris P. E., Haar J. L., Wani M. A., Lingrel J. B., Gaensler K. M., Lloyd J. A. (2005) Blood 106, 2566–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu P., Lung T. K., Lemsaddek W., Sargent T. G., Williams D. C., Jr., Basu M., Redmond L. C., Lingrel J. B., Haar J. L., Lloyd J. A. (2007) Blood 110, 3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan J. Y., Han X. L., Kan Y. W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11366–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chénais B. (1998) Biochem. Biophys. Res. Commun. 253, 883–886 [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Wang C., Tsui H. W., Las Heras F., Cheng E. Y., Iscove N. N., Chiu B., Inman R. D., Pritzker K. P., Tsui F. W. (2007) Exp. Cell Res. 313, 4120–4129 [DOI] [PubMed] [Google Scholar]
- 44.Snow J. W., Abraham N., Ma M. C., Abbey N. W., Herndier B., Goldsmith M. A. (2002) Blood 99, 95–101 [DOI] [PubMed] [Google Scholar]
- 45.Brunet de la Grange P., Armstrong F., Duval V., Rouyez M. C., Goardon N., Romeo P. H., Pflumio F. (2006) Blood 108, 2998–3004 [DOI] [PubMed] [Google Scholar]
- 46.Schuh A. H., Tipping A. J., Clark A. J., Hamlett I., Guyot B., Iborra F. J., Rodriguez P., Strouboulis J., Enver T., Vyas P., Porcher C. (2005) Mol. Cell. Biol. 25, 10235–10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elagib K. E., Xiao M., Hussaini I. M., Delehanty L. L., Palmer L. A., Racke F. K., Birrer M. J., Shanmugasundaram G., McDevitt M. A., Goldfarb A. N. (2004) Mol. Cell. Biol. 24, 7779–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu R., Trainor C. D., Nishikawa K., Kobayashi M., Ohneda K., Yamamoto M. (2007) J. Biol. Chem. 282, 15862–15871 [DOI] [PubMed] [Google Scholar]
- 49.Garçon L., Rivat C., James C., Lacout C., Camara-Clayette V., Ugo V., Lecluse Y., Bennaceur-Griscelli A., Vainchenker W. (2006) Blood 108, 1551–1554 [DOI] [PubMed] [Google Scholar]
- 50.Hafid-Medheb K., Augery-Bourget Y., Minatchy M. N., Hanania N., Robert-Lézénès J. (2003) Blood 101, 2575–2583 [DOI] [PubMed] [Google Scholar]
- 51.Ghaffari S., Dougherty G. J., Lansdorp P. M., Eaves A. C., Eaves C. J. (1995) Blood 86, 2976–2985 [PubMed] [Google Scholar]
- 52.Deguchi T., Komada Y., Sugiyama K., Zhang X. L., Azuma E., Yamamoto H., Sakurai M. (1999) Exp. Hematol. 27, 542–552 [DOI] [PubMed] [Google Scholar]
- 53.Deguillien M., Huang S. C., Morinière M., Dreumont N., Benz E. J., Jr., Baklouti F. (2001) Blood 98, 3809–3816 [DOI] [PubMed] [Google Scholar]
- 54.Yang G., Huang S. C., Wu J. Y., Benz E. J., Jr. (2008) Blood 111, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rana A. P., Ruff P., Maalouf G. J., Speicher D. W., Chishti A. H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6651–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickenhauser C., Pérez F., Siebolts U., Lorenzen J., Varus E., Frimpong S., Thiele J. (2003) Int. J. Oncol. 23, 437–443 [PubMed] [Google Scholar]
- 57.Ferrari B., Pavia A. A. (1986) Int. J. Pept. Protein Res. 28, 456–461 [DOI] [PubMed] [Google Scholar]
- 58.Kudo S., Fukuda M. (1994) J. Biol. Chem. 269, 22969–22974 [PubMed] [Google Scholar]
- 59.Morceau F., Chénais B., Gillet R., Jardillier J. C., Jeannesson P., Trentesaux C. (1996) Cell Growth Differ. 7, 1023–1029 [PubMed] [Google Scholar]
- 60.Li C. Y., Zhan Y. Q., Xu C. W., Xu W. X., Wang S. Y., Lv J., Zhou Y., Yue P. B., Chen B., Yang X. M. (2004) Cell Death Differ. 11, 1299–1308 [DOI] [PubMed] [Google Scholar]
- 61.Jacobs-Helber S. M., Abutin R. M., Tian C., Bondurant M., Wickrema A., Sawyer S. T. (2002) J. Biol. Chem. 277, 4859–4866 [DOI] [PubMed] [Google Scholar]
- 62.Fukuchi Y., Kizaki M., Yamato K., Kawamura C., Umezawa A., Hata Ji, Nishihara T., Ikeda Y. (2001) Oncogene 20, 704–713 [DOI] [PubMed] [Google Scholar]
- 63.Wiener E., Shiels A., Wickramasinghe S. N., Avent N. D. (1998) Br. J. Haematol. 103, 259–267 [DOI] [PubMed] [Google Scholar]
- 64.Daniels G., Green C. (2000) Vox Sang. 78, Suppl. 2, 149–153 [PubMed] [Google Scholar]
- 65.Boulanger L., Sabatino D. E., Wong E. Y., Cline A. P., Garrett L. J., Garbarz M., Dhermy D., Bodine D. M., Gallagher P. G. (2002) J. Biol. Chem. 277, 41563–41570 [DOI] [PubMed] [Google Scholar]
- 66.Gallagher P. G., Sabatino D. E., Romana M., Cline A. P., Garrett L. J., Bodine D. M., Forget B. G. (1999) J. Biol. Chem. 274, 6062–6073 [DOI] [PubMed] [Google Scholar]
- 67.Furukawa Y. (2002) Leuk. Lymphoma 43, 225–231 [DOI] [PubMed] [Google Scholar]
- 68.Kawano T., Horiguchi-Yamada J., Saito S., Iwase S., Furukawa Y., Kano Y., Yamada H. (2004) Leuk. Res. 28, 623–629 [DOI] [PubMed] [Google Scholar]
- 69.Yen J., Wisdom R. M., Tratner I., Verma I. M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5077–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferretti E., Villaescusa J. C., Di Rosa P., Fernandez-Diaz L. C., Longobardi E., Mazzieri R., Miccio A., Micali N., Selleri L., Ferrari G., Blasi F. (2006) Mol. Cell. Biol. 26, 5650–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slade N., Zaika A. I., Erster S., Moll U. M. (2004) Cell Death Differ. 11, 357–360 [DOI] [PubMed] [Google Scholar]
- 72.Grob T. J., Novak U., Maisse C., Barcaroli D., Lüthi A. U., Pirnia F., Hügli B., Graber H. U., De Laurenzi V., Fey M. F., Melino G., Tobler A. (2001) Cell Death Differ. 8, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 73.Stiewe T., Theseling C. C., Pützer B. M. (2002) J. Biol. Chem. 277, 14177–14185 [DOI] [PubMed] [Google Scholar]
- 74.Lau L. M., Nugent J. K., Zhao X., Irwin M. S. (2008) Oncogene 27, 997–1003 [DOI] [PubMed] [Google Scholar]
- 75.Orkin S. H. (1992) Blood 80, 575–581 [PubMed] [Google Scholar]
- 76.Sbisà E., Catalano D., Grillo G., Licciulli F., Turi A., Liuni S., Pesole G., De Grassi A., Caratozzolo M. F., D'Erchia A. M., Navarro B., Tullo A., Saccone C., Gisel A. (2007) BMC Bioinformatics 8, Suppl. 1, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishimura S., Takahashi S., Kuroha T., Suwabe N., Nagasawa T., Trainor C., Yamamoto M. (2000) Mol. Cell. Biol. 20, 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Socolovsky M., Nam H., Fleming M. D., Haase V. H., Brugnara C., Lodish H. F. (2001) Blood 98, 3261–3273 [DOI] [PubMed] [Google Scholar]
- 79.Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 80.De Laurenzi V., Raschellá G., Barcaroli D., Annicchiarico-Petruzzelli M., Ranalli M., Catani M. V., Tanno B., Costanzo A., Levrero M., Melino G. (2000) J. Biol. Chem. 275, 15226–15231 [DOI] [PubMed] [Google Scholar]
- 81.Stiewe T. (2007) Nat. Rev. Cancer 7, 165–168 [DOI] [PubMed] [Google Scholar]
- 82.Sherr C. J. (2004) Cell 116, 235–246 [DOI] [PubMed] [Google Scholar]
- 83.Morena A., Riccioni S., Marchetti A., Polcini A. T., Mercurio A. M., Blandino G., Sacchi A., Falcioni R. (2002) Blood 100, 96–106 [DOI] [PubMed] [Google Scholar]
- 84.Silva M., Benito A., Sanz C., Prosper F., Ekhterae D., Nuñez G., Fernandez-Luna J. L. (1999) J. Biol. Chem. 274, 22165–22169 [DOI] [PubMed] [Google Scholar]
- 85.Cantor A. B., Orkin S. H. (2002) Oncogene 21, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 86.Wu H., Liu X., Jaenisch R., Lodish H. F. (1995) Cell 83, 59–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.