Abstract
Sialidase Neu4 is reported to be dominantly expressed in the mouse brain, but its functional significance is not fully understood. We previously demonstrated that sialidase Neu3, also rich in mouse brain, is up-regulated during neuronal differentiation with involvement in acceleration of neurite formation. To elucidate physiological functions of Neu4, as well as Neu3, we determined expression during mouse brain development by quantitative RT-PCR. Expression was relatively low in the embryonic stage and then rapidly increased at 3–14 days after birth, whereas Neu3 demonstrated high levels in the embryonic stage and down-regulation after birth. Murine Neu4 was found to possess two isoforms differing in expression levels, developmental pattern, and enzymatic character. Distinct from the human isoforms, the murine forms, to a different extent, both catalyzed the removal of sialic acid from gangliosides as well as glycoproteins, and one isoform seemed to act on polysialylated NCAM efficiently, despite the low activity toward ordinary substrates. In situ hybridization demonstrated Neu4 mRNA to be present mainly in the hippocampus in which NCAM is rich and decreases after birth. During retinoic acid-induced differentiation, Neu4 expression was down-regulated in Neuro2a cells. Overexpression of Neu4 resulted in suppression of neurite formation, and its knockdown showed the acceleration. Thin layer chromatography of the glycolipids from Neu4-transfected cells showed ganglioside compositions to be only slightly affected, although lectin blot analysis revealed increased binding to Ricinus communis agglutinin (RCA) lectin of a ∼95-kDa glycoprotein, which decreased with cell differentiation. These results suggest that mouse Neu4 plays an important regulatory role in neurite formation, possibly through desialylation of glycoproteins.
Sialidases catalyze the removal of sialic acid from non-reducing ends of glycoproteins and glycolipids. In mammals, four types of sialidases have so far been cloned, classified according to their subcellular localization and enzymatic properties (abbreviated to Neu1, Neu2, Neu3, and Neu4) (1–3). Studies have provided strong evidence that these sialidases play crucial roles in various physiological functions such as cell differentiation, cell growth, and malignant transformation. Among these sialidases, Neu4 is unique in its tissue expression pattern and enzymatic properties. In the mouse, it is dominantly expressed in brain, but its sialidase activity is very weak compared with other mouse sialidases (4). In contrast, human NEU4 is expressed not only in brain, but also in liver, kidney, and colon (5–7). We have demonstrated that NEU4 has two isoforms, differing in the N-terminal 12-amino acid residues that act as a mitochondrial-targeting sequence (7). Except for the subcellular localization, enzymatic properties are very similar. The short form of NEU4 (NEU4S) suppresses malignancy in colon cancer cells, mainly through desialylation of some glycoproteins, whereas the long form of NEU4 (NEU4L) may be involved in apoptosis with hydrolysis of ganglioside GD3 in mitochondria (8). Recently, Neu4 knockout mice (Neu4−/−) were generated for pathological analysis (9). Neu4−/− grew normally with a normal lifespan and proved fertile, but vacuolization of the lung and spleen was observed with a lysosomal storage phenotype, and the GM1/GD1a ratio was decreased in the brain. The observations on Neu4−/− are very interesting, but there is some ambiguity in the available previous reports, because, as mentioned above, mouse Neu4 has been reported to have weak sialidase activity in vitro, and its expression is restricted in brain. To clarify this ambiguity and further understand the physiological functions of Neu4, we examined expression in the mouse brain and observed a possible involvement in neural differentiation in connection with another sialidase, Neu3, which greatly increases during differentiation of neuroblastoma cells (10, 11) and causes acceleration of neurite formation (10–13).
In the GenBankTM data base, nucleotide sequences of mouse Neu4 have been submitted as AY258421 and AK034236. The former contains a complete coding sequence of 1506 bp, with two ATGs at positions 1 and 70, and AK034236 encodes only the second ATG (4). The gene from AY258421 has been reported to encode Neu4, showing weak sialidase activity, but there is no information on whether the gene based on AK034236 encodes Neu4 with sialidase activity toward natural substrates. We have now extended our studies to the existence of different mouse Neu4 isoforms, focusing on their significance in neuronal cells by measuring expression levels during cell differentiation. We present, here, evidence that two murine Neu4 isoforms contribute to neurite formation.
EXPERIMENTAL PROCEDURES
Animals and Tissue Preparation
Adult and pregnant C57BL/6J mice were purchased from Japan CREA (Tokyo, Japan). For in situ hybridization, adult brains were immediately fixed in 4% paraformaldehyde at 4 °C for 4 h and then soaked in 30% sucrose/RNase-free PBS2 at 4 °C for 16 h. Brain tissues were also obtained from embryos at gestational days 18 and postnatal days at 0, 3, 7, 14, and 49 days. They were pooled from 3–4 embryonic and postnatal mice and kept frozen at −80 °C until use.
Antibodies
Antibodies for NCAM and polysialylated NCAM (PSA-NCAM) were obtained from Chemicon International. The antibody for β-tubulin was from Sigma.
Cell Culture
Mouse neuroblastoma Neuro2a cells (American Type Culture Collection: ATCC) and human embryonic kidney HEK293T cells (a gift from M. Sugai, Kyoto University School of Medicine, Kyoto, Japan) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS). Rat pheochromocytoma PC12 cells (ATCC) were maintained in DMEM with 5% inactivated FBS and 10% inactivated horse serum. PC12 cells were cultured in collagen-coated dishes.
Identification of Neu4 Isoforms
A set of primers (5′-CGGAGCCTGATATTGCTTTAC-3′) and (5′-GTTCTTGCCAGTGGCGATTTGC-3′) was prepared based on the nucleotide sequence (AY258421), and RT-PCR amplifications were performed with mouse brain cDNA under the following conditions; 10 s at 96 °C, 5 s at 58 °C and 4 min at 60 °C for 30 cycles, preceded by 3 min at 96 °C. The PCR product contained two isoforms: one having the first and the second ATGs (Neu4a, longer form) and the other with the second ATG only (Neu4b, shorter form). After subcloning these products into the pBluescript vector, DNA sequences were analyzed with the 3130 Genetic analyzer (Applied Biosystems).
Plasmid DNA and Transfection
To obtain mouse Neu4 cDNA, total RNA was isolated from adult mouse brain with an RNeasy kit (Qiagen), as recommended by the manufacturer. First-strand cDNAs were synthesized with oligo-dT primers by reverse transcription and used as templates for PCR, followed by PCR for Neu4a with primers of (5′-GTGAATTCACCATGGAGACTGGAGCT-3′) and (5′-GTGAATTCTCAAGAGGGCCAGCAATGCCC-3′), and for Neu4b with primers of (5′-GTGAATTCACCATGGGGCCCACGTGTT-3′) and (5′-GTGAATTCTCAAGAGGGCCAGCAATGCCC-3′). Expression plasmids for Neu4 were constructed by inserting the entire open reading frame of the mouse gene into pcDNA3.1 (Invitrogen) (pcDNA-Neu4) or into pCAGGS vector (pCA-Neu4), generously provided by Dr. Jun-ichi Miyazaki, Osaka University School of Medicine. The expression plasmid, pCA-Neu4, was transiently transfected into HEK293T or Neuro2a, and pcDNA-Neu4 was stably transfected into PC12 cells using Effectene (Qiagen) as recommended by the manufacturer. Stable transfectants were selected with G418 (500 μg/ml for PC12 cells). The expression plasmid for ST8 siaIV, α-2,8-sialyltransferase, was constructed by inserting the entire open reading frame of the human gene into the pcDNA3.1 vector.
For targeting mouse Neu3, Neu3 siRNAs, number 1 (CCAACUAGCAAGAGGAAGAGGAUUA) and number 2 (CCUGAUGAUCUACAGUGAUGACUUU) beginning at nucleotides 997 and 594, respectively, of the Neu3 open reading frame, were obtained from iGENE (Tsukuba, Japan) and transfected into Neuro2a cells using Lipofectamine 2000 (Invitrogen). To target mouse Neu4 for both Neu4a and Neu4b, Neu4 siRNAs number 1 (L-055343-01) and number 2 (GCUCCUACGGAAGGUUAU) obtained from Dharmacon RNA Technologies, Inc. (Lafayette, CO) were used.
Sialidase Activity
The sialidase activity was measured with various substrates, and released sialic acid was determined by a modification of the thiobarbituric acid method as described previously (14). Cells were sonicated in PBS containing 1 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 0.5 μg/ml pepstatin, and centrifuged at 1000 × g for 10 min. The supernatant was then used for the measurement of sialidase activity as cell homogenates. A unit of activity was defined as the amount of enzyme that cleaved 1 nmol of sialic acid/h from substrates. Protein concentration was determined by the dye binding assay (Bio-Rad). To assess polysialic acid degradation, 200 nmol of colominic acid was used as substrate in the sialidase assay. For degradation of polysialylation on PSA-NCAM, the reduced level was evaluated by Western blotting with specific antibodies. As a positive control, cell homogenates were incubated with 0.1 μg of Endo-N (Abcys, Paris) at 37 °C for 30 min.
In Situ Hybridization
The selected mouse Neu4 cDNA fragment (nucleotides 231–638 from the start codon) was cloned into the Bluescript vector in the correct orientation. To generate sense and antisense probes, the plasmids were then linearized by HindIII and XbaI and transcribed by T3 and T7 RNA polymerase, respectively, in the presence of digoxigenin-labeled UTP (Roche Applied Science). Probes were fragmented to ∼150 bp in length by alkaline treatment before use and were designed to recognize both Neu4a and Neu4b. Hybridization was performed as described previously (10). Positive signals were detected by immunoassay with anti-digoxigenin-alkaline phosphatase conjugates and NBT/X-phosphate as the substrate.
Quantitative Reverse Transcription-PCR Analysis
Expression levels of Neu1, Neu2, Neu3, and Neu4 in mouse tissues were evaluated by quantitative RT-PCR (real-time PCR, LightcyclerR, Roche Diagnostics) as described previously (15). Total RNA was isolated, and first-strand cDNAs were synthesized by reverse transcription and used as templates. Mouse Neu1 primers were sense (5′-TTCATCGCCATGAGGAGGTCCA-3′, nucleotide 304–325 from start codon) and antisense (5′-AAAGGGAATGCCGCTCACTCCA-3′, nucleotides 499–520). Mouse Neu2 primers were sense (5′-AGGAAGCTACAACGAAGCCACA-3′, nucleotides 168–189) and antisense (5′-TTCTGAGCAGGGTGCAGTTTCC-3′, nucleotides 545–566). Mouse Neu3 primers were sense (5′-CTCAGTCAGAGATGAGGATGCT-3′, nucleotides 138–159) and antisense (5′-GTGAGACATAGTAGGCATAGGC-3′, nucleotides 532–553). Total mouse Neu4 primers were sense (5′-AGGAGAACGGTGCTCTTCCAGA-3′, nucleotides 91–112) and antisense (5′-GTTCTTGCCAGTGGCGATTTGC-3′, nucleotides 408–429). Mouse Neu4a primers were sense (5′-GCTGGAGCTCCCTTCTGCTTCCA)-3′, nucleotides 10–32) and antisense (5′-GTTCTTGCCAGTGGCGATTTGC-3′, nucleotides 408–429). To obtain Neu4b expression, we used Neu4 cDNA as a standard and calculated values by subtracting Neu4a expression from total Neu4 expression. Rat Neu3 primers were sense (5′-TGTCTACCTGGTGTTCAGACGA-3′, nucleotides 159–180) and antisense (5′-GGAAAGCAGAGAAACCAGCATG-3′, nucleotides 551–572). Rat Neu4 primers were sense (5′-TCGGGTACCATCTTCCTCTTCT-3′, nucleotide 364–385) and antisense (5′-TAGAGAAAGTCTCCATCCACCG-3′, nucleotide 761–782). To compare the expression levels among sialidases, standard curves for respective sialidase cDNAs were generated by serial dilution of the pBluescript vector containing the gene encoding the entire open reading frame as described (7, 16). To normalize for sample variation, expression of glyceraldehyde-3-phosphate dehydrogenase was determined as an internal control.
Neurite Formation
Neuro2a cells were seeded in 6-well plates at a density of 2 × 104/cm2. After overnight incubation in the DMEM with 2% FBS, they were exposed for various periods to 20 μm all-trans-retinoic acid (Sigma). PC12 cells were seeded in 6-well plates at a density of 1 × 104/cm2 and exposed for various periods to 50 ng/ml NGF (nerve growth factor) in DMEM medium without serum. The morphology of the cells was examined, and photographs were taken at ×100 magnification. To quantify neuritogenesis, cells bearing neurites at least 2.0-fold the length of the soma diameter were considered as differentiated and counted in five randomly chosen fields in each well. All the cells collected were assayed for Neu3 and Neu4 mRNA levels by quantitative RT-PCR. Acetylcholine esterase activity was measured as described (10), and a unit of activity is defined as 1 nmol of product formed per minute.
Determination of Cellular Sialic Acids
Cells harvested at subconfluence were washed with PBS and lyophilized. The glycolipids were extracted with chloroform/methanol (C/M, 1:1, v/v), C/M (2:1, v/v), and C/M (1:2, v/v), followed by evaporation to dryness. Glycolipid extracts (lipid-bound sialic acids) and the residues after extraction (protein-bound sialic acids) were treated with 0.1 n H2SO4 at 80 °C for 1 h and assayed for sialic acid by fluorometric high-performance liquid chromatography with malononitrile (17).
Lectin Blotting
Lectin blot analyses of SSA (Sambucus sieboldiana agglutinin), MAM (Maackia amurens mutagen), PNA (peanut agglutinin), and RCA (Ricinus communis agglutinin, Honen, Tokyo, Japan) were conducted as described previously (18). Briefly, cell homogenates were resolved on an 8% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. Blots were blocked with 1% bovine serum albumin and then incubated with biotinylated lectins. After washing, lectin-binding molecules were visualized with horseradish peroxidase-streptavidin (Vector, Burlingame, CA).
Thin Layer Chromatography
Glycolipids were extracted from cells as described elsewhere (19), fractionated by thin layer chromatography on HPTLC plates (Baker, Phillipsburg, NJ) in chloroform, methanol, 0.5% CaCl2 (60:40:9, v/v/v), and visualized with orcinol-H2SO4.
Statistical Analysis
Results were expressed as mean ± S.D. All values were compared using the Student's t test or the Welch's t test.
RESULTS
Identification and Characterization of Two Forms of Mouse Neu4
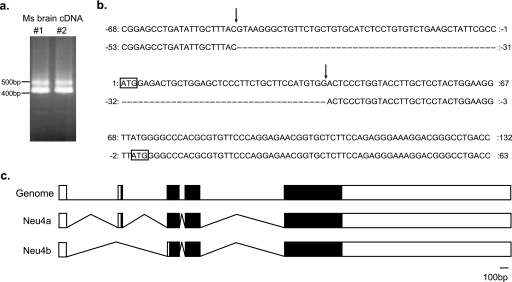
Mouse Neu4 was first identified using the brain cDNA by Comelli et al. (4) (AY258421), and transfection of the cDNA exhibited only a weak sialidase activity. On the other hand, other sequences including AK034236 submitted in GenBankTM show the shortage of N-terminal 23 amino acids, although its information about enzymatic properties is unavailable. We then investigated whether the two forms are actually expressed. After PCR amplification of the sequence containing the first and the second ATG, two PCR products of 497 and 413 bp were obtained using two different mouse brain cDNAs (Fig. 1a). Sequence analysis revealed that the former contained the two ATGs (Neu4a, long form) and the latter only the second ATG (Neu4b, short form), indicating that mouse Neu4 possesses two splicing variants (Fig. 1, b and c). We also confirmed exon/intron boundaries at the 5′-site of the first ATG of the long form, consistent with the boundary prediction (Splice-view). The length of intron between the first exon and the second exon was 7740 nucleotides.
FIGURE 1.
Identification of murine Neu4 isoforms. a, amplification of the fragment of murine Neu4 isoforms by RT-PCR. cDNAs obtained from different mouse brain tissues (1 and 2) were used, and PCR products were subjected to agarose electrophoresis. An upper band (Neu4a) and lower band (Neu4b) were obtained. b, nucleotide sequence of Neu4 isoforms containing the first and second ATGs. Nucleotide sequences of PCR products in a were analyzed. The upper sequence (Neu4a) possessed an additional exon with 84 bases compared with the lower sequence (Neu4b). The arrow indicates the boundary of the exon/intron. The square shows the first methionine in each sequence. c, splicing structure of murine Neu4. White bar, non-coding region; black bar, coding region.
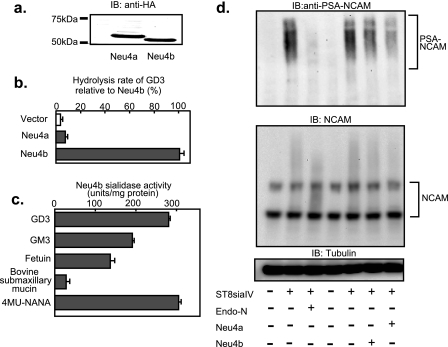
Neu4a was found to contain an additional 23 amino acid residues at the N terminus, compared with Neu4b. PSORT II sever analysis predicted no targeting signals with the additional 23 amino acids, in contrast to the N terminus of human NEU4L that acts as a mitochondrial-targeting signal (7). To examine whether these Neu4 isoforms, Neu4a and Neu4b, encode sialidases, the genes were transfected into HEK293T cells. Immunoblotting revealed expression of a C-terminal HA-tagged 55.0-kDa protein for Neu4a and a 53.3-kDa protein for Neu4b, as expected by calculation of molecular masses (Fig. 2a). Co-transfection of a luciferase vector showed similar expression levels of the two isoforms, irrespective of the presence or absence of the HA tag (data not shown). However, tagged Neu4 expression plasmids yielded lower sialidase activity in the transfected cells than those without the HA tag (a decrease in the activity by ∼30–50%). The Neu4 plasmids without HA tag, therefore, were employed for all experiments except immunodetection of the isoform proteins. Neu4b showed a remarkable increase of sialidase activity to GD3, whereas Neu4a showed only a slight increase relative to mock transfectants (Fig. 2b), consistent with the previous report (4). In the substrate specificity studies using cell homogenates (Fig. 2c), Neu4b was found to act on glycolipids and glycoproteins as well as 4MU-sialic acid (4-methylumbelliferyl sialic acid), which is similar to human NEU4 but in contrast to Neu1 and Neu3. Like human NEU4, Neu4b removed sialic acids from bovine submaxillary mucin. Neu4a also reacted with these substrates although the hydrolytic rate was low. We then studied sialo-glycoconjugates having α-2,8-sialyl linkages as the substrates, including colominic acids (α-2,8-sialic acid polymer) and PSA-NCAM. With colominic acids, both Neu4 isoforms showed a low but significant activity (1.5 and 2.9 units/mg protein for Neu4a and Neu4b, respectively). In comparison with the activities toward GD3, the value with colominic acids divided by that with GD3 was 6.7-fold higher for Neu4a than that for Neu4b, indicating much more efficient hydrolysis of colominic acids by Neu4a. To produce PSA-NCAM, known to be involved in cell differentiation and cell attachment, ST8 siaIV, α-2,8-sialyltransferase, was introduced into Neuro2a cells. ST8 siaIV synthesized PSA on NCAM, and the treatment with Endo-N, as a positive control, decreased PSA. Interestingly, consistent with the results with colominic acids, co-transfection of the Neu4 gene with ST8 siaIV revealed Neu4a to effectively remove sialic acids from PSA-NCAM (Fig. 2d), even though possessing only slight activity toward GD3 and GM3, as described above. As the protein amount for Neu4a expressed by the transfection was almost equivalent to that for Neu4b, efficient hydrolysis of PSA by Neu4a indicates that PSA-NCAM is possibly one of natural substrates for Neu4a.
FIGURE 2.
Expression of the murine Neu4 sialidase. a, immunoblotting of the two isoforms of the HA-tagged Neu4 protein. Expression plasmids encoding HA-tagged Neu4a and Neu4b were transfected into HEK293T cells, and homogenates were subjected to SDS-PAGE and immunoblotted with anti-HA antibody. Transfection efficiency was adjusted using the luciferase vector as a control. Bands for 55 kDa and 53.5 kDa were obtained for Neu4a and Neu4b, respectively. b, sialidase activity in the transfected cells using GD3 as the substrate. c, substrate specificity of Neu4b expressed in HEK293T cells. Sialidase activity toward various sialic acid-containing glycoconjugates was examined using cell homogenates. d, PSA-NCAM change in Neu4-transfected cells. Neuro2a cells were co-transfected with ST8siaIV and Neu4 genes. PSA-NCAM, NCAM, and tubulin were assessed by immunoblotting.
With regard to subcellular localization, we found the Neu4 isoforms on intracellular membranes in Neuro2a cells transfected with C-terminal HA-tagged Neu4a and Neu4b by indirect immunofluorescence microscopy. Co-localization studies with organelle markers including anti-LAMP1 for lysosomes, Mitotracker for mitochondria, anti-calnexin for ER, anti-EEA for early endosomes, and anti-58K Golgi protein for Golgi did not show clear results (data not shown), although Neu4a and Neu4b seemed to partly co-localize with calnexin. It is uncertain whether this partial co-localization in ER is physiological, because Neu4 does not have an N-terminal signal sequence, and, furthermore, it may be as a result of the improper folding caused by overexpression. There is no typical GPI-anchor consensus sequence in mouse Neu4, but a potential C-terminal cleavage site does exist (4), suggesting that like human NEU4S, mouse Neu4 is an intracellular membrane protein.
Neu4 Expression during Mouse Brain Development
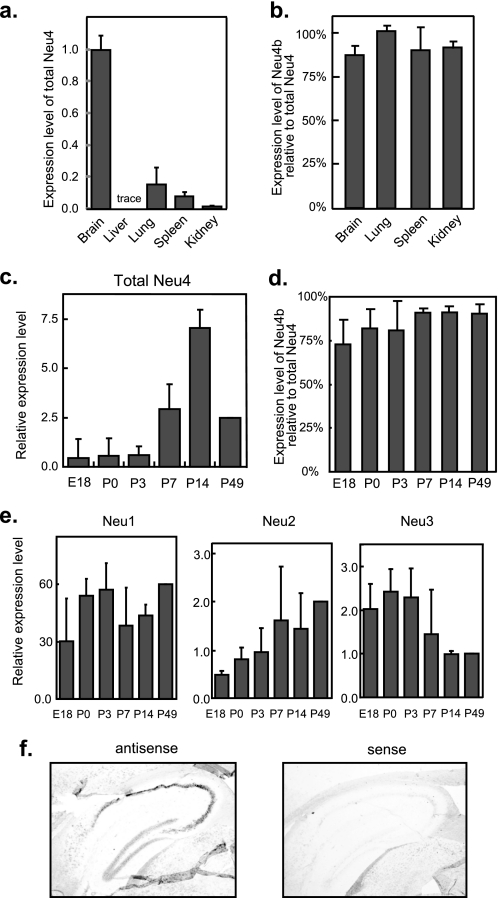
The expression level of Neu4 was then determined in various tissues of the mouse by RT-PCR. Levels were high in adult brain as previously reported with Northern blot analysis (4). However, Neu4 expression was also evident in other tissues, especially in kidney and lung (Fig. 3a). The expression ratio of Neu4b relative to total Neu4 was about 0.9 in tissues (Fig. 3b), although the ratio was less than 0.75 in the embryonic stage of brain (Fig. 3d). Evaluation during brain development using standard plasmids, in comparison with three other sialidases, indicated the expression level of Neu4 to be almost the same as those of Neu2 and Neu3 but lower than that of Neu1 (Fig. 3, c and e). The total Neu4 expression level was found to be relatively low in the late embryonic stage and postnatal 3 days, and then showed a rapid increase at postnatal 7 days (Fig. 3c), reaching a maximum at day 14 and then a decrease. These results are not contradictory with other reports in which Neu4 was drastically increased at postnatal 10 days (20). The ratio of Neu4a to total Neu4 showed a gradual increase during development (Fig. 3d). With regard to other sialidases, the expression patterns in mouse brain were similar to our previous report on rat (16) and another on mouse brain (21). Neu1 showed a slight increase after birth and then decreased and remained steady (Fig. 3e). Neu2 expression was moderately increased after birth, whereas Neu3 expression showed a sharp decrease at postnatal day 7, in contrast to Neu4. These results suggest that Neu4 may play a role in brain development in contrast to Neu3. Next, we observed spatial localization of Neu4 mRNA in mouse brain by in situ hybridization. As our attempts to prepare two distinct RNA probes for Neu4 isoforms were unsuccessful because the length of the nucleotide difference was too small (69 nucleotides), total Neu4 expression was estimated. Positive signals were mainly in the hippocampus (Fig. 3f) and weak. Significant signals in Purkinje cells were found in the cerebellum, but not the cerebral cortex (data not shown).
FIGURE 3.
Neu4 expression in murine tissues. a, expression levels of Neu4 gene in murine tissues, as estimated by quantitative RT-PCR. The expression of β-actin was also examined as an internal reference. b, relative expression of the short form (Neu4b) compared with the total Neu4 level is shown. c, expression profile of the Neu4 gene in developing mouse brain determined by quantitative RT-PCR. E18, embryonic day 18; P0, P3, P7, P14, and P49, postnatal days 0, 3, 7, 14, and 49. Total Neu4 mRNA levels were expressed relative to the P49 mouse value. d, ratios of Neu4b to total Neu4 during the brain development. e, relative Neu1, Neu2, and Neu3 levels in the developing brain mouse. The data given are mean values from three experiments ± S.D. In c and e, quantities of the transcripts encoding Neu4 and other murine sialidases were determined by standard curves obtained from plasmids containing respective sialidase cDNAs as described under “Experimental Procedures.” The values are based on the assumption that the efficiency in PCR is apparently comparable among these sialidase genes. f, frozen parasaggital sections of a C57BL/6J mouse brain were hybridized with digoxigenin-labeled Neu4 antisense and sense RNAs. Note positive signals in the hippocampus.
Down-regulation of Neu4 during Neuro2a Cell Differentiation
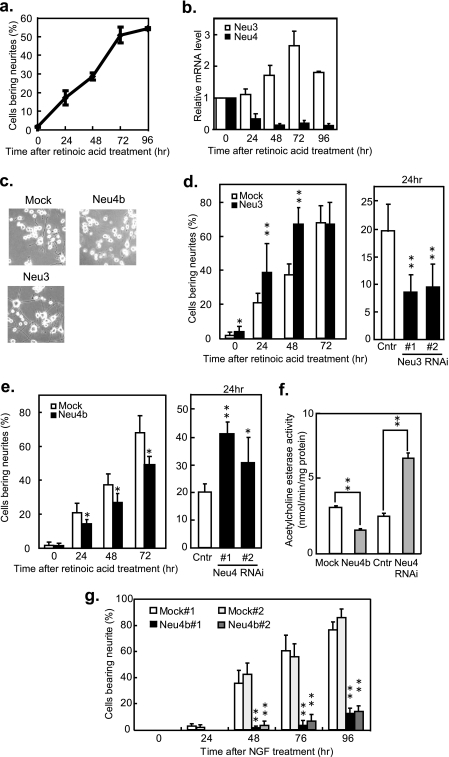
Sialidases have been implicated in neuronal cell differentiation (10–13, 22, 23). To clarify the significance of the developmental change of Neu4 expression in mouse brain, mouse neuroblastoma Neuro2a cells were employed, because regulation of axonal growth by Neu3 was previously reported (10–13, 23). During retinoic acid-induced cell differentiation, the proportion of cells bearing neurites rose to 50% over 72 h, and the Neu3 mRNA level increased 2.8-fold, while the total Neu4 level decreased to 10% of the value for non-differentiated cells (Fig. 4, a and b). As Neuro2a cells were found to dominantly express Neu4b but less Neu4a (only less than 1% of total Neu4) in either untreated or differentiated cells, Neu4b may be essentially involved in the differentiation. 5-Bromodeoxyuridine also caused the decrease of the Neu4 expression level during differentiation (data not shown).
FIGURE 4.
The regulatory role of Neu4 in neurite formation during retinoic acid-induced differentiation. a, neurite formation during retinoic acid stimulation of Neuro2a cells. Cells were cultured in medium containing 20 μm retinoic acid, and cells bearing neurites at least 2.0-fold the length of the soma diameter were counted at the times indicated. b, quantitative assessment of endogenous Neu3 and Neu4 mRNA in Neuro2a cells. After retinoic acid stimulation, cells were harvested for RNA preparation and estimation of mRNA expression by quantitative RT-PCR. White bars, Neu3 mRNA; black bars, Neu4 mRNA. c, photographs of Neuro2a cells treated with retinoic acid for 48 h at ×100 magnification. Upper left, vector transfectant; upper right, Neu4 transfectant; lower left, Neu3 transfectant. d, positive regulatory role of Neu3 in neurite formation. Neuro2a cells transiently transfected with Neu3 (left panel) or with the siRNAs (right panel), and the cells bearing neuritis were counted during retinoic acid treatment. White bar, control cells; black bar, Neu3 or the siRNA transfectants. e, negative regulatory role of Neu4 in neurite formation. Neuro2a cells were transiently transfected with Neu4b (left panel) or with Neu4 siRNAs (right panel) and observed for neurite formation. White bar, control cells; black bar, Neu4b or Neu4 siRNA transfectants. f, acetylcholine esterase activity at 72 h in Neuro2a cells after retinoic acid stimulation. White bar, control cells; gray bar, Neu4b, or Neu4 siRNA transfectants. In d–f, *, p < 0.05, and **, p < 0.01 compared with the vector. The data given are mean values from three experiments ± S.D. p values were evaluated by the Student's t test. g, suppression of neurite formation in Neu4 stable transfectants of PC12 cells. Two independent positive clones (Neu4b#1, Neu4b#2) and empty vector-transfected (Mock#1, Mock#2) clones of PC12 cells were cultured in medium containing NGF, and cells bearing neurites were counted as described above. White bar, Mock#1 cells; Light gray bar, Mock#2 cells; Black and gray bars, Neu4b#1 and Neu4b#2 cells, respectively. **, p < 0.01 compared with the Mock.
Suppression of Neurite Formation by Neu4
To further elucidate the function of Neu4 in neurite formation, Neuro2a cells were transiently transfected with the expression plasmids (PCA-Neu4a, Neu4b, or Neu3) or with the siRNAs. The Neu3 gene was used as the positive control that enhances neurite formation. After 24 h of transfection, differentiation was induced by 20 μm all-trans-retinoic acid treatment, and cells bearing neurites were assessed thereafter (Fig. 4, c–e). As shown in the photographs (Fig. 4c), cells treated with retinoic acid for 48 h bore neurites in mock- and Neu3 transfectants but not in Neu4b transfectants. After retinoic acid stimulation, neurite formation was significantly increased with Neu3 transfectants (200 units/mg protein toward GD3), but decreased with Neu3 siRNA-transfected cells (75.1 and 73.0% suppression in the mRNA level with nos. 1 and 2 Neu3 siRNAs, respectively) (Fig. 4d). When Neu4b-transfected cells showing an increase in sialidase activity (104.9 units/mg protein toward GD3) were treated with retinoic acid, neurites were statistically low, compared with mock transfectants (3.9 units/mg protein) of which 70% bore neurites by 3 days (Fig. 4e, left panel). On the other hand, Neu4b-silencing cells (83.9 and 70.8% suppression in Neu4 mRNA level with nos. 1 and 2 Neu4 siRNAs, respectively) showed enhancement of neurite formation (Fig. 4e, right panel), indicating that suppression of Neu4b possibly leads to increasing neurites. Neu4a transfection resulting in a slight increase of sialidase activity (9.0 units/mg toward GD3) did not cause significant suppression of neurite formation compared with mock transfectants, although it was confirmed by Western blotting that transfection efficiency for Neu4a was almost the same as that for Neu4b in Neuro2a cells (data not shown). Under these conditions, acetylcholine esterase activity was decreased by Neu4b overexpression and increased by the knockdown compared with the control, concomitantly with these changes in neurites (Fig. 4f). Endogenous Neu3 and Neu4 mRNA levels were not altered by transfection of the Neu4 and Neu3, respectively. Suppression of neurite formation by Neu4 transfection was confirmed in rat pheochromocytoma PC12 cells with undetectable Neu4 expression. The cells underwent differentiation upon NGF treatment, and the endogenous Neu3 level increased 4.2-fold compared with non-differentiated cells after 96 h, while the proportion of cells bearing neurites rose to 60% (data not shown). Some 85 and 75% of the mock transfectants, mock nos. 1 and 2, with activity 1.4 and 0.8 units/mg protein, respectively, showed neurite formation at 96 h after NGF stimulation, whereas Neu4b stable transfectants, Neu4 nos. 1 and 2, having 25.6 and 20.0 units/mg protein, respectively, showed suppression of NGF-stimulated neurite formation to levels of 13 and 15%, respectively (Fig. 4g). The suppression was not caused by cell damage on Neu4b transfection, because there was no difference between Mock and Neu4 transfectants in cell growth assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays (data not shown). These results clearly indicate that mouse Neu4b acts as a suppressor in neurite formation, in direct contrast to the stimulation observed with Neu3.
Changes in Glycosylation by Neu4 Transfection
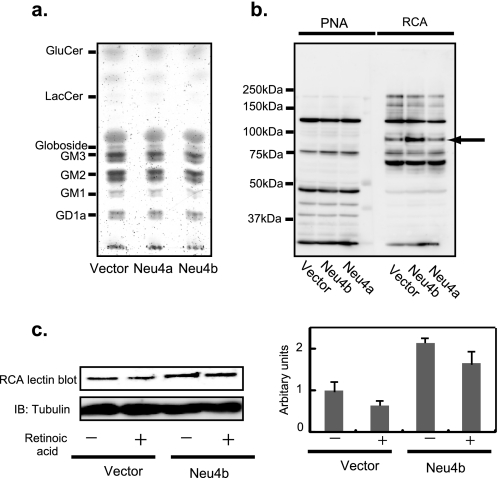
To obtain the molecular basis for actions of Neu4 on neurite formation described above, we assessed changes in cellular sialic acid contents and qualitative changes in cellular glycolipids and glycoproteins by thin layer chromatography and lectin blotting, respectively. Lipid-bound sialic acids in 106 cells were determined to be 0.82 and 0.74 nmol, whereas protein-bound sialic acids were 1.08 and 0.85 nmol in mock and Neu4b transfectants. No significant changes were observed with Neu4a transfectants. When the glycolipids from Neuro2a cells were subjected to thin layer chromatography, there was little change between mock- and Neu4-transfected cells (Fig. 5a). Among SSA, MAM, RCA, and PNA lectins, RCA demonstrated clearly increased binding to a 95-kDa glycoprotein on Neu4b transfection, indicating Neu4b cleaves sialic acids from Gal-β1-4-GlcNAc (Fig. 5b). Furthermore, the RCA lectin binding of the 95-kDa molecule was decreased by retinoic acid-induced differentiation after 3 days (Fig. 5c). There was no difference in the protein levels in this area between Neu4 transfectants and the mock cells, when the two-dimensional electrophoresis gel was silver-stained. These results suggest that desialylation of a 95-kDa glycoprotein may contribute to suppression of Neu4b-mediated cell differentiation, although identification of the core protein is necessary for further understanding.
FIGURE 5.
Analysis of glycoproteins and glycolipids of Neu4-transfected cells. a, glycolipids were extracted from Neu4-transfected Neuro2a cells, subjected to thin layer chromatography, and visualized with orcinol-H2SO4. b, glycoproteins of Neuro2a cells were analyzed by lectin blotting with PNA or RCA and compared with those of vector transfectants. Numbers indicate molecular sizes of the standard markers. The arrow indicates the “95-kDa protein.” c, lectin blotting of the 95-kDa protein with RCA, with or without 3 days of retinoic acid stimulation of Neuro2a cells.
DISCUSSION
In the present study, we demonstrated that murine Neu4 possesses two isoforms differing in enzymatic properties and expression pattern. In particular, a dominantly expressing isoform, Neu4b, seems to play an essential role in negative regulation of neurite formation. Comelli et al. (4) reported that murine Neu4 is dominantly expressed in brain, and a cDNA cloned from the brain encoded a protein with low sialidase activity. We found that Neu4 is expressed not only in brain but also in other mouse tissues including lung and spleen, sites shown by Seyrantepe et al. (9) to exhibit vacuolization in Neu4 knockout mice. We also showed that murine Neu4 possesses two splicing variants. The shorter form (Neu4b) expresses sufficient sialidase activity toward various substrates, but the long one (Neu4a), particularly highly expressed in the brain, shows weak sialidase activity as Comelli et al. (4) reported. We do not know the reason for the low activity of Neu4a compared with Neu4b. From the predicted three-dimensional structure of human NEU4, the 23 N terminus amino acid residues in Neu4a are not likely to be related to the active site (24). Despite the low activity toward ordinary substrates easily cleavable by Neu4b, interestingly, Neu4a was able to hydrolyze colominic acids and polysialylated NCAM more efficiently than Neu4b. In situ hybridization showed Neu4 mRNA in hippocampus (Fig. 3f), in which polysialylated NCAM is known to be rich and to decrease after birth. As Neu4 expression increases at this period, Neu4, especially Neu4a, may participate in the decrease. It should be noted here that the mouse Neu4 isoforms are different from those of human NEU4, which exhibit similar enzymatic properties but distinct subcellular localization, as we previously reported (7).
During mouse brain development, Neu4 mRNA levels appear to be quite low in the embryonic stage, becoming up-regulated in the postnatal period, just as Neu3 mRNA is down-regulated. We previously observed that hippocampus neurons have enhanced axonal growth with increases in Neu3 expression, and this can be suppressed with NeuAc2en, a Neu3 inhibitor (12). Furthermore, neuroblastoma Neuro2a and NB-1 cells similarly show axonal growth upon Neu3 up-regulation (10, 11). Directly contrasting with Neu3, Neu4 is down-regulated with neuronal differentiation. Neu4b overexpression caused actual suppression of neurite formation. In addition to hydrolysis of PSA-NCAM, desialylation of 95-kDa glycoprotein, as reflected by increases in RCA binding in Neu4b-transfected cells, may also be important events during cell differentiation. Although an attempt was made to identify the 95-kDa core protein by RCA affinity column and two-dimensional electrophoresis, it was unsuccessful because of its low expression. Seyrantepe et al. (9) described detection of Neu4 in scattered cells similar to microglia cells in adult mouse brain and suggested that their migration might be related to Neu4 expression because of the decrease of the GM1/GD1a ratio observed in Neu4−/− mouse. Therefore, ganglioside modulation by Neu4 could occur in vivo and affect neuronal functions simultaneously with desialylation of glycoproteins. Inconsistent with our present and previous observations, Neu3 knockdown in Neuro2a cells was recently described to induce axonal differentiation induced by retinoic acid (23). We observed here that Neu3 knockdown decreased and its overexpression enhanced neurite formation in the same cells shown in Fig. 4. When two different siRNAs for Neu3 knockdown were used to exclude the possibility of the off-targeting effect, their introduction resulted in a significant decrease in neurite formation. Detailed investigations should solve the inconsistency between the reports. Taken together, Neu4 expression dynamically changed during mouse brain development, and the sialidase possibly plays a regulatory role for neuronal function together with Neu3 through desialylation of gangliosides and glycoproteins.
Acknowledgment
We thank Kazuaki Sato for expert animal handling.
This work was supported in part by Grants-in-aid for Exploratory Research and Scientific Research on Priority Areas Cancer from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AB510404.
- PBS
- phosphate-buffered saline
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- MAM
- maackia amurens mutagen
- NCAM
- neuronal cell adhesion molecule
- NGF
- nerve growth factor
- PNA
- peanut agglutinin
- RCA
- Ricinus communis agglutinin
- RT-PCR
- reverse transcription polymerase chain reaction
- SSA
- Sambucus sieboldiana agglutinin
- 4MU-NANA
- 4-methylumbelliferyl sialic acid
- siRNA
- short interfering RNA
- HA
- hemagglutinin
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Miyagi T., Yamaguchi K. (2007) Comprehensive Glycoscience :Biochemistry of Glycans (Kamerling J. P., Boons G., Lee Y. C., Suzuki A., Taniguchi N., Voragen A. G. J., eds) pp. 297–322, Elsevier BV, Amsterdam [Google Scholar]
- 2.Miyagi T., Wada T., Yamaguchi K. (2008) Biochim. Biophys. Acta 1780, 532–537 [DOI] [PubMed] [Google Scholar]
- 3.Monti E., Preti A., Venerando B., Borsani G. (2002) Neurochem. Res. 27, 649–663 [DOI] [PubMed] [Google Scholar]
- 4.Comelli E. M., Amado M., Lustig S. R., Paulson J. C. (2003) Gene 321, 155–161 [DOI] [PubMed] [Google Scholar]
- 5.Monti E., Bassi M. T., Bresciani R., Civini S., Croci G. L., Papini N., Riboni M., Zanchetti G., Ballabio A., Preti A., Tettamanti G., Venerando B., Borsani G. (2004) Genomics 83, 445–453 [DOI] [PubMed] [Google Scholar]
- 6.Seyrantepe V., Landry K., Trudel S., Hassan J. A., Morales C. R., Pshezhetsky A. V. (2004) J. Biol. Chem. 279, 37021–37029 [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K., Hata K., Koseki K., Shiozaki K., Akita H., Wada T., Moriya S., Miyagi T. (2005) Biochem. J. 390, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasegawa T., Sugeno N., Takeda A., Matsuzaki-Kobayashi M., Kikuchi A., Furukawa K., Miyagi T., Itoyama Y. (2007) FEBS Lett. 581, 406–412 [DOI] [PubMed] [Google Scholar]
- 9.Seyrantepe V., Canuel M., Carpentier S., Landry K., Durand S., Liang F., Zeng J., Caqueret A., Gravel R. A., Marchesini S., Zwingmann C., Michaud J., Morales C. R., Levade T., Pshezhetsky A. V. (2008) Hum. Mol. Genet. 17, 1556–1568 [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa T., Yamaguchi K., Wada T., Takeda A., Itoyama Y., Miyagi T. (2000) J. Biol. Chem. 275, 8007–8015 [DOI] [PubMed] [Google Scholar]
- 11.Proshin S., Yamaguchi K., Wada T., Miyagi T. (2002) Neurochem. Res. 27, 841–846 [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez J. A., Piddini E., Hasegawa T., Miyagi T., Dotti C. G. (2001) J. Neurosci. 21, 8387–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Silva J. S., Hasegawa T., Miyagi T., Dotti C. G., Abad-Rodriguez J. (2005) Nat. Neurosci. 8, 606–615 [DOI] [PubMed] [Google Scholar]
- 14.Kato K., Shiga K., Yamaguchi K., Hata K., Kobayashi T., Miyazaki K., Saijo S., Miyagi T. (2006) Biochem. J. 394, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueno S., Saito S., Wada T., Yamaguchi K., Satoh M., Arai Y., Miyagi T. (2006) J. Biol. Chem. 281, 7756–7764 [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa T., Feijoo Carnero C., Wada T., Itoyama Y., Miyagi T. (2001) Biochem. Biophys. Res. Commun. 280, 726–732 [DOI] [PubMed] [Google Scholar]
- 17.Yamanami H., Shiozaki K., Wada T., Yamaguchi K., Uemura T., Kakugawa Y., Hujiya T., Miyagi T. (2007) Cancer Sci. 98, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato T., Wang Y., Yamaguchi K., Milner C. M., Shineha R., Satomi S., Miyagi T. (2001) Int. J. Cancer 92, 797–804 [DOI] [PubMed] [Google Scholar]
- 19.Shiozaki K., Yamaguchi K., Sato I., Miyagi T. (2009) Cancer Sci. 100, 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngamukote S., Yanagisawa M., Ariga T., Ando S., Yu R. K. (2007) J. Neurochem. 103, 2327–2341 [DOI] [PubMed] [Google Scholar]
- 21.Ishii A., Ikeda T., Hitoshi S., Fujimoto I., Torii T., Sakuma K., Nakakita S., Hase S., Ikenaka K. (2007) Glycobiology 17, 261–276 [DOI] [PubMed] [Google Scholar]
- 22.Fanzani A., Colombo F., Giuliani R., Preti A., Marchesini S. (2004) FEBS Lett. 566, 178–182 [DOI] [PubMed] [Google Scholar]
- 23.Valaperta R., Valsecchi M., Rocchetta F., Aureli M., Prioni S., Prinetti A., Chigorno V., Sonnino S. (2007) J. Neurochem. 100, 708–719 [DOI] [PubMed] [Google Scholar]
- 24.Magesh S., Suzuki T., Miyagi T., Ishida H., Kiso M. (2006) J. Mol. Graph. Model 25, 196–207 [DOI] [PubMed] [Google Scholar]