Abstract
The excision of specific DNA sequences from integrated transgenes in insects permits the dissection in situ of structural elements that may be important in controlling gene expression. Furthermore, manipulation of potential control elements in the context of a single integration site mitigates against insertion site influences of the surrounding genome. The cre–loxP site-specific recombination system has been used successfully to remove a marker gene from transgenic yellow fever mosquitoes, Aedes aegypti. A total of 33.3% of all fertile families resulting from excision protocols showed evidence of cre–loxP-mediated site-specific excision. Excision frequencies were as high as 99.4% within individual families. The cre recombinase was shown to precisely recognize loxP sites in the mosquito genome and catalyze excision. Similar experiments with the FLP/FRT site-specific recombination system failed to demonstrate excision of the marker gene from the mosquito chromosomes.
INTRODUCTION
An important feature of a transformation system is its utility in the analysis of isolated genes and control DNA sequences. Molecular dissection of the primary structure of control DNA sequences relies on the ability to assay functionally altered promoters. While it is possible to evaluate promoter fragments in a number of assays in vitro, genes whose functions are manifest in complex physiological or developmental processes, or involve more than one type of cell or tissue often can be studied only in the background of the whole organism. This requires the ability to put an altered promoter back into the genome of the organism. However, few transformation systems allow targeted integration of a gene into the exact chromosomal environment from which it was derived. Targeted gene replacement in insects was achieved first in Drosophila melanogaster (1), and techniques developed by Golic and colleagues have been adapted widely for use in this species (2). However, these techniques are best suited for gene knockout experiments that result in lines lacking the function of a specific gene.
Insertion of a transgene into random or non-homologous chromosome regions introduces experimental variation due to position effects. The local environment of the insertion site may contain enhancer or silencing DNA sequences (3) or chromatin domains (4) that modify the expression of the introduced transgene. To account for this variability, transformation experiments in D.melanogaster are designed to recover five to 10 independent insertion events and establish them as separate lines. Independent lines could come from primary transformation experiments in which a transgene is inserted into a previously untransformed recipient strain, or from mobilizations and reinsertions of pre-existing, integrated transgenes (5). The activity of the inserted transgene is evaluated in each line and an average picture of the gene function is derived. It is presumed that this averaged function represents the properties of the inserted element.
Position effects on inserted transgenes also confound direct comparisons among different promoter constructs. Often a number of different promoter-reporter genes will be made by varying the amount of putative control DNA contained in the constructs. The intent of these experiments is to answer questions about the function of specific DNA sequence domains on the expression of the reporter gene. However, since these constructs most likely will integrate into different sites in the chromosomes, the analyses must include efforts to account for these effects. Subtle effects of introduced sequence difference most likely will not be observed because they are below the background variation imposed by the position effects. Matrix or scaffold-attachment DNA can insulate the expression of a gene from the effects of the surrounding chromatin (6,7). DNA sequences that insulate transgenes from local insertion site effects have been used in analyses of gene expression in D.melanogaster (8,9). However, primary transformation experiments in mosquitoes are not as efficient as those for D.melanogaster and significant effort would be required to demonstrate rigorously that insulating sites function in the mosquito as they do in the fruit fly. Therefore, we used a technique based on the cre–loxP and FLP/FRT site-specific recombination systems derived from bacteriophage P1 and the yeast Saccharomyces cerevisiae, respectively (10,11), that will permit comparisons of different promoter constructs integrated into the same site of the genome.
We tested a modification of the ‘coplacement’ technique developed in D.melanogaster (12) in which DNA flanked by loxP or FRT sites can be excised following expression of the homologous recombinase. Here we show that a marker gene, a wild-type copy of the D.melanogaster cinnabar gene, can be excised with the cre–loxP system at a high frequency from mosquito chromosomes restoring the mutant white-eye phenotype of the recipient strain. Constructs expressing FLP did not catalyze the excsion of FRT-flanked DNA, and thus the coplacement scheme is not feasible with existing tools. However, the applications of cre–loxP techniques for manipulating the mosquito genome will help in both basic and applied problems in vector biology.
MATERIALS AND METHODS
Homozygous mosquitoes of the khw strain (13) were used in these experiments. This strain, originally designated white-eye (14), has a mutation in the gene encoding kynurenine monooxygenase and this results in a recessive white-eye color phenotype (15). Mosquitoes were reared using standard protocols (16).
The donor plasmid used in this study was derived by ligation of a BglI–EcoRI fragment of pMS109 (17) carrying loxP and FRT recognition sites for the cre and Flp site-specific recombinases, respectively, into the BglI–EcoRI site of pSL1180 (Amersham-Pharmacia). The nucleotide sequence of the minimal loxP recombination site consists of 13 nt inverted repeat sequences flanking an 8 nt asymmetric middle region, 5′-ATAACTTCGTATA GCATACAT TATACGAAGTTAT-3′ (10).
This cloning results in a plasmid with the site-specific recognition sequences inserted into a multiple cloning site (polylinker). In a separate reaction, pBSMos1, containing the mariner transposable element, Mos1 (18,19), was digested with SacI and the vector backbone fragment containing the Mos1 right-hand region with the terminal inverted repeat (TIR) was purified by electrophoresis in agarose, as was the 1.2 kb fragment containing the Mos1 left-hand TIR. The two fragments were ligated, thus removing much of the 5′-end flanking genomic region. The EcoRI and HindIII sites in the original multiple cloning site were destroyed by digestion, blunt-ending and religation (consecutively for each site) to generate p-Δ-M. The EcoRI–SacII fragment from the pMS109-pSL1180 product was blunt-end ligated into the SalI site of p-Δ-M to create p-Δ-M-SL. In a separate reaction, an XbaI–SacII fragment from pH[Cn] (13,20) was ligated into the XbaI–SacII site of pBCKS+ (Stratagene, San Diego, CA). The SacII-Blunt–XhoI fragment from this construct was excised and ligated into the PmlI–XhoI site of p-Δ-M-SL. The XhoI–NotI fragment from pHSHH1.9 (21), which contains the Hermes transposase under control of a heat-shock promoter, was ligated into the XhoI–NotI sites of pBCKS+. This sequence was included for a set of experiments not related to this study. Finally, the KpnI–NotI fragment from this construct was ligated into the KpnI–NotI site of p-Δ-M-SL, to yield the donor plasmid, p[loxP/FRT-cn-Hsphermes] (Fig. 1). This plasmid along with the Mos1 transposase helper plasmid, pKhsp82MOS (22), was used to produce primary transformed lines. The helper plasmid, pMLS104 (a gift from M. L. Siegal, Stanford University), encoding the cre recombinase under the control of the D.melanogaster heat shock protein 70 (hsp70) gene was used for the excision experiments.
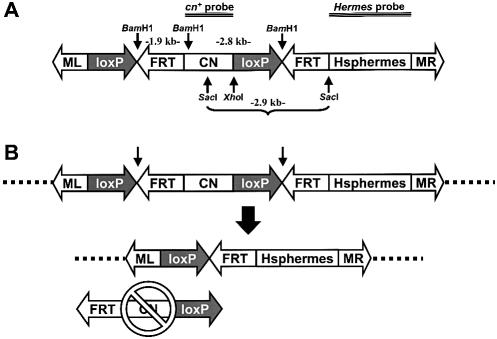
Figure 1.
Structure of the donor element, p[loxP/FRT-cn-Hsphermes] before and after excision. (A) Schematic representation of the plasmid. The boxed figures represent DNA sequences contained in the transformation construct. The left- (ML) and right-hand (MR) Mos1 TIRs have arrowheads to denote the polarity of the repeat sequences, and flank a sequence containing two adjacent loxP and FRT recognition sites (loxP and FRT, respectively), each arranged in the same respective orientation (also denoted by arrowheads). The recognition sites flank a wild-type copy of the D.melanogaster cinnabar gene (CN). The Hermes transposase gene (Hsphermes) is also shown. The location of diagnostic restriction endonuclease cleavage sites, BamHI, SacI and XhoI, are shown as vertical arrows with labels, and the lengths of the anticipated fragments are listed. The extent of the DNA fragments used as probes in Southern analyses are indicated as triple horizontal lines and labeled (cn+ probe and Hermes probe). (B) Schematic representations of the transformation construct integrated into a mosquito chromosome and the expected products following site-specific excision. All labels are the same as in (A). The dotted heavy line represents A.aegypti chromosomal DNA. Excision of the fragment between the arrows (top) leads to a product that is missing the marker gene (bottom). Animals with the intact construct have purple eyes, and those with the excised construct have white eyes.
Microinjection procedures to produce the primary transformed lines containing the p[loxP/FRT-cn-Hsphermes] transgene were essentially the same as in Jasinskiene et al. (20). Crosses with individual microinjected males and multiple untreated females were used to establish single-founder families (G0 generations). Following standard procedures (20), G0 females were mated in pools with untreated males. Positive (colored eyes) G1 progeny were used to establish transgenic families. Genomic DNA was isolated from these individuals, prepared for Southern analysis and probed with radiolabeled DNA complementary to the D.melanogaster cn+ gene (cn+ probe) and/or with sequences complementary to the Hermes transposase (Hermes probe).
Excision experiments were performed by injecting embryos transformed with p[loxP/FRT-cn-Hsphermes] with the cre helper plasmid, pMLS104, at 0.8 mg/ml in transformation buffer (20). Animals were heat-shocked 18–24 h after injection at 41°C for 1 h to induce recombinase expression. These procedures maximize embryo survival and permit recombination and transposition in germ cells (20,23). Surviving adults were mated individually to multiple khw members of the opposite sex to produce a G0. Adult progeny resulting from these matings were scored for the presence of a white-eye phenotype. White-eye siblings were mated to produce a G1 and to establish a line. Gene amplification of control and putative excised DNA was carried out using primers ML4 (5′-GGGAATGTCGGTTCGAACAT-3′), cnF (5′-ATCCCATTTGGTCTACGAG-3′) and hspout (5′-TTATACTCCGGCGCTCTTTT-3′), and the following reaction conditions: one cycle at 94°C for 3 min; 29 cycles at 94°C for 40 s, 63°C for 45 s and 72°C for 1 min; followed by one cycle at 72°C for 7 min. Samples and marker DNA were resolved on agarose gels and photographed. Similar experiments were performed using the FLP helper plasmid, pP[ry+ hsFLP], which has the FLP recombinase under the control of the D.melanogaster hsp70 gene promoter (22,24). All injection, rearing, mating and screening protocols were identical to the cre–loxP experiments.
RESULTS
Transformation of Aedes aegypti with p[loxP/FRT-cn-Hsphermes]
A total of 1382 khw/khw A.aegypti embryos were injected with donor plasmid, p[loxP/FRT-cn-Hsphermes] and the Mos1 helper plasmid, pkhsp82MOS. Of the injected animals, 295 matured to the adult stage, and 73 single-male founder families and 12 pooled lines of females were set up from the survivors. Five independent primary transformed lines were recovered (Table 1) for an absolute transformation frequency of 1.3% (1/73) and an adjusted transformation frequency of 5.8% (5/85) (23). Southern blot analyses revealed that all five lines had integrated the transformation construct (data not shown). Analysis of line P11 showed that it likely contained an integration of a single transformation construct (Fig. 2). In addition to the 1.9 and 2.8 kb fragments of the BamHI digestion, a large ∼9.0 kb fragment in the XhoI digestion is evident and this results from hybridization of the cn+ probe to a fragment produced by a restriction site within the plasmid and one in the genome. The D.melanogaster cn+ probe does not cross-hybridize with the endogenous orthologous mosquito gene. The P11 line was made homozygous and used for further analysis.
Table 1. G0 families producing G1 progeny with colored eyesa.
| G0 family | Total no. of G1 progeny screened | No. of G1 progeny with colored eyes (%) |
|---|---|---|
| 128 | 431 | 32 (7.4) |
| P3b | 375 | 1 (0.3) |
| P3.1 | 375 | 3 (0.8) |
| P11 | 188 | 4 (2.0) |
| P10 | 190 | 10 (5.2) |
aThese lines have insertions of the p[loxP/FRT-cn-Hsphermes] construct.
b‘P’ indicates a line derived from pooled G1 females (20).
Figure 2.
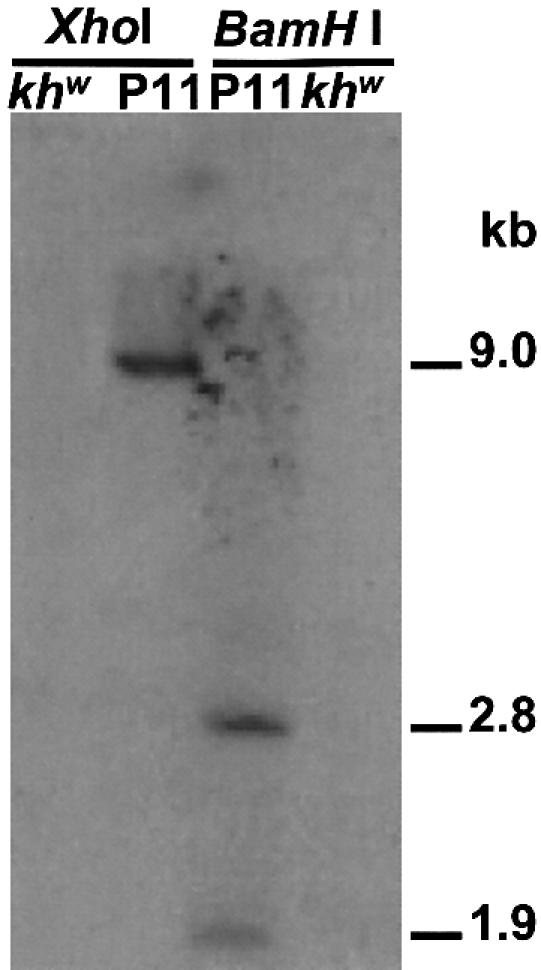
The transformed line, P11, has a single insertion of the transformation construct, p[loxP/FRT-cn-Hsphermes]. Genomic DNA from line P11 was digested with either XhoI or BamHI and probed with the cn+ gene. Genomic DNA from the recipient strain, khw was used as a control. In addition to the diagnostic fragments at 1.9 and 2.8 kb in the BamH1 pattern, an additional fragment of ∼9.0 kb in the XhoI digestion is consistent with a single insertion of the transgene.
Excision of the D.melanogaster wild-type cinnabar gene from line P11
A total of 443 homozygous P11 embryos were microinjected with the recombinase-encoding plasmid, pMLS104. Follow ing individual matings of the 39 surviving adults to 15 members of the opposite sex of the khw recipient strain, all progeny were checked for eye color. Seven of 21 (33.3%) fertile families produced some progeny with white eyes. Furthermore, analysis of the number of progeny in the individual lines showing white-eyes revealed that the excision efficiency within a line could be as low as 20% and as high as 99.4% (Table 2). All seven families were set up as sublines of the original P11 line and designated P1, 6, 15, 18, 24, 25 and 29.
Table 2. Distribution of putative excision progeny in individual families.
| Family | Phenotypic class | Number of animals | Percentage of white-eye phenotypes |
|---|---|---|---|
| 24 | Purple-eye | 3 | 99.4 |
| White-eye | 499 | ||
| 6 | Purple-eye | 28 | 45 |
| White-eye | 23 | ||
| 25 | Purple-eye | 4 | 48 |
| White-eye | 3 | ||
| 15 | Purple-eye | 8 | 20 |
| White-eye | 2 | ||
| 18 | Purple-eye | 16 | 24 |
| White-eye | 5 | ||
| 29 | Purple-eye | 4 | 50 |
| White-eye | 2 | ||
| 30 | Purple-eye | 2 | 60 |
| White-eye | 3 |
Molecular analysis of putative excision events
Three types of analyses were performed to confirm the molecular structure of a putative cre-mediated excision event. Gene amplification with oligonucleotide primers, ML4 and hspout, flanking the putative excised region showed that white-eyed individuals from all seven sublines produced an ∼600 bp fragment whose size is consistent with the loss of the cn+ gene and flanking loxP site (Fig. 3). Furthermore, Southern analysis of two of the P11 sublines, 6 and 24 using the cn+ probe and Hermes probe, revealed that they lack a 2.9 kb fragment that is diagnostic in a SacI digestion of genomic DNA for the presence of a portion of the cn+ gene and flanking loxP site (Fig. 4). Two of the larger fragments, 6.0 and 9.0 kb, in the control P11 digestion likely represent the adjacent genomic fragments while the origin of the third fragment is unknown. The presence of the 9.0 kb fragment in lines 24 and 6 support the conclusion that it is the fragment containing Hsphermes and MR (Fig. 1). Finally, genomic DNA amplified from line 24 was sequenced to determine the primary structure of the product of the putative excision event (Fig. 5). This analysis verified that the Mos1 left-hand repeat sequence was adjacent to a loxP site that was contiguous with DNA that adjoined the Hsphermes sequences. The recombination event leading to excision is precise with no loss or addition of any DNA other than what would be predicted based on the function of the cre–loxP system.
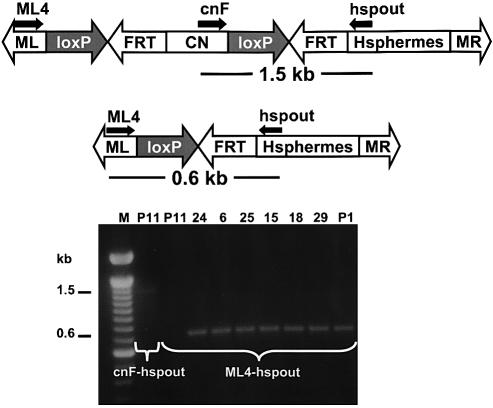
Figure 3.
Gene amplification analysis of genomic DNA from putative excision events. Genomic DNA from G1 individuals was used as a template for amplification with specific primers, ML4, cnF and hspout (small horizontal arrows). A 1.5 kb DNA fragment is expected from the primers cnF and hspout in the P11 parental line. A 0.6 kb fragment is anticipated in excised lines, 24, 6, 25, 15, 18, 29 and P1, using the primers ML4 and hspout. The ML4-hspout fragment in the parental line, P11 is too large to amplify efficiently in these reactions.
Figure 4.
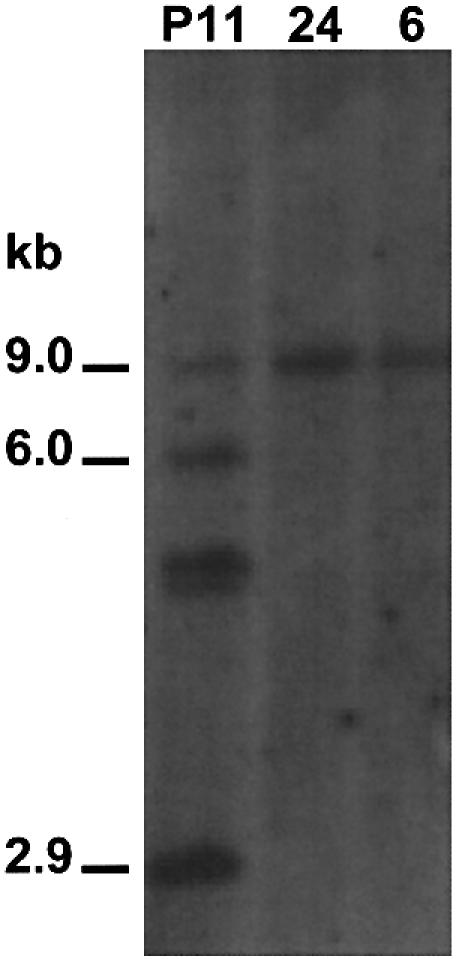
Southern blot analysis of excision lines. Genomic DNA was prepared from G1 animals, digested with SacI and probed with a mixture of the cn+ and Hermes probes (Fig. 1). Fragments of 2.9 and ∼9.0 kb are expected if no excision occurs (P11; Fig. 1).
Figure 5.
Sequence analyses of the excision site in line 24. The DNA flanking the remaining loxp site in line 24 was sequenced. Sequence corresponding to the Mos1 left-hand TIR repeat domain and D.melanogaster Hsp70 promoter now flank a single loxP site (bold). The adjacent BamHI site is shown in italics.
FLP/FRT site-specific excision experiments
Three separate experiments were performed to determine if the FLP/FRT site-specific recombination system could function as efficiently as the cre–loxP system in excising DNA from mosquito chromosomes. In the first experiment, 6539 progeny derived from 28 fertile matings of line P3 mosquitoes injected with the FLP helper plasmid and mated with the khw/khw recipient line yielded no progeny with white eyes. In a second experiment, 16 270 progeny from 88 fertile matings of injected P11 animals were screened, and four white-eyed adults were recovered. However, none of these animals bred true, and preliminary molecular analyses failed to demonstrate any evidence of excision (data not shown). A new isolate of the helper plasmid was used in the third experiment and 12 892 progeny from 69 families derived from injections of line P11 were screened. None of these mosquitoes had white eyes. Sequence analyses of the construct DNA inserted in line P11 confirmed that the FRT sites were intact structurally, and did not contain any mutations, additions or deletions (data not shown). We interpret these experiments to indicate that FLP-mediated excision of a marker gene flanked by FRT sites is not as efficient as the cre–loxP system in the conditions used here.
DISCUSSION
Site-specific excision of the D.melanogaster cn+ gene from transgenic A.aegypti is relatively efficient with the cre–loxP system and procedures used here. The efficiencies, 33% of all lines showing excision with as many as 99.4% of individuals within a line having an excised fragment, although lower than the 100% reported for D.melanogaster (12), still qualify this as a powerful tool for gene analysis in A.aegypti. Indeed, excision efficiencies likely could be increased if an inducible source of cre recombinase was integrated into the mosquito genome as was done with the fruit fly experiments. Furthermore, stable lines of mosquitoes expressing the recombinase controlled by constitutive or tissue-specific promoters could provide powerful tools for dissecting important gene functions. The lower levels of efficiency of excision within lines came from those crosses in which for convenience only a few progeny were counted. However, in each case, sufficient progeny were obtained to establish an excised line by mating siblings. This recovery rate coupled with the relatively low number of embryos injected make this a technique that could be used routinely for this species. It should be noted that coplacement methods can only compare two different constructs at a time and that several different transformed lines are needed to attribute significance to small differences in expression (25).
It remains to be demonstrated whether loxP sites in mosquito chromosomes can serve as ‘docking sites’ for site-specific integration following direct injection of a host strain with integrated loxP sites with an integration construct carrying a loxP site (23). Efforts to develop this specific strategy in D.melanogaster have yet to be successful [although other strategies for targeted gene replacement have been developed (2)], but the biology of the mosquito maybe be different enough to make this possible.
The survival of embryos in the primary and excision injection portions of the experiments, 21.3 and 8.8%, respectively, are within the range seen with other microinjection experiments with A.aegypti (20,22,26,27). In addition, fertility of surviving G0 adults is consistent with previous experiments (26). The transformation frequency of the primary integration experiment is consistent with what is seen with Mos1 transposition (22,26). Thus, the application of the cre–loxP system appears not to increase the difficulty of handling transgenic A.aegypti. These techniques may be considered straightforward and routine, although they still remain labor intensive and time consuming.
The failure to detect site-specific excision with the FLP/FRT system is puzzling. There is clear evidence that there are no inhibitory factors in mosquito embryos that would interfere with FLP function (23). Furthermore, integrated copies of marker genes flanked by FRT sites could be moved from one location in the D.melanogaster genome to a target FRT site present at another location in the genome (24). Perhaps the differences in results between the work done here and the other experiments have to do with using integrated or non-integrated donor elements and the specific timing of initiating the recombinant event. It is also possible that the FLP protein is not as stable as the cre recombinase. FLP-expressing constructs based on conditional promoters derived from the mosquito may also mitigate some of these effects. However, in the final analysis, the robust activity of the cre–loxP system makes this general type of tool available to mosquito researchers.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank their colleagues for comments and Lynn Olson for helping in preparing the manuscript. The research was supported by an award from the National Institutes of Health (AI-44238) to A.A.J.
REFERENCES
- 1.Gloor G.B., Nassif,N.A., Johnson-Schlitz,D.M., Preston,C.R. and Engels,W.R. (1991) Targeted gene replacement in Drosophila via P element-induced gap repair. Science, 253, 1110–1117. [DOI] [PubMed] [Google Scholar]
- 2.Rong Y.S. and Golic,K.G. (2000) Gene targeting by homologous recombination in Drosophila. Science, 288, 2013–2018. [DOI] [PubMed] [Google Scholar]
- 3.Wilson C., Bellen,H.J. and Gehring,W.J. (1990) Position effects on eukaryotic gene expression. Annu. Rev. Cell. Biol., 6, 679–714. [DOI] [PubMed] [Google Scholar]
- 4.Milot E., Strouboulis,J., Trimborn,T., Wijgerde,M., de Boer,E., Langeveld,A., Tan-Un,K., Vergeer,W., Yannoutsos,N., Grosveld,F. and Fraser,P. (1996) Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell, 4, 105–114. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J.D. (2002) Putting mobile DNA to work: the toolbox. In Craig,N.L. et al. (eds). Mobile DNA II. ASM Press, Washington DC, pp. 24–37. [Google Scholar]
- 6.Poljak L., Seum,C., Mattioni,T. and Laemmli,U.K. (1994) SARs stimulate but do not confer position independent gene expression. Nucleic Acids Res., 22, 4386–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attal J., Cajero-Juarez,M., Petitclerc,D., Theron,M.C., Stinnakre,M.G., Bearzotti,M., Kann,G. and Houdebine,L.M. (1995) The effect of matrix attached regions (MAR) and specialized chromatin structure (SCS) on the expression of gene constructs in cultured cells and in transgenic mice. Mol. Biol. Rep., 22, 37–46. [DOI] [PubMed] [Google Scholar]
- 8.Hagstrom K., Muller,M. and Schedl,P. (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev., 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Barolo,S., Szymanski,P. and Levine,M. (1996) The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- 10.Austin S., Ziese,M. and Sternberg,N. (1981) A novel role for site-specific recombination in maintenance of bacterial replicons. Cell, 25, 729–736. [DOI] [PubMed] [Google Scholar]
- 11.McLeod M., Craft,S. and Broach,J.R. (1986) Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Mol. Cell Biol., 6, 3357–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegal M.L. and Hartl,D.L. (1996) Transgene coplacement and high efficiency site-specific recombination with the cre/loxP system in Drosophila. Genetics, 144, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornel A.J., Benedict,M.Q., Salazar Rafferty,C., Howells A.J. and Collins,F.H. (1997) Transient expression of the Drosophila melanogaster cinnabar gene rescues eye color in the white-eye (WE) strain of Aedes aegypti. Insect Biochem. Mol. Biol., 12, 993–997. [DOI] [PubMed] [Google Scholar]
- 14.Bhalla S.C. (1968) White eye, a new sex-linked mutant of Aedes aegypti.Mosquito News, 28, 380–385. [Google Scholar]
- 15.Han Q., Calvo,E., Marinotti,O., Fang,J., Rizzi,M., James,A.A. and Li,J. (2003) Analysis of the wild-type and mutant genes encoding the enzyme kynurenine monooxygenase of the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol., 12, 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munsterman L.E. (1997) Care and maintenance of Aedes mosquitoes colonies. In Crampton,J.M., Beard,C.B. and Louis,C. (eds), The Molecular Biology of Insect Disease Vectors. Chapman and Hall, London, pp. 13–20. [Google Scholar]
- 17.Snaith M.R., Murray,J.A. and Boulter,C.A. (1995) Multiple cloning sites carrying loxP and FRT recognition sites for the cre and Flp site-specific recombinases. Gene, 166, 173–174. [DOI] [PubMed] [Google Scholar]
- 18.Medhora M.M., MacPeek,A.H. and Hartl D.L. (1988) Excision of the Drosophila transposable element mariner: identification and characterization of the Mos factor. EMBO J., 7, 2185–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medhora M., Maruyama,K. and Hartl,D.L. (1991) Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics, 128, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasinskiene N., Coates,C.J., Benedict,M.Q., Cornel,A.J., Salazar-Rafferty,C., James,A.A. and Collins,F.H. (1998) Stable, transposon-mediated transformation of the yellow fever mosquito, Aedes aegypti, using the Hermes element from the housefly. Proc. Natl Acad. Sci. USA, 95, 3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren W.D., Atkinson,P.W. and O’Brochta,D.A. (1994) The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac and Tam3 (hAT) element family. Genet. Res., 64, 87–97. [DOI] [PubMed] [Google Scholar]
- 22.Coates C.J., Jasinskiene,N., Miyashiro,L. and James,A.A. (1998) Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti.Proc. Natl Acad. Sci. USA, 95, 3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris A.C., Schaub,T.L. and James,A.A. (1991) FLP-mediated recombination in the vector mosquito, Aedes aegypti. Nucleic Acids Res., 19, 5895–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golic M.M., Rong,Y.S., Petersen,R.B., Lindquist,S.L. and Golic,K.G. (1997) FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res., 25, 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegal M.L. and Hartl,D.L. (1998) An experimental test for lineage-specific position effects on alcohol dehydrogenase (Adh) genes in Drosophila. Proc. Natl Acad. Sci. USA, 26, 15513–15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adelman Z.N., Jasinskiene,N. and James,A.A. (2002) Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti.Mol. Biochem. Parasitol., 121, 1–10. [DOI] [PubMed] [Google Scholar]
- 27.Coates C.J., Jasinskiene,N., Pott,G.B. and James,A.A. (1999) Promoter-directed expression of recombinant fire-fly luciferase in the salivary glands of Hermes-transformed Aedes aegypti.Gene, 226, 317–325. [DOI] [PubMed] [Google Scholar]