Abstract
It has been shown previously that ribosomal protein S3 (rpS3) has an endonuclease activity, which is increased by protein kinase Cδ (PKCδ)-dependent phosphorylation. However, the reciprocal mechanism for rpS3 dephosphorylation is not known. In this study, we examined phosphatases involved in rpS3 dephosphorylation, and we determined that rpS3 is specifically dephosphorylated by protein phosphatase 2A (PP2A). By immunoprecipitation assay, rpS3 only interacted with PP2Ac but not with protein phosphatase 1. The interaction between rpS3 and PP2Ac occurred only in the nuclear fraction. Moreover, the PP2Ac association with rpS3 was identified in cells transfected with wild-type rpS3 but not with mutant rpS3 lacking PKCδ phosphorylation sites. PP2A inhibition using okadaic acid induced rpS3 phosphorylation. The level of phosphorylated rpS3 in cells was decreased by the overexpression of PP2Ac and was increased by the down-regulation of PP2Ac. Taken together, these results suggest that oxidative stress regulates the phosphorylation status of nonribosomal rpS3 by both activating PKCδ and blocking the PP2A interaction with rpS3.
Protein phosphatase 2A (PP2A)2 is a major serine/threonine phosphatase that plays a variety of roles in mammalian cells (1). The PP2A holoenzyme is composed of three subunits, including a structural A subunit, a catalytic C subunit (PP2Ac), and one of several B regulatory subunits. The heterodimeric PP2A core enzyme, comprised of a C subunit and an A subunit, associates with a wide variety of B regulatory subunits. To date, 15 genes for the B subunit have been identified in the human genome (2), which regulate both the substrate specificity and the localization of the PP2A holoenzyme (3–5). It has been shown that PP2A has many interacting proteins containing signaling molecules, including kinases (1, 2, 6). Therefore, PP2A plays pivotal roles in many cellular processes, including transcription (7–9), translation (10), the cell cycle (11–13), and transformation (14–16).
Ribosomal protein S3 (rpS3) is a component of the 40 S small ribosomal subunit and therefore is associated with protein synthesis. However, it also has been determined that rpS3 is a multifunctional protein involved in DNA repair, transcription, metastasis, and apoptosis (17–22). In conjunction with its multiple functions, there are many signaling molecules or modifications that have effects on rpS3 function. To date, although no clear correlation has been found between these modifications and rpS3 function, it has been determined that rpS3 is post-translationally modified, including by phosphorylation, neddylation, sumoylation, and ubiquitination (22–28). Among these, rpS3 phosphorylation in particular has been studied in conjunction with the function of rpS3.
Extracellular signal-regulated kinase (ERK) interacts with rpS3 and phosphorylates threonine 42 on rpS3 (23). Hrr25-dependent phosphorylation of rpS3 affects the stable integration of rpS3 into the pre-40 S subunit and the maturation of 40 S ribosomal subunits in yeast (24). A similar situation has also been observed in mammalian cells where rpS3 is phosphorylated by PKCδ, and the phosphorylated form of rpS3 is only detected in the nonribosomal fraction (22).
In this study, we postulated that phosphorylated rpS3 was dephosphorylated by a phosphatase in the nucleus to be incorporated into the 40 S ribosomal subunit. We determined that PP2Ac interacted with the N terminus of nonribosomal rpS3 in the nucleus. Moreover, we found that PKCδ-dependent phosphorylation sites on rpS3 were involved in the interaction between them. These results indicate that the phosphorylation status of rpS3 is regulated by both PKCδ and PP2A.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against GFP, GST, poly(ADP-ribose) polymerase, RACK1, B55α, and JNK were from Santa Cruz Biotechnology (Santa Cruz, CA). Tubulin and FLAG antibodies were from Sigma. PP1/PP2A Toolbox (catalog number 17-301), PP2Ac, and PP6 antibody were from Upstate Biotechnology, Inc. (Lake Placid, NY); phospho-serine/threonine antibody was from BD Transduction Laboratories, and rpS3 antibody was from BioInstitute (Korea University, Seoul, South Korea). ECL reagents were purchased from Pierce, and protein A-Sepharose was purchased from Roche Applied Science. H2O2 was purchased from Sigma. Ara-C, U0126, PD98059, rottlerin, and okadaic acid were from Calbiochem, and glutathione-Sepharose 4B was from Amersham Biosciences.
Cell Culture and Transfection
HEK293T, NIH3T3, and HT1080 cells were maintained in Dulbecco's modified Eagle's medium (WelGENE Inc.) supplemented with 10% fetal bovine serum (WelGENE Inc.), 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected using Lipofectamine (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation and Immunoblotting
Cells were harvested and lysed on ice in TNN lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 50 mm NaF, 1 mm Na3VO4, and protease inhibitors 2 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A). Clarified lysates were incubated with 1–2 μg of the appropriate antibody for 2 h followed by 25 μl of a 50% slurry of protein A-agarose (Roche Applied Science) for 3–16 h at 4 °C. Precipitates were washed three times, eluted with SDS sample buffer, and subjected to SDS-PAGE.
For immunoblot analysis, proteins were separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (Millipore) or nitrocellulose (Bio-Rad) membrane. Membranes were blocked with 5% nonfat dry milk for 1 h. Blots were incubated with primary antibody in blocking solution for 1 h at 4 °C. Membranes were rinsed twice with TBST and incubated with horseradish peroxidase-conjugated secondary antibody (Chemicon) in blocking solution for 30 min. The bound complex was visualized using the ECL system (Roche Applied Science).
Metabolic Labeling
HT1080 cells were cultured in phosphate-free Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (WelGENE Inc.). After 1 h at 37 °C, the cells were labeled using the same medium containing 500 μCi/ml [32P]orthophosphate (PerkinElmer Life Sciences) for 2 h. The cells were pretreated with 10 nm okadaic acid (OA) for 2 h and then followed by 20 μm Ara-C treatment for 30 min. Labeling was stopped by washing the cells twice with ice-cold phosphate-buffered saline. The cells were harvested and lysed in 0.8 ml of TNN buffer containing protease inhibitors. Equal amounts of cell extracts were used for immunoprecipitation analysis with rpS3 antibody. The immunoprecipitates were washed with lysis buffer four times and subjected to SDS-PAGE. After the gel was dried, labeled proteins were visualized with a BAS2500 imaging analyzer (Fujifilm, Tokyo, Japan). Parallel gels were immunoblotted with rpS3 antibody to confirm equal amounts of immunoprecipitated rpS3.
Nuclear/Cytosol Fractionation
Cell fractionation was performed as described previously (22). For immunoprecipitation of the nuclear fraction, a nuclear pellet was resuspended in TNN buffer and mechanically disrupted by sonication. After centrifugation, equal amounts of nuclear proteins were subjected to immunoprecipitation with the indicated antibody.
Ribosome Fractionation
HT1080 cells were treated with the indicated drugs, and ribosome fractionation and immunoprecipitation were performed as described previously (25).
RNA Interference
AccuTargetTM human PP2Ac (1120864), PP1 (1120564), PP6 (1121117), PKCδ (1121871), and negative control siRNA (SN-1013) were purchased from Bioneer (Daejeon, Korea). HT1080 cells were reverse-transfected using LipofectamineTM RNAiMax (Invitrogen) according to the manufacturer's instructions.
In Vitro Dephosphorylation Assay
HT1080 cell extracts were immunoprecipitated with PKCδ antibody, after which PKCδ kinase assay was performed using His-rpS3 as a substrate (22). To verify the specific phosphatase of rpS3, phosphorylated His-rpS3 was mixed with PP1 (0.1 unit) or PP2A (0.05 unit) for indicated times at 30 °C. Equal amounts of proteins were separated by SDS-PAGE in 12% gels. After the gel was dried, labeled proteins were visualized with a BAS2500 imaging analyzer (Fujifilm, Tokyo, Japan).
RESULTS
Identification of the Interaction between rpS3 and PP2A in Vivo
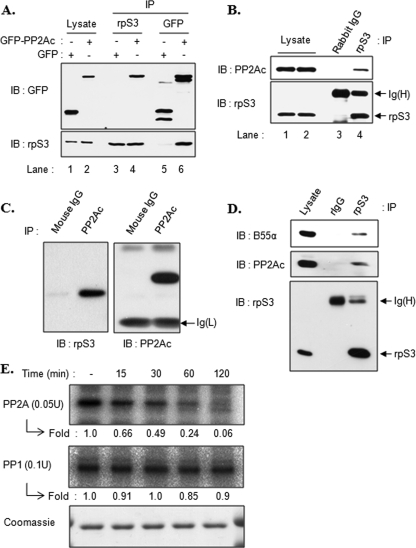
To determine the interaction of PP2Ac with rpS3, we first used a co-immunoprecipitation assay using overexpressed GFP-PP2Ac. HEK293T cells were transfected with either a GFP or a GFP-PP2Ac plasmid and then immunoprecipitated with GFP or rpS3 antibodies. As shown in Fig. 1A, an interaction between rpS3 and GFP-PP2A was verified (lanes 4 and 6) but not control GFP (lanes 3 and 5). We next investigated if this interaction occurred between endogenous PP2Ac and rpS3. We confirmed a specific interaction between endogenous proteins in HT1080 cells (Fig. 1B). Conversely, rpS3 protein was detected in immunoprecipitates derived from PP2Ac antibody (Fig. 1C). We also tested whether rpS3 interacted with PP1, but we detected no interaction between them by immunoprecipitation assays (data not shown). These results indicate that rpS3 specifically interacts with PP2Ac in vivo. To expand our data, we examined which regulatory B subunit specifically interacts with rpS3. Even though there are many isoforms of the B subunit, we tested with B55α because this was purified as the sole B subunit complexed with the PP2A C and A subunit in HT1080 cells (29). Fig. 1D shows that rpS3 was associated simultaneously with PP2Ac and B55α. These results suggest that rpS3 is a substrate of PP2A. Furthermore, we tested whether PP2A actually dephosphorylates phosphorylated rpS3 through the in vitro dephosphorylation assay. We first obtained phosphorylated His-rpS3 as a substrate from PKCδ kinase assay. Equal amounts of phosphorylated His-rpS3 were mixed with PP1 or PP2A and incubated for the indicated times. Phosphorylation level of rpS3 was time-dependently decreased only in the PP2A mixture (Fig. 1E, upper panel) but not in the PP1 (Fig. 1E, middle panel). These data indicate that PP2A regulates the phosphorylation status of rpS3.
FIGURE 1.
rpS3 interacts with PP2A. A, HEK293T cells were transfected with pEGFPc1 or pEGFPc1-PP2Ac plasmids. Cell extracts (lanes 1 and 2) were immunoprecipitated (IP) using rpS3 (lanes 3 and 4) or GFP (lanes 5 and 6) antibodies, and immunoprecipitated proteins were resolved by 10% SDS-PAGE and immunoblotted (IB) using the indicated antibodies. B, HT1080 cells were lysed, and extracts were immunoprecipitated with rabbit IgG (lane 3) or rpS3 antibody (lane 4). Cell extracts (lanes 1 and 2) and precipitated proteins (lanes 3 and 4) were resolved and immunoblotted with the indicated antibodies. C, HT1080 cell lysates were immunoprecipitated with mouse IgG or PP2Ac antibody, and proteins were detected using the indicated antibodies. D, HT1080 cells were lysed and extracts were immunoprecipitated with rabbit IgG (rIgG) or rpS3 antibody. Immunoprecipitates were resolved and immunoblotted with the indicated antibodies. E, PKCδ kinase assay was performed with purified His-rpS3 to make the phosphorylated form of rpS3 as described under “Experimental Procedures.” Phosphorylated His-rpS3 was incubated with PP2A (upper panel) or PP1 (middle panel) for the indicated times at 30 °C. Coomassie staining shows the equal amounts of His-rpS3 substrates (lower panel). Numbers indicate relative fold decrease of phosphorylated His-rpS3 compared with the control in the absence of phosphatase.
PP2A Interacts with N-terminal Region of rpS3
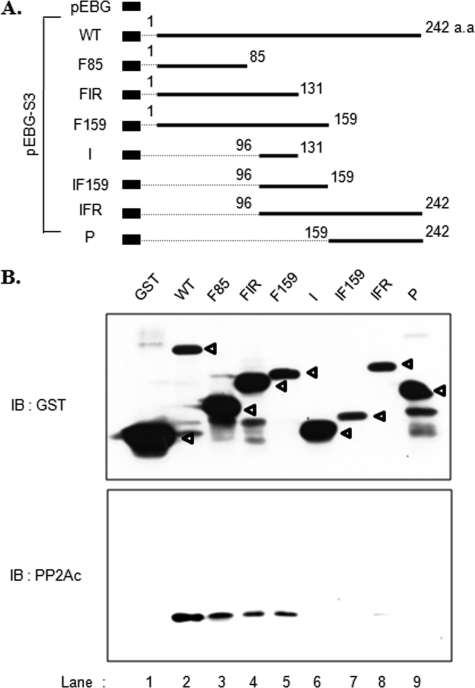
We tested which region of rpS3 interacted with PP2Ac. For this we generated GST-fused rpS3 deletion constructs as described in Fig. 2A (25). These GST fusion constructs were transfected into HEK293T cells, and the binding of these rpS3 fragments to PP2Ac was examined using GST pulldown assays. As expected, PP2Ac was pulled down by wild-type GST-rpS3 (Fig. 2B, lane 2) but not by GST alone (Fig. 2B, lane 1). Additionally, PP2Ac was also co-precipitated with the N terminus that contained GST-rpS3 deletion constructs (Fig. 2B, lanes 3–5). This result clearly indicates that the N terminus of rpS3 is essential for the interaction with PP2Ac.
FIGURE 2.
N-terminal domain of rpS3 interacts with PP2Ac. A, schematic representation of GST-rpS3 deletion constructs. The numbers indicate amino acid (a.a) positions in rpS3. B, HEK293T cells were transiently transfected with GST or GST-rpS3 deletion mutants as described in A. After 24 h, cells were lysed, and extracts were pulled down with glutathione-Sepharose beads. Isolated GST fusion proteins were resolved by 10% SDS-PAGE and immunoblotted (IB) with PP2Ac and rpS3 antibodies. Open arrowheads indicate the positions of GST and GST-rpS3 constructs and GST, respectively. WT, wild type.
Oxidative Stress Abrogates the Interaction between rpS3 and PP2Ac in the Nucleus
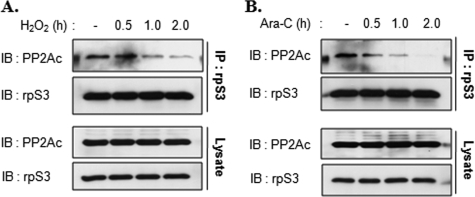
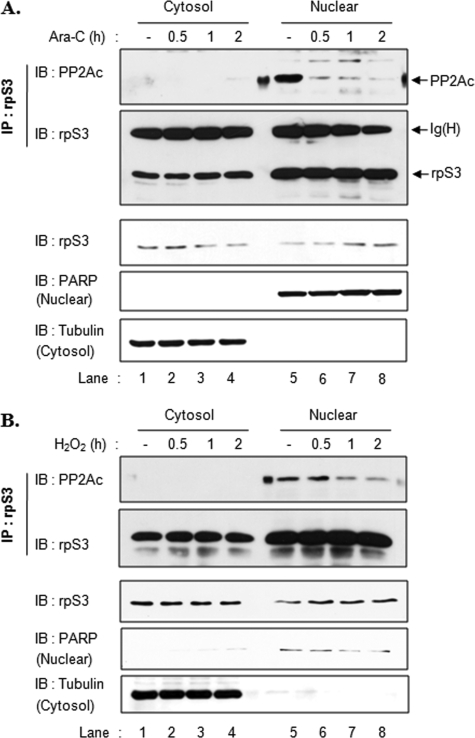
In previous studies, we demonstrated that rpS3 is phosphorylated by PKCδ and that the phosphorylation of rpS3 was increased by treatment with DNA-damaging agents, such as H2O2 and Ara-C (22). Thus, we first tested if the interaction between rpS3 and PP2Ac was affected by oxidative stress. HT1080 cells were treated with 100 μm H2O2 (Fig. 3A) or 10 μm Ara-C (Fig. 3B) for different periods of time, followed by immunoprecipitation assays using rpS3 antibody. We obtained similar results using H2O2 and Ara-C treatments indicating that the interaction between rpS3 and PP2Ac decreases in a time-dependent manner (Fig. 3). This result is in a good agreement with our previous results showing that rpS3 phosphorylation was increased by H2O2 and Ara-C (22). Next, we performed a subcellular fractionation to explore the location of this interaction in cells. HT1080 cells were treated with Ara-C (Fig. 4A) or H2O2 (Fig. 4B) for the indicated times and then fractionated into cytosol and nuclear fractions. Each fraction was immunoprecipitated with rpS3 to examine the rpS3 interaction with PP2A. The interaction between rpS3 and PP2A was observed only in the nucleus (Fig. 4, A, lane 5, and B, lane 5), and this was decreased by Ara-C or H2O2 treatment (Fig. 4, A, lanes 6–8, and B, lanes 6–8). This result was predicted, as we reported previously that rpS3 phosphorylation is promoted by oxidative stress and that phosphorylated rpS3 was accumulated in the nucleus (22). Accordingly, these results suggest that the phosphorylated form of rpS3 associates with PP2A in the nucleus. We also confirmed that the all constructs used in this study are localized both in the cytosol and nuclear fraction (supplemental Fig. 1).
FIGURE 3.
Interaction between rpS3 and PP2Ac is decreased by H2O2 or Ara-C treatment. A and B, HT1080 cells were treated with 100 μm H2O2 (A) or 10 μm Ara-C (B) for the indicated times, and extracts were immunoprecipitated (IP) with rpS3 antibody. rpS3 immunoprecipitates were resolved by 10% SDS-PAGE and immunoblotted (IB) with rpS3 and PP2Ac antibodies (upper panel in A and B). Equal amounts of protein were used for immunoprecipitation (lower panel in A and B).
FIGURE 4.
rpS3 interacts with PP2Ac in the nucleus. A and B, HT1080 cells were treated with 10 μm Ara-C (A) or 100 μm H2O2 (B) for the indicated times. Cells were fractionated into cytosol and nuclear fractions as described under “Experimental Procedures.” Equal amounts of fractionated proteins were immunoprecipitated (IP) with rpS3 antibody. Fractionated cell extracts (lower panel) and immunoprecipitates (upper panel) were resolved by 10% SDS-PAGE and subjected to immunoblotting (IB) with the indicated antibodies. Tubulin was used as a cytosolic marker and poly(ADP-ribose) polymerase (PARP) as a nuclear marker to estimate both correct fractionation and normalization.
Okadaic Acid Prevents the PP2A Interaction with rpS3
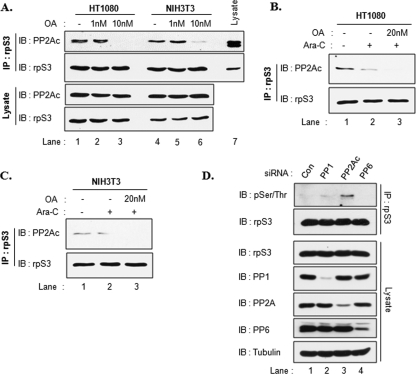
We next tested the effects of OA, a specific inhibitor of PP2A, on the PP2Ac association with rpS3. HT1080 or NIH3T3 cells were treated with 1 or 10 nm OA for 1 h, and then cell extracts were immunoprecipitated with rpS3 antibody. This showed that treatment with 10 nm OA significantly reduced the interaction between the two proteins in both cell lines (Fig. 5A, lanes 3 and 6). We also examined the interaction using a combination of Ara-C and OA. HT1080 cells were pretreated with or without 20 nm OA for 1 h and were then stimulated with Ara-C as described in Fig. 5B. As shown previously, Ara-C attenuated the rpS3-PP2A interaction (Fig. 5B, lane 2). Moreover, combined stimulation with OA and Ara-C showed greater disruption of the interaction compared with Ara-C alone (Fig. 5B, lane 3). We obtained the same results for NIH3T3 cells (Fig. 5C). These findings suggest that the phosphatase activity of PP2A is necessary for the association with rpS3.
FIGURE 5.
PP2Ac activity is critical for the association with rpS3. A, HT1080 (lanes 1–3) and NIH3T3 (lanes 4–6) cells were treated with OA for 1 h. The cells were lysed, and extracts were immunoprecipitated (IP) with rpS3 antibody. Cell extracts (lower panel and lane 7) and rpS3 immunoprecipitates (upper panel) were immunoblotted (IB) with PP2Ac and rpS3 antibodies. B and C, HT1080 (B) or NIH3T3 (C) cells were pretreated with OA for 3 h followed by Ara-C (10 μm) treatment for 30 min. Cell extracts were immunoprecipitated with rpS3 antibody. Immunoprecipitates were resolved and subjected to immunoblotting with rpS3 and PP2Ac antibodies. D, HT1080 cells were transfected with siRNAs against control (Con), PP1, PP2Ac, or PP6. After 48 h, cells were lysed, and extracts were immunoprecipitated with rpS3 antibody. Immunoprecipitates and cell extracts were resolved and subjected to immunoblotting with the indicated antibodies.
PP2A Is a Specific Phosphatase Regulating rpS3 Phosphorylation Status
In the previous data, we used OA as a specific inhibitor of PP2A. To strengthen our data, we tested other OA-sensitive phosphatases using siRNAs. HT1080 cells were transfected with siRNAs for control, PP1, PP2Ac, and PP6, respectively. As shown in Fig. 5D, rpS3 phosphorylation was increased only in the PP2Ac knockdown cells (Fig. 5D, lane 3). This result indicates that PP2A is a sole phosphatase modulating the phosphorylation status of rpS3.
Implications of PP2A Phosphatase Activity for Phosphorylated rpS3 Levels
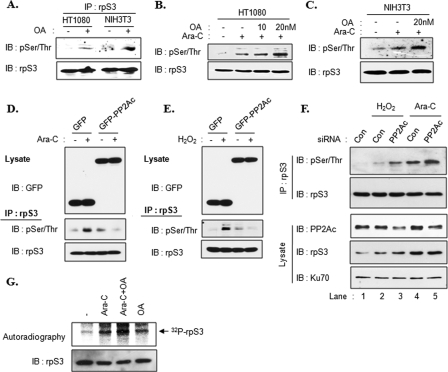
We previously verified that the interaction between rpS3 and PP2A was decreased by OA treatment. Therefore, we tested rpS3 phosphorylation under this condition. HT1080 and NIH3T3 cells were treated with OA for 3 h, and then rpS3 phosphorylation was determined using rpS3 immunoprecipitates. As shown in Fig. 6A, increased phosphorylation was detected in both of the OA-treated cell lines. To explore a connection with previous results that used genotoxins, we stimulated HT1080 or NIH3T3 cells with Ara-C in the presence or absence of OA pretreatment. rpS3 phosphorylation was not only increased by Ara-C treatment (Fig. 6, B, 2nd lane, and C, 2nd lane) but also increased to a greater extent by OA pretreatment than by Ara-C alone (Fig. 6, B, 3rd and 4th lanes, and C, 3rd lane). These results suggest that PP2A regulates the level of rpS3 phosphorylation. To strengthen these results, we transfected HT1080 cells with either a GFP or a GFP-PP2Ac expression plasmid to observe the effects on rpS3 phosphorylation after Ara-C (Fig. 6D) and H2O2 (Fig. 6E) treatments. We obtained the same results using both Ara-C and H2O2. Both agents promoted rpS3 phosphorylation in cells that expressed GFP (Fig. 6, D and E, 2nd lanes), but there was no induction in cells expressing GFP-PP2Ac (Fig. 6, D and E, 4th lanes). To explore a possible reciprocal relationship, we examined rpS3 phosphorylation in PP2Ac knockdown cells using siRNA. As shown in Fig. 6F, the level of phosphorylated nuclear rpS3 was increased by H2O2 (2nd lane) or Ara-C treatment (4th lane). Moreover, these phosphorylation levels were increased to a greater extent in PP2Ac knockdown cells (Fig. 6F, 3rd and 5th lanes, respectively). This also confirmed previous reports that the amounts of nuclear rpS3 are increased by genotoxic stress (22, 30) indicating that PP2A regulates the rpS3 phosphorylation status in cells. We next performed metabolic labeling using [32P]phosphoric acid to confirm the previous results in vivo. Metabolically labeled cells were treated with Ara-C and OA as indicated, and cell extracts were immunoprecipitated with rpS3 antibody. As expected, rpS3 phosphorylation was induced by Ara-C or OA treatment (Fig. 6G, 2nd and 4th lanes), and phosphorylation was synergistically increased in cells treated with a combination of OA and Ara-C (Fig. 6G, 3rd lane). Taken together, these results clearly show that PP2A phosphatase activity regulates the phosphorylation level of rpS3.
FIGURE 6.
Phosphorylation level of rpS3 is regulated by PP2A. A, HT1080 and NIH3T3 cells were treated with 10 nm OA for 3 h. Cells were lysed, and extracts were immunoprecipitated (IP) with rpS3 antibody. Immunoprecipitates were resolved by 10% SDS-PAGE and subjected to immunoblotting (IB) with the indicated antibodies. B and C, HT1080 (B) and NIH3T3 (C) cells were pretreated with OA for 3 h followed by Ara-C treatment for 30 min. D and E, HT1080 cells were transiently transfected with GFP or GFP-PP2Ac plasmids. After 24 h, cells were treated with 20 μm Ara-C for 3 h (D) or 100 μm H2O2 for 1 h (E). Cell extracts were immunoprecipitated with rpS3 antibody, and rpS3 immunoprecipitates were resolved by 10% SDS-PAGE and subjected to immunoblotting with phospho-Ser/Thr antibody. F, HT1080 cells were transfected with control (Con) siRNAs or siRNAs against PP2Ac. After 48 h, cells were stimulated with 100 μm H2O2 or 10 μm Ara-C for 30 min as indicated. Nuclear extracts were immunoprecipitated with rpS3 antibody. Immunoprecipitates (upper panel) and nuclear extracts (lower panel) were resolved and subjected to immunoblotting with the indicated antibodies. Ku70 protein was used as a loading control for the nuclear fraction. G, HT1080 cells were metabolically labeled with [32P]orthophosphate for 2 h. Cells were pretreated with 10 nm OA for 2 h followed by 20 μm Ara-C treatment for 30 min. The labeled rpS3 proteins were immunoprecipitated with rpS3 antibody, resolved by 10% SDS-PAGE, and analyzed by autoradiography and immunoblot analysis.
PP2A Regulates the Phosphorylation Level of Nonribosomal rpS3
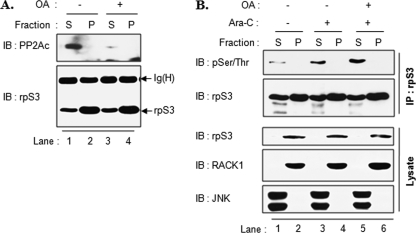
We previously reported that phosphorylated rpS3 was detected only in free nonribosomal rpS3 but not in ribosome-associated rpS3 (22). Therefore, we tested if the interaction between rpS3 and PP2A occurs in the supernatant fraction with existing phosphorylated rpS3. OA-treated or untreated HT1080 cells were fractionated into ribosome fractions and nonribosome fractions by ultracentrifugation as described under “Experimental Procedures.” As shown in Fig. 7A, PP2A was co-purified with rpS3 immunoprecipitates only in the soluble fraction (Fig. 7A, lane 1). Moreover, the amounts of co-purified PP2A were significantly decreased in OA-treated cells (Fig. 7A, lane 3). These results suggest that PP2A interacts with nonribosomal free rpS3 and that OA abrogates the interaction. Subsequently, we examined rpS3 phosphorylation in the nonribosomal fraction. HT1080 cells were treated with Ara-C in the presence or absence of OA pretreatment, and then cell extracts were fractionated into a ribosomal pellet and a supernatant fraction as in Fig. 7A. The phosphorylation level of rpS3 was examined using rpS3 immunoprecipitates. As in our previous report (22), rpS3 phosphorylation was detected only in the nonribosomal fraction (Fig. 7B, lane 1) and was increased by Ara-C treatment (Fig. 7B, lane 3). Furthermore, a large increase in rpS3 phosphorylation was observed in cells treated with both Ara-C and OA (Fig. 7B, lane 5). These results show that PP2A regulates the level of phosphorylated rpS3 through an interaction with nonribosomal free rpS3 but not with ribosome-associated rpS3.
FIGURE 7.
Nonribosomal rpS3 is regulated by PP2Ac. A, HT1080 cells were treated with 10 nm OA for 3 h. Cell extracts were fractionated with nonribosomal soluble (S) and ribosomal pellet fractions (P) using ultracentrifugation as described under “Experimental Procedures.” Each fraction was immunoprecipitated with rpS3 antibody. B, HT1080 cells were treated with 10 nm OA for 3 h followed by 10 μm Ara-C treatment for 30 min. Fractionated samples, nonribosomal (S) and ribosomal pellet fractions (P), were immunoprecipitated with rpS3 antibody (upper panel). Immunoprecipitates were resolved and immunoblotted (IB) with the indicated antibodies. RACK1 and JNK were used as ribosomal and nonribosomal fraction markers, respectively.
PKCδ-dependent Phosphorylation on rpS3 Is Critical for the Interaction between rpS3 and PP2A
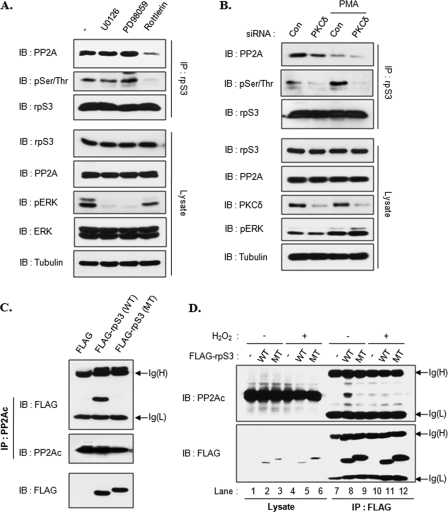
Based on our previous and current results, the phosphorylation status of rpS3 appears to be bi-directionally regulated by PKCδ and PP2A. To confirm this supposition, HT1080 cells were treated with inhibitors for MEK (U0126 and PD98059) or PKCδ (rottlerin). The result showed that the phosphorylation status of rpS3 is decreased by the PKCδ inhibitor but not by the MEK inhibitors. Furthermore, association of PP2A with rpS3 also decreased only in the rottlerin-treated cells (Fig. 8A). We next used PKCδ siRNA to confirm the previous result. Control or PKCδ siRNA-transfected HT1080 cells were untreated or treated with PMA. PMA induced the level of rpS3 phosphorylation in control cells (Fig. 8B, 3rd lane) compared with untreated cells (Fig. 8B, 1st lane). However, PKCδ knockdown cells showed a remarkable decrease of rpS3 phosphorylation (Fig. 8B, 2nd and 4th lanes). Therefore, the phosphorylation status of rpS3 appears to be correlated with PP2A interaction. PP2A-rpS3 interaction was decreased by PKCδ down-regulation (Fig. 8B, compare 1st with 2nd lane) and decreased more by PMA treatment (Fig. 8B, 3rd and 4th lanes), suggesting that stress-induced rpS3 phosphorylation is increased both by PKCδ activation and PP2A dissociation. These results indicate that the level of phosphorylated rpS3 is regulated not only by PKCδ-dependent phosphorylation but also PP2A accessibility. Next, we tested whether PKCδ-dependent phosphorylation sites affect the interaction between these two proteins using immunoprecipitation assay. As shown in Fig. 8C, PP2A interacted only with wild-type FLAG-rpS3 but not with mutant FLAG-rpS3 (S6A,T221A). Subsequently, we tested if this interaction was affected by H2O2 treatment. Wild-type or mutant FLAG-rpS3-transfected HT1080 cells were treated with or without H2O2, and then cell extracts were immunoprecipitated with FLAG antibody. We clearly reconfirmed that PP2A only interacted with wild-type FLAG-rpS3 (Fig. 8D, 8th lane) and that the PP2A interaction with FLAG-rpS3 was interrupted by H2O2 treatment (Fig. 8D, 11th lane). These results indicate that the PKCδ phosphorylation sites on rpS3 are required for interaction with PP2A and that both PKCδ and PP2A regulate the status of rpS3 phosphorylation.
FIGURE 8.
PKCδ-dependent phosphorylation sites on rpS3 are critical for the interaction with PP2Ac. A, HT1080 cells were untreated or treated with inhibitors against MEK (PD98059 and U0126) or PKCδ (rottlerin) for 3 h. Cell extracts were immunoprecipitated (IP) with rpS3 antibody. B, HT1080 cells were transfected with control (Con) or PKCδ siRNAs. After 48 h, cells were treated with PMA (100 ng/ml) for 30 min. Cell extracts and rpS3 immunoprecipitates were resolved by 10% SDS-PAGE and immunoblotted (IB) with the indicated antibodies. C, HEK293T cells were transfected with FLAG vector, FLAG-tagged rpS3 wild-type (WT), or S6A/T221A double point mutant (MT). After 24 h, cells were lysed, and extracts were immunoprecipitated with PP2Ac monoclonal antibody. PP2Ac immunoprecipitates were resolved by 10% SDS-PAGE and immunoblotted with FLAG and PP2Ac antibodies. D, HEK293T cells were transfected with FLAG (lanes 1 and 4), FLAG-rpS3 wild-type (lanes 2 and 5), or double point mutant (lanes 3 and 6). After 24 h, cells were treated with H2O2 (250 μm) for 1 h (lanes 4–6). Cell extracts were immunoprecipitated with FLAG antibody (lanes 7–12). Cell lysates (lanes 1–6) and immunoprecipitates (lanes 7–12) were resolved by 10% SDS-PAGE and immunoblotted with the indicated antibodies.
DISCUSSION
We previously reported that PKCδ phosphorylated two residues on rpS3, serine 6 and threonine 221, and that this phosphorylation was only detected in nonribosomal rpS3 and not in ribosome-associated rpS3 (22). Therefore, we reported that the rpS3 function between the ribosomal component and the extra-ribosomal protein was regulated by its phosphorylation status. These results indicated that there would be a phosphatase that negatively regulates the endonuclease activity of rpS3 function. We have confirmed that the phosphatase is PP2A in this study.
We initially identified an interaction between rpS3 and the catalytic C subunit of the PP2A complex. Then we also verified that rpS3 is associated with B55α, a regulatory B subunit of the PP2A holoenzyme (Fig. 1D). Although we did not certify the association of PP2A A subunit in this study, rpS3 appears to be a novel substrate of PP2A.
As described previously, there are three major classes of regulatory B subunits, including B/PR55, B′/PR61, and B″/PR72, and each of these subunits has several isoforms (31, 32). These different B subunits regulate the enzyme activity, localization, and substrate specificity of PP2A (33–36). Consequently, we can conclude from our previous results that the PP2A-dependent regulation of rpS3 influences the enzyme activity of rpS3 (22).
Further experiments strengthened our results that the status of phosphorylated rpS3 is bidirectionally regulated by PKCδ and PP2A. The basal increment of rpS3 phosphorylation was induced only in the PP2A siRNA-treated cells (Fig. 5D). We also demonstrated by using the in vitro dephosphorylation assay that PP2A specifically dephosphorylates rpS3 (Fig. 1E). Collectively, these results indicate that PP2A is the only phosphatase for rpS3 dephosphorylation.
It has been previously been shown that PP2A generally interacted with a phosphorylated form rather than an intact form of target proteins to induce its dephosphorylation (37–40). We identified that PP2A interacts with wild-type rpS3, but not with mutants (S6A/T221A) (Fig. 8), and that it associates with the N-terminal region of rpS3 (Fig. 2). From our results presented here, we conclude that PP2A is involved in the dephosphorylation of phosphorylated rpS3 by PKCδ, and that serine 6 on the N-terminal region of rpS3 appears to mediate the PP2A recruitment.
In our previous report, we demonstrated that the phosphorylated form of rpS3 was increased and accumulated in the nucleus under oxidative stress conditions (22). These results perfectly coincide with the present results. We demonstrated that rpS3 interacted with PP2A only in the nucleus and not in the cytoplasm (Fig. 4). Moreover, the interaction between rpS3 and PP2A in the nucleus was abrogated by H2O2 or Ara-C treatment. Although the effects of reactive oxygen species on PP2A activity are not precisely known, some reports indicate that reactive oxygen species, such as H2O2, inhibits the serine/threonine phosphatase activity, including PP2A (41–45). Therefore, it can be collectively explained that incremental changes of nuclear rpS3 phosphorylation are due both to PKCδ activation and PP2A inactivation, which induces PP2A dissociation from rpS3 in the nucleus during oxidative stress.
Moreover, we also used PMA to activate PKCδ in Fig. 8B. The result showed that rpS3 phosphorylation is increased by PMA, and the interaction between rpS3 and PP2A is decreased in cells transfected with control siRNA (Fig. 8B, 1st and 3rd lanes). Although there was no direct evidence for PMA-dependent inactivation of PP2A, we identified two possibilities from previous reports. One is the PP2Ac regulatory proteins, such as IEX-1 or SET, which suppress PP2Ac activity by PMA treatment (46, 47). The other is a PP2Ac phosphorylation at Tyr-307, which inactivates PP2A phosphatase activity (48, 49). It was previously reported that this phosphorylation is mediated by p60v-Src, p56Lck, or JAK, and these kinases are activated by PMA treatment (32, 47, 48, 50). Therefore, we tested whether the tyrosine phosphorylation of PP2Ac is affected by PMA treatment, and we identified an increased tyrosine phosphorylation in PMA-treated cells (supplemental Figs. 2 and 3).
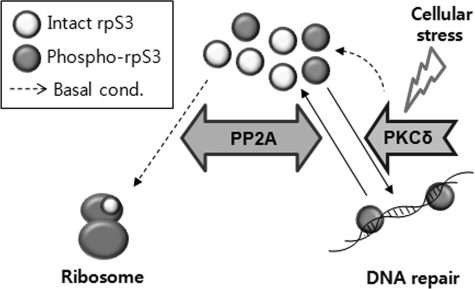
In this study, we demonstrated that PP2A interacted with rpS3 and regulated the phosphorylation status of rpS3. This interaction occurred only in the nucleus, and it was decreased under conditions of oxidative stress. Furthermore, we confirmed that PP2A interacted with wild-type FLAG-rpS3 but not with mutant FLAG-rpS3 (S6A/T221A). The interaction was abrogated by OA treatment, indicating that active PP2A interacted with the N-terminal region of rpS3 in the nucleus. Taken together, our results show that the phosphorylation status of rpS3 is regulated reciprocally by PP2A and PKCδ in cells (Fig. 9).
FIGURE 9.
Schematic diagram of bidirectional regulation of rpS3. Intact or basally phosphorylated rpS3 existed in cells. Intact rpS3 is incorporated in the ribosome complex, whereas the phosphorylated rpS3 is localized in the nucleus to process damaged DNA. After finishing the repair processing, the increased level of phosphorylated rpS3 will be down-regulated by PP2A-specific dephosphorylation to be used as ribosomal component.
Supplementary Material
This work was supported in part by Proteomics Grant FPR05C2-390 and Korea Science and Engineering Foundation Grant 09-74777.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- PP2A
- protein phosphatase 2A
- PP1
- protein phosphatase 1
- PP6
- protein phosphatase 6
- rpS3
- ribosomal protein S3
- OA
- okadaic acid
- Ara-C
- 1-β-d-arabinofuranosylcytosine
- PKCδ
- protein kinase Cδ
- siRNA
- small interfering RNA
- PMA
- phorbol 12-myristate 13-acetate
- JNK
- c-Jun NH2-terminal kinase
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
REFERENCES
- 1.Janssens V., Goris J. (2001) Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichhorn P. J., Creyghton M. P., Bernards R. (2009) Biochim. Biophys. Acta 1795, 1–15 [DOI] [PubMed] [Google Scholar]
- 3.Janssens V., Goris J., Van Hoof C. (2005) Curr. Opin. Genet. Dev. 15, 34–41 [DOI] [PubMed] [Google Scholar]
- 4.Westermarck J., Hahn W. C. (2008) Trends Mol. Med. 14, 152–160 [DOI] [PubMed] [Google Scholar]
- 5.Schönthal A. H. (2001) Cancer Lett. 170, 1–13 [DOI] [PubMed] [Google Scholar]
- 6.Lechward K., Zolnierowicz S., Hemmings B. A. (1999) Biochemistry 64, 1373–1381 [PubMed] [Google Scholar]
- 7.Yan L., Lavin V. A., Moser L. R., Cui Q., Kanies C., Yang E. (2008) J. Biol. Chem. 283, 7411–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeling J. M., Miller J. R., Gil R., Moon R. T., White R., Virshup D. M. (1999) Science 283, 2089–2091 [DOI] [PubMed] [Google Scholar]
- 9.Lacroix I., Lipcey C., Imbert J., Kahn-Perlès B. (2002) J. Biol. Chem. 277, 9598–9605 [DOI] [PubMed] [Google Scholar]
- 10.Andjelković N., Zolnierowicz S., Van Hoof C., Goris J., Hemmings B. A. (1996) EMBO J. 15, 7156–7167 [PMC free article] [PubMed] [Google Scholar]
- 11.Voorhoeve P. M., Hijmans E. M., Bernards R. (1999) Oncogene 18, 515–524 [DOI] [PubMed] [Google Scholar]
- 12.Kawabe T., Muslin A. J., Korsmeyer S. J. (1997) Nature 385, 454–458 [DOI] [PubMed] [Google Scholar]
- 13.Bennin D. A., Don A. S., Brake T., McKenzie J. L., Rosenbaum H., Ortiz L., DePaoli-Roach A. A., Horne M. C. (2002) J. Biol. Chem. 277, 27449–27467 [DOI] [PubMed] [Google Scholar]
- 14.Pallas D. C., Shahrik L. K., Martin B. L., Jaspers S., Miller T. B., Brautigan D. L., Roberts T. M. (1990) Cell 60, 167–176 [DOI] [PubMed] [Google Scholar]
- 15.Sablina A. A., Chen W., Arroyo J. D., Corral L., Hector M., Bulmer S. E., DeCaprio J. A., Hahn W. C. (2007) Cell 129, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neviani P., Santhanam R., Trotta R., Notari M., Blaser B. W., Liu S., Mao H., Chang J. S., Galietta A., Uttam A., Roy D. C., Valtieri M., Bruner-Klisovic R., Caligiuri M. A., Bloomfield C. D., Marcucci G., Perrotti D. (2005) Cancer Cell 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 17.Kim J., Chubatsu L. S., Admon A., Stahl J., Fellous R., Linn S. (1995) J. Biol. Chem. 270, 13620–13629 [DOI] [PubMed] [Google Scholar]
- 18.Kim S. H., Lee J. Y., Kim J. (2005) Biochem. Biophys. Res. Commun. 328, 962–967 [DOI] [PubMed] [Google Scholar]
- 19.Jang C. Y., Lee J. Y., Kim J. (2004) FEBS Lett. 560, 81–85 [DOI] [PubMed] [Google Scholar]
- 20.Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 21.Kim S. H., Kim J. (2006) Biochim. Biophys. Acta 1763, 823–832 [DOI] [PubMed] [Google Scholar]
- 22.Kim T. S., Kim H. D., Kim J. (2009) Biochim. Biophys. Acta 1793, 395–405 [DOI] [PubMed] [Google Scholar]
- 23.Kim H. D., Lee J. Y., Kim J. (2005) Biochem. Biophys. Res. Commun. 333, 110–115 [DOI] [PubMed] [Google Scholar]
- 24.Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]
- 25.Kim T. S., Jang C. Y., Kim H. D., Lee J. Y., Ahn B. Y., Kim J. (2006) Mol. Biol. Cell 17, 824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto M., Hatakeyama S., Oyamada K., Oda Y., Nishimura T., Nakayama K. I. (2005) Proteomics 5, 4145–4151 [DOI] [PubMed] [Google Scholar]
- 27.Xirodimas D. P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R. T. (2008) EMBO Rep. 9, 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wykoff D. D., O'Shea E. K. (2005) Mol. Cell. Proteomics 4, 73–83 [DOI] [PubMed] [Google Scholar]
- 29.Junttila M. R., Saarinen S., Schmidt T., Kast J., Westermarck J. (2005) Proteomics 5, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 30.Yadavilli S., Hegde V., Deutsch W. A. (2007) DNA Repair 6, 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechward K., Awotunde O. S., Swiatek W., Muszyńska G. (2001) Acta Biochim. Pol. 48, 921–933 [PubMed] [Google Scholar]
- 32.Sontag E. (2001) Cell. Signal. 13, 7–16 [DOI] [PubMed] [Google Scholar]
- 33.Kamibayashi C., Estes R., Lickteig R. L., Yang S. I., Craft C., Mumby M. C. (1994) J. Biol. Chem. 269, 20139–20148 [PubMed] [Google Scholar]
- 34.Tehrani M. A., Mumby M. C., Kamibayashi C. (1996) J. Biol. Chem. 271, 5164–5170 [DOI] [PubMed] [Google Scholar]
- 35.McCright B., Rivers A. M., Audlin S., Virshup D. M. (1996) J. Biol. Chem. 271, 22081–22089 [DOI] [PubMed] [Google Scholar]
- 36.Strack S., Zaucha J. A., Ebner F. F., Colbran R. J., Wadzinski B. E. (1998) J. Comp. Neurol. 392, 515–527 [PubMed] [Google Scholar]
- 37.Magenta A., Fasanaro P., Romani S., Di Stefano V., Capogrossi M. C., Martelli F. (2008) Mol. Cell. Biol. 28, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shouse G. P., Cai X., Liu X. (2008) Mol. Cell. Biol. 28, 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horn V., Thélu J., Garcia A., Albigès-Rizo C., Block M. R., Viallet J. (2007) Mol. Biol. Cell 18, 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiely P. A., Baillie G. S., Lynch M. J., Houslay M. D., O'Connor R. (2008) J. Biol. Chem. 283, 22952–22961 [DOI] [PubMed] [Google Scholar]
- 41.Hashigasako A., Machide M., Nakamura T., Matsumoto K., Nakamura T. (2004) J. Biol. Chem. 279, 26445–26452 [DOI] [PubMed] [Google Scholar]
- 42.Rao R. K., Clayton L. W. (2002) Biochem. Biophys. Res. Commun. 293, 610–616 [DOI] [PubMed] [Google Scholar]
- 43.Whisler R. L., Goyette M. A., Grants I. S., Newhouse Y. G. (1995) Arch. Biochem. Biophys. 319, 23–35 [DOI] [PubMed] [Google Scholar]
- 44.Foley T. D., Armstrong J. J., Kupchak B. R. (2004) Biochem. Biophys. Res. Commun. 315, 568–574 [DOI] [PubMed] [Google Scholar]
- 45.Howe C. J., Lahair M. M., McCubrey J. A., Franklin R. A. (2004) J. Biol. Chem. 279, 44573–44581 [DOI] [PubMed] [Google Scholar]
- 46.Letourneux C., Rocher G., Porteu F. (2006) EMBO J. 25, 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samanta A. K., Chakraborty S. N., Wang Y., Kantarjian H., Sun X., Hood J., Perrotti D., Arlinghaus R. B. (2009) Oncogene 28, 1669–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J., Martin B. L., Brautigan D. L. (1992) Science 257, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 49.Bos C. L., Kodach L. L., van den Brink G. R., Diks S. H., van Santen M. M., Richel D. J., Peppelenbosch M. P., Hardwick J. C. (2006) Oncogene 25, 6447–6456 [DOI] [PubMed] [Google Scholar]
- 50.Begum N., Ragolia L. (1999) Biochem. J. 344, 895–901 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.