Abstract
Natural killer (NK) cell recognition and formation of a conjugate with target cells, followed by intracellular signal pathway activation and degradation of cytolytic granules, are essential for NK cell cytotoxicity. In this study, NK92 cells were used to investigate synapse formation and subsequent signaling after binding to the target cell. The binding rate of the NK92-target cell was associated with NK92 cell cytotoxicity. Confocal results showed that adhesion molecules, LFA-1 (CD11a) and CD2, accumulated at the interface of the NK92-K562 contact. Ligation with K562 cells activated the Erk1/2 signal pathway of NK92 cells. The blocking of the NK-target conjugate by EDTA or anti-CD11a or/and anti-CD2 antibody decreased the phosphorylation of Erk1/2 and NK cell cytotoxicity. Inhibition of Erk1/2 phosphorylation by the chemical inhibitor U0126 suppressed the cytolytic activity of NK92 cells, but had no effect on NK-target conjugate formation. Thus, conjugate formation of the NK92-target cell was prerequisite to NK cell activation, and subsequent signal transduction was also required for NK cell cytotoxicity.
Natural killer (NK)3 cells are a population of granular lymphocytes that play an essential role in cellular immune defense against a variety of tumor cells, virus-infected cells, or allogeneic cells (1–3). NK cells are critical for host immunity for their ability for a quick cytotoxic response and to produce a wide variety of cytokines and chemokines to modulate other cellular components of the immune system (4, 5). NK cells express two functional types of receptors: activating and inhibitory receptors (6–8). The effector function of NK cells is regulated by a balance between opposite signals delivered by the MHC class I-specific inhibitory receptors and the activating receptors responsible for NK cell triggering to permit elimination of pathogens (6).
NK cell recognition and binding to target cells, as well as formation of conjugates, are essential for NK cell cytotoxicity (9). Conjugate formation by the NK cell with a target cell is a process mediated by integrins and immunoglobulin superfamily molecules including CD2, CD11a (LFA-1), CD11b, CD11c, and CD28, which also participate in the promotion of NK cell function (10–12). They participate in adhesion between the NK cell and the target cell, and blocking antibodies suppress the adhesion. In addition to possessing an adhesive role, ligation of CD2 induces kinase function and lipid raft polarization (11), whereas ligation of CD11a, CD11b, and CD11c induces phosphorylation-dependent NK cell activation (13, 14). The interaction of specific cell surface receptors with their ligands on a target cell at their interface forms specific activating NK cell immunological synapses and leads to the activation of a cascade of intracellular signals, resulting in Ca2+ flux, polarization of granules, and subsequent release of lytic molecules (13, 15, 16). The Erk1/2 (p44/42 mitogen-activated protein kinase) pathway plays an important role in NK cell cytotoxicity (17–21). Inhibition of Erk1/2 might block NK cell cytolytic activity by compromising the release of perforin (22). In this study, the roles of adhesion molecules in NK92-target cell conjugate formation of immunological synapse, and subsequent Erk1/2 activation in NK92 cells was investigated.
EXPERIMENTAL PROCEDURES
Cell Culture
The IL-2-dependent NK cell line NK92 was maintained in α-MEM (Invitrogen) containing 12.5% heat-inactivated fetal bovine serum (Invitrogen), 12.5% equine serum (HyClone), 2 mm l-glutamate, 100 units/ml penicillin, 100 μg/ml streptomycin, and supplemented with 100 units/ml rhIL-2 (Changchun Institute of Biological Products Ministry of Public Health People's Republic of China). The target cell lines of NK92, K562, and HL60, were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cell culture was performed at 37 °C in a 5% CO2-humidified atmosphere. Cells from a mid-log phase culture were harvested.
Antibodies
FITC-conjugated CD2, CD28, CD56, and CD85j, Alexa Fluor 488-conjugated CD11b; PE-conjugated CD56, CD11a, CD11c, CD54, HLA-ABC, phospho-p44/42, and PE-Cy5-CD56 were purchased from BD PharMingen; anti-CD11a (LFA-1a) mAb (clones TS1/22) and anti-CD2 (clone TS2/18) were from Pierce; p44/42 MAPK (Erk1/2) antibody, phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody, and β-actin antibody were from Cell Signaling Technology; horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody was from Wuhan Boster Biological Technology, LTD.
Flow Cytometry
Cells were washed twice, blocked, and incubated with saturating concentrations of the appropriate mAbs for 30 min at 4 °C. Thereafter, cells were washed twice and analyzed using FACSCalibur (Becton Dickinson).
CFSE Staining
Target cells were washed and suspended in 1 ml of PBS, 1% bovine serum albumin at a final concentration of 5 × 106/ml, then labeled for 10 min at 37 °C with CFSE (2 μm, Sigma). Quench-staining was performed on ice for 5 min by adding 5 volumes of ice-cold RPMI 1640/10% fetal bovine serum. Then the cells were washed three times with ice-cold PBS, 1% bovine serum albumin and cultured under appropriate conditions.
Cell Conjugation Assay
NK92 cells at a concentration of 5 × l06 cells per milliliter were mixed with 5 × l06 CFSE-labeled target cells at an effector to target ratio of 1:1, allowing the formation of effector-target conjugates. The NK92-K562 cell mixture was centrifuged at 1,000 rpm for 1 min and incubated in 37 °C for the indicated time points. Then the cell mixture was gently resuspended and fixed with 2% paraformaldehyde. The cells were blocked, stained with PE-CD56 at 4 °C for 30 min, and analyzed by flow cytometry. The conjugation ratio was calculated as the portion of FITC/PE double-positive events within the PE-positive events. For antibody-blocking experiments, 5 × l06 NK92 cells were incubated with 10 μg/ml mAbs for 30 min or mouse IgG as a negative control in advance, and then the conjugation ratio was detected by flow cytometry (23).
Intracellular Erk1/2 Detection by Flow Cytometry
NK92 and target cells were cultured with serum-free medium for 4 h to reduce the background phosphorylation of Erk1/2. NK92 cells at a concentration of 5 × l06 cells/ml were incubated with equal numbers of CFSE-labeled target cells at an E/T of 1:1 for indicated times. The cell mixture was gently resuspended and fixed with 2% paraformaldehyde for 15 min at room temperature in the dark. After washing twice with PBS, the cells were stained with PE-Cy5-CD56 at 4 °C for 30 min. Then the cells were permeabilized and stained with PE-conjugated phospho-p44/42 MAP kinase for 1 h at room temperature in the dark. Cells were washed twice and analyzed by flow cytometry.
Confocal Microscopy
NK92 cells at a concentration of 5 × l06 cells/ml were conjugated to K562 cells at a 1:1 ratio for 5 min at 37 °C in suspension. The cell mixture was resuspended gently and adhered to poly-l-lysine-coated (Sigma) glass slides for 15 min at 37 °C. The cells were fixed with 2% paraformaldehyde and incubated with antibodies against cell surface markers. For intracellular phospho-Erk1/2 detection, adherent cell mixtures were permeabilized with 0.1% saponin and 0.1% Triton X-100 and stained with PE-phospho-p44/42 or PE-IgG2a control in PBS for 1 h after cell surface staining with FITC-CD56. Slides were washed twice with PBS and visualized using a Zeiss LSM 510 laser-scanning confocal microscope. Protein clustering was considered when the fluorescence intensity around the effector-target interface was at least twice the sum of the fluorescence in the unconjugated membranes.
Cytotoxicity Assay
The cytotoxic activity of NK92 cells against target cells was measured by a standard 4-h 51Cr release assay, as previously described (24). Briefly, K562 or HL60 were labeled with 200 μCi of sodium [51Cr]chromate (PerkinElmer Life Sciences) per 106 for 1 h at 37 °C. NK92 cells were incubated with K562/HL60 in 96-well round bottom plates for 4 h at 37 °C in 5% CO2. The percentage of specific 51Cr release was calculated by the formula (A − B)/(C − B) × 100%, where A is 51Cr release in the presence of effector cells, B is the spontaneous release in the absence of effector cells, and C is the total 51Cr release from K562 incubated with 1% Triton X-100. Spontaneous release did not exceed 10% of the maximum release.
Western Blotting
After stimulation, cells were collected, washed in ice-cold PBS, and lysed in lysis buffer containing 20 mm Tris-HCl, pH 7.5, 1% Nonidet P-40 (v/v), 10 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 10 mg/ml each aprotinin and leupeptin, 1 mm phenylmethylsulfonyl fluoride (Sigma), 1 mm Na3VO4, 50 mm NaF, and 1 mm EDTA (Sigma). Lysates were centrifuged at 13,000 rpm for 20 min at 4 °C to remove nuclei and cell debris. The supernatants were retained. Equivalent amounts of total protein were resolved on 12% SDS-polyacrylamide gels followed by electrophoretic transfer to polyvinylidene difluoride membranes (Millipore). The membranes were blocked in Tris-buffered saline supplemented with 5% nonfat milk and 0.1% Tween 20 and incubated with the primary antibody at 1:1000 dilution for 1 h at room temperature. The membrane was then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody. Immunoblots were developed using ECL Western blotting detection reagents (Pierce).
Statistical Analysis
Statistical analysis was performed using the Student's t test. All p values were two-tailed, and p < 0.05 was taken as statistically significant.
RESULTS
The Binding Rate of NK92 Target Cells Is Associated with Natural Cytotoxicity
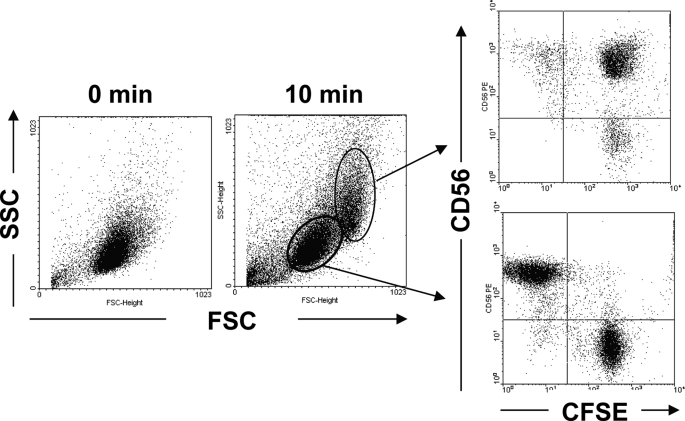
NK92 cells were mixed with CFSE-labeled K562 cells and incubated at 37 °C for 10 min. Then the cells were fixed and stained with PE-CD56 and analyzed by flow cytometry. As shown in Fig. 1, one cell population exhibited higher forward scatter, and side scatter was observed after 10 min of incubation. Phenotypic analysis showed that the cell population was almost CD56 and CFSE double-positive, while the original cell population was CD56 or CFSE single-positive. This result indicated that NK92 cells could bind with K562 cells and form conjugates.
FIGURE 1.
Detection of NK92 cell binding with the K562 cell by flow cytometry. NK92 cells and CFSE-labeled K562 cells were mixed at an E/T of 1:1, centrifuged at 1,000 rpm for 1 min, and incubated at 37 °C for 10 min. Then the cells were harvested, fixed, stained with PE-CD56 mAb, and analyzed by flow cytometry. One representative example is shown.
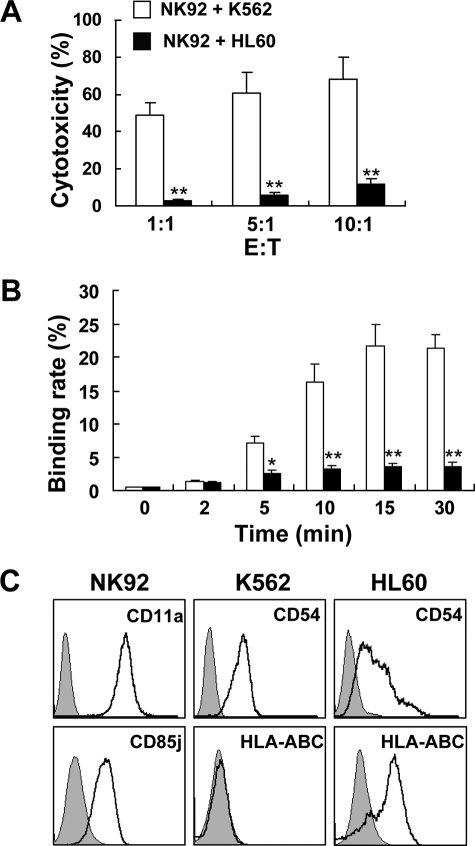
NK cell recognition of a target cell is a complex process: binding with a potential target cell, interaction between ligand-receptor, activating and inhibitory signal transduction and integration, and effector cytolysis (6, 25). K562 and HL60 cells are two tumor cell lines with different sensitivities to NK92 cells (Fig. 2A), and the results of conjugate formation showed that NK92-K562 conjugates are significantly greater than those of NK92 with HL60 cells during the 30-min incubation period (Fig. 2B). As adhesion molecule CD11a (LFA-1) played an important role in NK cell adhesion, we wondered if the different binding rates of NK92 to K562 and HL60 cells were because of the different expression levels of CD54, the ligand of CD11a, in these two target cell lines. The results showed that NK92 cells expressed CD11a, and both K562 and HL60 cells expressed high levels of CD54 (Fig. 2C). It was reported that NK cell adhesion to target cell could be disrupted by MHC I molecule-specific KIR (23). We found that NK92 cells expressed the inhibitory NK cell receptor CD85j, which recognized the HLA class I molecules (26–28). The phenotypic analysis of target cells showed that HLA-A, -B, -C were not detected on K562 cells but were highly expressed on HL60 cells (Fig. 2C). The inhibitory signal transmitted by CD85j disrupted the conjugation of NK92 and HL60, which might be the major reason for low NK92 cytotoxicity against HL60 cells.
FIGURE 2.
Binding rate of NK92-target cells was associated with NK92 natural cytotoxicity. A, cytotoxicity of NK92 cells against K562 or HL60 was detected by a 4-h 51Cr-release assay. B, NK92- and CFSE-labeled K562 or HL60 were mixed and incubated in 37 °C for different times. Then the cells were harvested, fixed, stained with PE-CD56 mAb, and analyzed by flow cytometry. Data were collected from at least three independent experiments and analyzed by Student's t test; *, p < 0.05; **, p < 0.01. C, NK92, K562, and HL60 cells were stained with the indicated mAb and analyzed by flow cytometry.
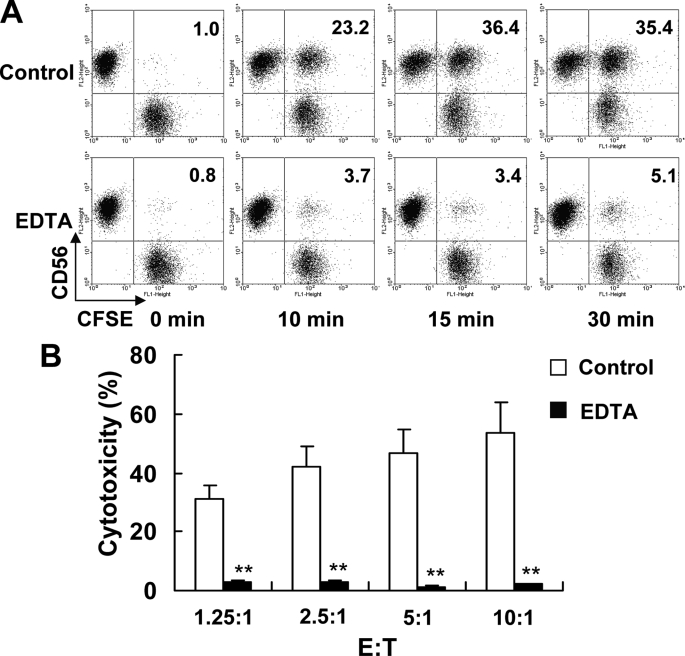
The binding of the NK cell with the target cell was essential to NK cytolysis. In our study, the conjugation of NK92 with K562 cells was significantly inhibited by EDTA (Fig. 3A), similar to the previous report (29, 30). Accordingly, the cytolytic activity of NK92 cells pretreated with EDTA against K562 cells was significantly alleviated (Fig. 3B). These results suggest that NK92 cell binding with the target cell and formation of the cell conjugate are prerequisite to NK92 cell cytotoxicity.
FIGURE 3.
The decrease of binding rate impaired the cytotoxicity of NK92 cells against K562 cells. A, NK92 and CFSE-labeled K562 were mixed and incubated in 37 °C for different times with or without the presence of 1% EDTA. Then the cells were harvested, fixed, stained with PE-CD56 mAb, and analyzed by flow cytometry. One representative example is shown from three independent experiments. B, cytotoxicity of NK92 cells against K562 cells with or without the presence of 1% EDTA was detected by 4-h 51Cr-release assay. Data were collected from three independent experiments and analyzed by Student's t test, **, p < 0.01.
CD2 and CD11a Play Important Roles in NK92 Cell Cytotoxicity
At the early stage of the cytolysis process, cell surface adhesion molecules, such as integrins and the immunoglobulin superfamily molecules, participated in effector-target recognition, interaction, and stable conjugate formation. The formation of cell conjugate and immunological synapse is a dynamic process, and the accumulation of adhesion molecules plays critical roles in the process (31).
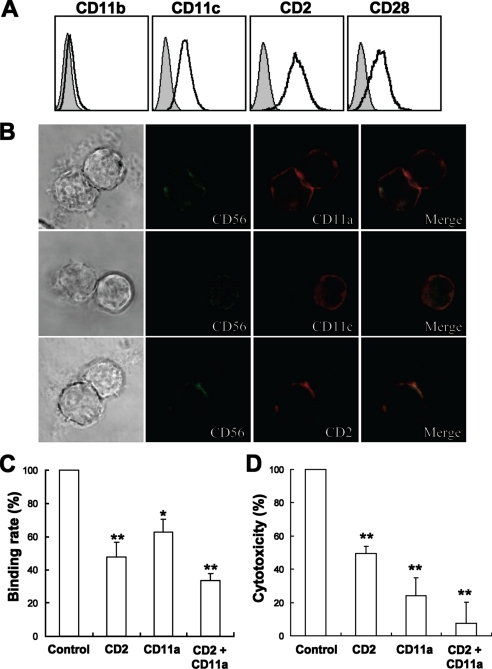
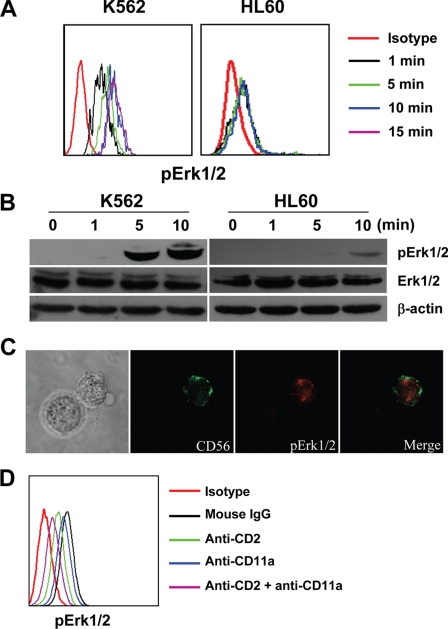
To explore the possible adhesion molecules involved in the conjugate formation, NK92 cells were screened for the expression of CD2, CD11a, CD11b, CD11c, and CD28. As shown in Figs. 2C and 4A, CD2, CD11a, CD11c, and CD28 were expressed, while CD11b was not detected on NK92 cells. Meanwhile, the detection of their reciprocal ligands demonstrated that little or no CD80 or CD86 (ligands of CD28) but high levels of CD54 (ligand of CD11a and CD11c) and CD58 (ligand of CD2) were expressed on K562 cells (data not shown) (19, 28, 32). Confocal microscopy was used to detect the accumulation of CD2, CD11a, and CD11c at the interface of the NK92-K562 interaction. As shown in Fig. 4B, CD2 and CD11a were accumulated at the interface of the NK92-K562 cell contact, but CD11c was not. The results suggested CD2 and CD11a might play critical roles in NK92-K562 interaction. The blocking mAbs were then used to determine the potential effects of CD2 and CD11a in NK92 cell cytotoxicity. These results showed that effector-target conjugate formation and cytotoxicity of NK92 against K562 were modestly inhibited by anti-CD2 and anti-CD11a mAbs, and the combination of these two mAbs exhibited a stronger effect.
FIGURE 4.
CD2 and CD11a played important roles in NK92 cell cytotoxicity. A, expression of some adhesion molecules on NK92 cells. NK92 cells were stained with indicated mAbs and analyzed by flow cytometry. B, confocal microscopic observation of accumulation of CD2, CD11a, and CD11c at the interface of NK92-K562 interaction (400×). C, blockade of CD2 or/and CD11a affected NK92-K562 conjugate formation. NK92 cells were preincubated with 10 μg/ml mouse IgG, anti-CD2, anti-CD11a, or anti-CD2 + anti-CD11a for 30 min and were mixed with equal numbers of CFSE-labeled K562 at 37 °C for 15 min. Then the cells were harvested, fixed, stained with PE-CD56 mAb, and analyzed by flow cytometry. D, blockade of CD2 or/and CD11a affected NK92 cell cytotoxicity against K562 targets. NK92 cells were preincubated with 20 μg/ml mouse IgG, anti-CD2, anti-CD11a, or anti-CD2 + anti-CD11a for 30 min, then equal volumes of K562 cells were added for a 4-h 51Cr-release assay. For C and D, data of control group were taken as 100%, and the relative ratio of other groups against control were presented. Data were collected from three independent experiments and analyzed by Student's t test, *, p < 0.05; **, p < 0.01 compared with control.
Erk1/2 Is Involved in NK92 Cell Cytotoxicity
After the synapse of the NK-target cells formed, the signal pathways of NK cells were activated (9, 15, 17). It was already reported that Erk1/2 was one of the important signal pathways for NK cell cytotoxicity (17). In our study, NK92-K562 interactions caused rapid phosphorylation of Erk1/2, and the signal activation was peaked at about 10 min after their binding, but when HL60 were used as target cells, the phosphorylation of Erk1/2 in NK92 cells was greatly decreased (Fig. 5, A and B). To confirm that Erk1/2 phosphorylation was in the activated NK92 cells but not in K562 cells, NK92-K562 conjugates were observed by confocal microscopy. Images of NK92-K562 conjugates indicated that the phosphorylation of Erk1/2 was mainly located in NK92 cells (Fig. 5C). When NK92 cells were pretreated with anti-CD2 and anti-CD11a mAbs, the phosphorylation levels of Erk1/2 in NK92 cells and NK92-K562 conjugates were greatly decreased (Fig. 5D).
FIGURE 5.
NK92-K562 interaction caused phosphorylation of Erk1/2 in NK92 cells. A, flow cytometric analysis of Erk1/2 activation after binding with target cells. NK92 cells were cultured with serum-free medium for 4 h to reduce the background phosphorylation of Erk1/2. Target cells were stained with CFSE and starved from serum for equal time with NK92. NK92 cells and target cells were mixed at equal numbers for different times. The cell mixture was fixed, stained with PE-Cy5-CD56 and PE-phospho-p44/42, and analyzed by flow cytometry. NK92 cells that conjugated with K562 cells were gated and analyzed. B, Western blotting analysis of Erk1/2 activation after binding with target cells. NK92 and target cells were starved for 4 h, mixed with an equal number for different time points and lysed. Equal amounts of total cell lysate were loaded onto SDS-PAGE gels for Western blot analysis. C, confocal microscopic (400×) analysis of Erk1/2 activation in NK92 cells after stimulation with target cells. D, flow cytometric analysis of Erk1/2 activation in NK92 cells that pretreated with anti-CD2 or/and anti-CD11a mAbs. NK92 and K562 cells were starved for 4 h, and NK92 cells were preincubated with 10 μg/ml mouse IgG, anti-CD2, anti-CD11a, or anti-CD2 + anti-CD11a for 30 min. Then they were mixed at equal numbers for 10 min and stained as described in A. NK92 cells (CD56+) were gated and analyzed the phosphorylation of Erk1/2.
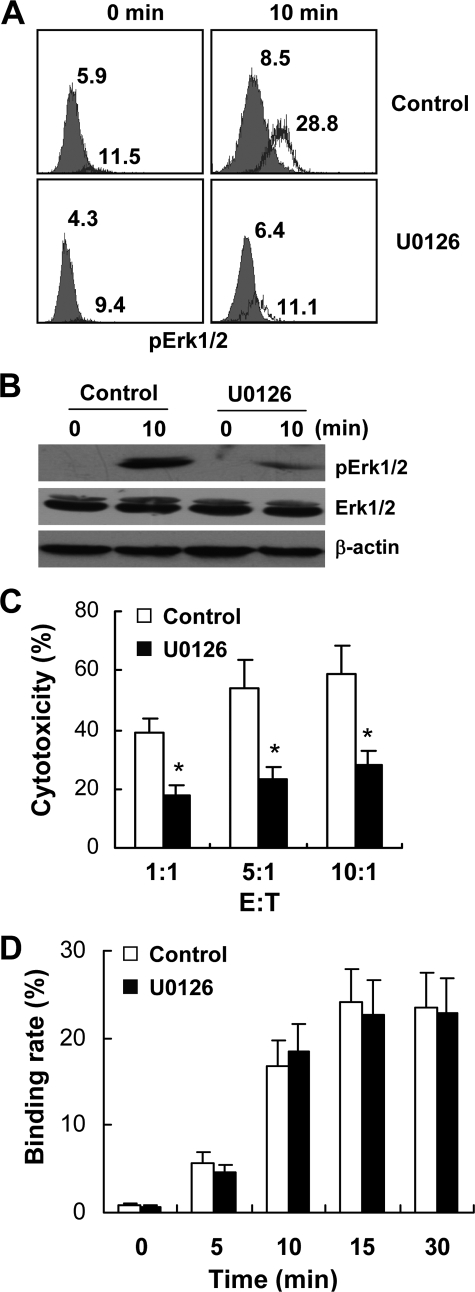
To further analyze the role of Erk1/2 in NK92 cytolysis, U0126, one of the chemical inhibitors of the Erk1/2 pathway, was used. As shown in Fig. 6, A and B, U0126 inhibited the phosphorylation of Erk1/2 in NK92 cells that conjugated with K562 cells and thus directly decreased the cytotoxicity of NK92 cells (Fig. 6C). However, U0126 pretreatment did not down-regulate NK92-K562 conjugate formation (Fig. 6D).
FIGURE 6.
Erk1/2 inhibitor U0126 directly decreased cytotoxicity of NK92 cells without influencing NK92-K562 binding. NK92 cells and CFSE-labeling K562 cells were starved of serum for 4 h and mixed with equal numbers with or without the presence of 10 μm U0126. A, phosphorylation of Erk1/2 in NK92 cells was analyzed by flow cytometry. NK92 cells ligated with K562 cells were gated and analyzed. The number indicates the mean fluorescence intensity (MFI) of phospho-Erk1/2 in NK92 cells. B, mixture was lysed, and equal amounts of total cell lysate were loaded onto SDS-PAGE gels for Western blot analysis of phosphorylation of Erk1/2. C, cytotoxicity of NK92 cells against K562 with or without the presence of 10 μm U0126 was examined by 4-h 51Cr release assay. D, NK92- and CFSE-labeled K562 cells were mixed and incubated at 37 °C for different times with or without the presence of 10 μm U0126. Then the cells were fixed and stained with CD56 mAb and analyzed by flow cytometry. For C and D, data were collected from three independent experiments and analyzed by Student's t test; *, p < 0.05.
DISCUSSION
NK cells are well known for their recognition and cytolysis of tumor cells without prior sensitization. During the triggering stage of cytolysis, it is necessary for NK cells to bind with target cells and form conjugates. In this study, we demonstrated by using flow cytometry that NK92, one of the important NK cell lines that is commonly used to study NK cell function and as an immunotherapeutic agent in clinical therapy (28, 33), was capable of rapidly binding with target cells. The binding rates of NK92 cells against different target cells were correlated with their cytolysis activities. The cytotoxicity of the NK cell could be affected by the factors influencing cell conjugate formation, including NK inhibitory receptors (23). Although most of the inhibitory receptors were deficient on NK92 cells (28), CD85j was expressed, which can recognize MHC I molecules on target cells and might inhibit effector-target conjugate formation (27). In addition, EDTA also greatly decreased the conjugate formation of NK92 with K562 and inhibited NK92 cell cytotoxicity. Altogether, NK cell ligation with target cell was the first and essential step for NK cell cytolysis of the target cell.
The formation of the stable NK-target cell conjugate is a complicated and dynamic process. In the process of conjugate formation, integrins and immunoglobulin superfamily molecules were crucial for NK cell adhesion with target cells (13, 20, 34). And different adhesion molecules were employed by different effector cells to adhesion with target cells (13, 20, 35). In our study, CD2 and CD11a, but not CD11c, were accumulated at the interface of NK92-K562 interaction (Fig. 4). Blocking mAbs of anti-CD2 and anti-CD11a could decrease the effector-target conjugate formation and the following NK92 cell cytotoxicity (Fig. 4).
NK cell receptors recognized their ligands on target cells and led to a series of intracellular signal activation (30). The different NK cell receptors/ligands could activate distinct signal pathways including Erk1/2 and JNK (9, 17, 20, 25, 36). If the NK-target cell conjugate was impaired, the signal pathways would not be activated, and NK cell cytotoxicity would be inhibited. However, when the signal pathways were inhibited, NK cell cytotoxicity was suppressed, but no influence was found on NK-target conjugate formation. In this study, Erk1/2 could be quickly activated when NK92 ligation with K562, but not HL60 cells (Fig. 5). U0126, the inhibitor of Erk1/2, inhibited NK92 cytotoxicity, but did not decrease NK92-K562 conjugate formation (Fig. 6). So, the successful signal transduction induced by effector-target interaction was also observed to be critical for NK cell cytotoxicity (16, 17).
Thus, NK cell cytolysis required a series of cell cascade events: NK cell ligated with the target cell and formed the activating immunological synapse; the ligand-receptor interaction-induced intracellular signal transduction; the degradation of lytic granules and cytolysis target cell. The blockade of any stage including NK-target ligation or signal pathways would inhibit NK cell cytotoxicity.
This work was supported in part by the Natural Science Foundation of China (Grants 30630059, 30721002, and 30801007), Ministry of Science & Technology of China (973 Basic Science Projects 2007CB512405 and 2006CB806504), and the China Postdoctoral Science Foundation (Grant 200801237).
- NK
- natural killer
- FITC
- fluorescein isothiocyanate
- PBS
- phosphate-buffered saline
- mAb
- monoclonal antibody
- Erk
- extracellular signal-regulated kinase
- CFSE
- carboxyfluorescein diacetate succinimidyl ester
- PE
- phycoerythrin.
REFERENCES
- 1.Trinchieri G. (1989) Adv. Immunol. 47, 187–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretta A., Bottino C., Mingari M. C., Biassoni R., Moretta L. (2002) Nat. Immunol. 3, 6–8 [DOI] [PubMed] [Google Scholar]
- 3.Hamerman J. A., Ogasawara K., Lanier L. L. (2005) Curr. Opin. Immunol. 17, 29–35 [DOI] [PubMed] [Google Scholar]
- 4.Roder J. C., Pross H. F. (1982) J. Clin. Immunol. 2, 249–263 [DOI] [PubMed] [Google Scholar]
- 5.Moretta L., Ciccone E., Mingari M. C., Biassoni R., Moretta A. (1994) Adv. Immunol. 55, 341–380 [DOI] [PubMed] [Google Scholar]
- 6.Lanier L. L. (2005) Annu. Rev. Immunol. 23, 225–274 [DOI] [PubMed] [Google Scholar]
- 7.Lanier L. L. (1998) Adv. Exp. Med. Biol. 452, 13–18 [DOI] [PubMed] [Google Scholar]
- 8.Raulet D. H., Held W. (1995) Cell 82, 697–700 [DOI] [PubMed] [Google Scholar]
- 9.Davis D. M., Dustin M. L. (2004) Trends Immunol. 25, 323–327 [DOI] [PubMed] [Google Scholar]
- 10.Helander T. S., Timonen T. (1998) Curr. Top. Microbiol. Immunol. 230, 89–99 [DOI] [PubMed] [Google Scholar]
- 11.Inoue H., Miyaji M., Kosugi A., Nagafuku M., Okazaki T., Mimori T., Amakawa R., Fukuhara S., Domae N., Bloom E. T., Umehara H. (2002) Eur. J. Immunol. 32, 2188–2198 [DOI] [PubMed] [Google Scholar]
- 12.Dustin M. L., Starr T., Coombs D., Majeau G. R., Meier W., Hochman P. S., Douglass A., Vale R., Goldstein B., Whitty A. (2007) J. Biol. Chem. 282, 34748–34757 [DOI] [PubMed] [Google Scholar]
- 13.Orange J. S., Harris K. E., Andzelm M. M., Valter M. M., Geha R. S., Strominger J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rezzonico R., Chicheportiche R., Imbert V., Dayer J. M. (2000) Blood 95, 3868–3877 [PubMed] [Google Scholar]
- 15.Bromley S. K., Burack W. R., Johnson K. G., Somersalo K., Sims T. N., Sumen C., Davis M. M., Shaw A. S., Allen P. M., Dustin M. L. (2001) Annu. Rev. Immunol. 19, 375–396 [DOI] [PubMed] [Google Scholar]
- 16.Vyas Y. M., Maniar H., Dupont B. (2002) Immunol. Rev. 189, 161–178 [DOI] [PubMed] [Google Scholar]
- 17.Djeu J. Y., Jiang K., Wei S. (2002) Clin. Cancer Res. 8, 636–640 [PubMed] [Google Scholar]
- 18.Visse E., Inostroza J., Cabello G., Parra E. (2003) Biol. Res. 36, 263–278 [DOI] [PubMed] [Google Scholar]
- 19.Chen X., Trivedi P. P., Ge B., Krzewski K., Strominger J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6329–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Allan D. S., Krzewski K., Ge B., Kopcow H., Strominger J. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10346–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei S., Gamero A. M., Liu J. H., Daulton A. A., Valkov N. I., Trapani J. A., Larner A. C., Weber M. J., Djeu J. Y. (1998) J. Exp. Med. 187, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez O. D., Mitchell D., Jager G. C., Nolan G. P. (2004) Blood 104, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 23.Burshtyn D. N., Shin J., Stebbins C., Long E. O. (2000) Curr. Biol. 10, 777–780 [DOI] [PubMed] [Google Scholar]
- 24.Friberg D. D., Bryant J. L., Whiteside T. L. (1996) Methods 9, 316–326 [DOI] [PubMed] [Google Scholar]
- 25.Bottino C., Castriconi R., Moretta L., Moretta A. (2005) Trends Immunol. 26, 221–226 [DOI] [PubMed] [Google Scholar]
- 26.Kirwan S. E., Burshtyn D. N. (2005) J. Immunol. 175, 5006–5015 [DOI] [PubMed] [Google Scholar]
- 27.Banham A. H., Colonna M., Cella M., Micklem K. J., Pulford K., Willis A. C., Mason D. Y. (1999) J. Leukoc. Biol. 65, 841–845 [DOI] [PubMed] [Google Scholar]
- 28.Maki G., Klingemann H. G., Martinson J. A., Tam Y. K. (2001) J. Hematother. Stem Cell Res. 10, 369–383 [DOI] [PubMed] [Google Scholar]
- 29.Trotta R., Puorro K. A., Paroli M., Azzoni L., Abebe B., Eisenlohr L. C., Perussia B. (1998) J. Immunol. 161, 6648–6656 [PubMed] [Google Scholar]
- 30.Barber D. F., Faure M., Long E. O. (2004) J. Immunol. 173, 3653–3659 [DOI] [PubMed] [Google Scholar]
- 31.Orange J. S. (2007) Adv. Exp. Med. Biol. 601, 225–233 [DOI] [PubMed] [Google Scholar]
- 32.Gong J. H., Maki G., Klingemann H. G. (1994) Leukemia 8, 652–658 [PubMed] [Google Scholar]
- 33.Tonn T., Becker S., Esser R., Schwabe D., Seifried E. (2001) J. Hematother. Stem Cell Res. 10, 535–544 [DOI] [PubMed] [Google Scholar]
- 34.Selvaraj P., Plunkett M. L., Dustin M., Sanders M. E., Shaw S., Springer T. A. (1987) Nature 326, 400–403 [DOI] [PubMed] [Google Scholar]
- 35.Chong A. S., Boussy I. A., Jiang X. L., Lamas M., Graf L. H., Jr. (1994) Cell Immunol. 157, 92–105 [DOI] [PubMed] [Google Scholar]
- 36.Li C., Ge B., Nicotra M., Stern J. N., Kopcow H. D., Chen X., Strominger J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]